Abstract
The complexity of the pathogenic cascade in lysosomal storage disorders suggests that combination therapy will be needed to target various aspects of pathogenesis. The standard of care for Pompe disease (glycogen storage disease type II), a deficiency of lysosomal acid alpha glucosidase, is enzyme replacement therapy (ERT). Many patients have poor outcomes due to limited efficacy of the drug in clearing muscle glycogen stores. The resistance to therapy is linked to massive autophagic buildup in the diseased muscle. We have explored two strategies to address the problem. Genetic suppression of autophagy in muscle of knockout mice resulted in the removal of autophagic buildup, increase in muscle force, decrease in glycogen level, and near-complete clearance of lysosomal glycogen following ERT. However, this approach leads to accumulation of ubiquitinated proteins, oxidative stress, and exacerbation of muscle atrophy. Another approach involves AAV-mediated TSC knockdown in knockout muscle leading to upregulation of mTOR, inhibition of autophagy, reversal of atrophy, and efficient cellular clearance on ERT. Importantly, this approach reveals the possibility of reversing already established autophagic buildup, rather than preventing its development.
Keywords: autophagy, muscle proteostasis, lysosomal storage diseases, metabolome, enzyme replacement therapy
Conventional enzyme replacement therapy for Pompe disease is of limited effectiveness at least in part because of the presence of large areas of autophagic debris in the diseased muscle. Here, Lim et al. explore pros and cons of different ways to manipulate autophagic process in order to improve the therapy. The authors also investigate protein synthesis and degradation in Pompe muscle and suggest a strategy to improve altered proteostasis.
Introduction
Pompe disease, an inherited deficiency of lysosomal acid alpha-glucosidase (GAA), belongs to a group of more than 60 lysosomal storage diseases (LSDs). The enzyme deficiency leads to progressive accumulation of glycogen in the lysosomal compartment in multiple tissues, including musculoskeletal, cardiac, respiratory, vascular, gastrointestinal, and nervous systems.1, 2, 3, 4, 5 Skeletal and cardiac muscles are most profoundly affected. The condition presents as a spectrum of disease severity largely dependent on the level of residual enzyme activity.6 The most severe infantile-onset form manifests within the first months of life with muscle weakness, respiratory impairment, and rapidly progressive hypertrophic cardiomyopathy that is fatal by 1 to 2 years of age. Later-onset forms are characterized by a gradually progressive proximal muscle weakness (with little or no cardiac involvement) that eventually causes significant morbidity, respiratory failure, and early mortality in children and adults.7
The only available therapy for Pompe disease is enzyme replacement therapy (ERT) with human recombinant GAA (alglucosidase alfa; Myozyme and Lumizyme, Genzyme, a Sanofi Company). This therapy restores cardiac function, thus significantly extending the lifespan of infantile-onset patients,8 but the effect of ERT in skeletal muscle is much less robust. Although the therapy stabilizes neuromuscular and respiratory function in late-onset patients, the effect is often short lived and is followed by a decline.9, 10, 11, 12 Furthermore, most surviving infants suffer from residual muscle weakness, and many experience progressive worsening of motor and respiratory function; those who develop high antibody titers to the recombinant enzyme (the cross-reactive immunologic material [CRIM]-negative) have particularly poor treatment outcomes.13, 14, 15, 16, 17 Since the introduction of ERT, diagnostic methods have improved, allowing for early therapeutic intervention.18 However, a recent study indicates that muscle damage progresses even when ERT is initiated within days after birth in infants diagnosed through a newborn screening program.19
A number of factors contribute to skeletal muscle resistance to therapy, including muscle mass, inefficient uptake of the drug by skeletal muscle, and immune response in CRIM-negative patients (reviewed in Lim et al.20). We have shown that on top of that, defective autophagy, a process that brings cytosolic material to lysosomes for degradation and recycling,21 contributes to resistance to therapy.22 As in other LSDs, lysosomal dysfunction in Pompe disease leads to incomplete autophagic flux and accumulation of autophagic debris that are particularly prominent in muscle tissue.23, 24 The massive autophagic buildup in Pompe skeletal muscle negatively affects the trafficking and lysosomal delivery of the recombinant enzyme.22, 25, 26, 27 The removal of this autophagic buildup allows for efficient lysosomal glycogen clearance following ERT.28
Defective autophagy is also linked to muscle atrophy,26, 29, 30, 31, 32, 33 a hallmark of Pompe disease. Muscle atrophy occurs when the balance between protein synthesis and degradation is shifted in favor of protein degradation. Surprisingly, little is known about these processes in Pompe skeletal muscle. Here, we have identified the mechanisms involved in protein homeostasis by analyzing mechanistic target of rapamycin (mTOR)-dependent and independent protein synthesis and proteasomal degradation in muscle of GAA knockout (KO) mice. These data served as reference points for evaluation of the consequences of different ways to inhibit autophagy and render muscle cells more responsive to ERT.
Results
Genetic Suppression of Autophagy in KO Muscle
The enlargement and rupture of glycogen-filled lysosomes, buildup of autophagic debris, and atrophy are the major morphological changes in skeletal muscle in Pompe disease.7, 24 We have previously shown that the removal of autophagic buildup by genetic suppression of autophagy in skeletal muscle of KO mice resulted in reduction of lysosomal glycogen accumulation, and, most importantly, the remaining lysosomal glycogen was efficiently cleared following ERT.28 These experiments were done using autophagy-deficient KO mice (referred to as DKO), in which a critical autophagic gene, Atg7, is inactivated in skeletal muscle. Suppression of autophagy in skeletal muscle of wild-type (WT) mice was shown to have negative consequences, including oxidative stress, muscle weakness, and atrophy.34, 35, 36 To evaluate potential problems associated with this therapeutic approach in Pompe disease, we first assessed muscle proteostasis in KO mice, of which little, if anything, is known.
Both Pompe patients and KO mice develop progressive muscle atrophy, indicating that the normal equilibrium between protein synthesis and degradation is disturbed in the diseased muscle, leading to a net loss of contractile proteins and reduced muscle function. The regulation of muscle protein synthesis is largely mediated by the rapamycin-sensitive mTOR complex 1 (mTORC1), a component of the evolutionarily conserved TOR kinase. Muscle proteolysis is controlled by the ubiquitin-proteasome system (UPS), autophagy-lysosomal, and calcium-dependent pathways.37, 38, 39
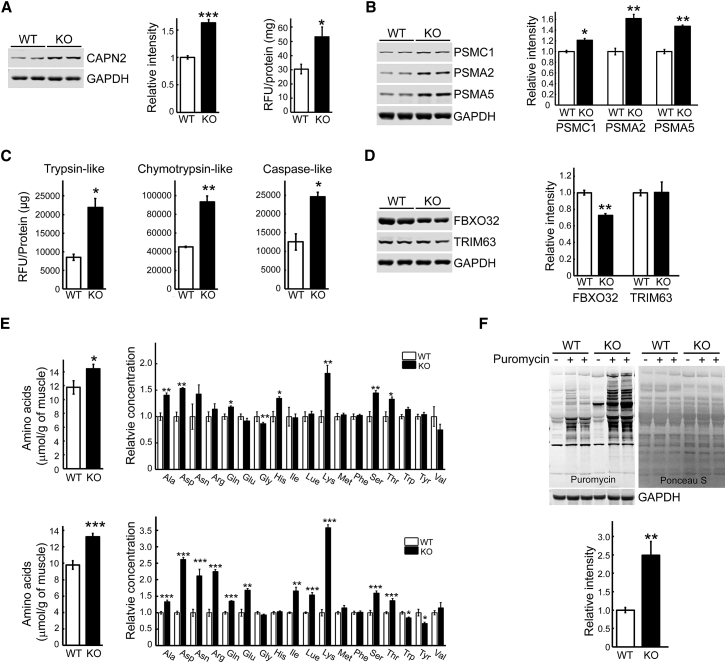
Consistent with our previous data on the increase in intracellular calcium in Pompe muscle,40 we have found an increase in the level and activity (30.5 ± 3.3 relative fluorescence units [RFU]/mg versus 53.2 ± 6.9 RFU/mg in WT and KO, respectively) of calpain-2 (CAPN2)—a ubiquitous calcium-dependent protease involved in limited proteolysis of sarcomeric proteins, followed by their degradation by the proteasome system37 (Figure 1A). We have also found a significant increase in the levels of several proteasomal subunits (PSMC1, PSMA2, and PSMA5) by western blot and an increase in all three proteasome activities, chymotrypsin-, trypsin- and caspase-like, in proteasome-enriched fraction from muscle extracts of KO mice compared to WT (Figures 1B and 1C). Furthermore, analysis of our previously reported mRNA sequencing (mRNA-Seq) of muscle samples from KO and WT mice40 (GEO: GSE57980) revealed sets of upregulated genes (≥2.5-fold relative to WT controls; p < 0.05) coding for proteins involved in protein degradation pathways (Tables S1–S6; Supplemental Information).
Figure 1.
Markers of Muscle Catabolism, Amino Acid Content, and the Rate of Protein Synthesis in KO Muscle
(A) Immunoblot analysis of total lysates from WT and KO muscle shows increased level of CAPN2 in the KO (n = 6 for each condition). Calpain activity was measured in tissue extracts from WT and KO muscle (prepared in RIPA buffer without protease and phosphatase inhibitors), and the results are presented in relative fluorescence units (RFU)/mg protein. (B) Immunoblot analysis of total lysates from WT and KO muscle shows increased levels of proteasome 26S subunit, ATPase 1 (PSMC1), and proteasomal alpha 2 (PSMA2) and alpha 5 (PSMA5) subunits (components of the 20S core proteasome complex) in the diseased muscle (n = 3 for each condition). (C) The proteasome activity was significantly increased in KO muscle lysates compared to WT controls. The activity was measured in proteasome-enriched fractions isolated from WT and KO muscle extracts. The results are displayed in relative fluorescence units (RFU)/mg protein. Six-month-old female mice were used for the experiments (n = 6 for each condition). (D) Immunoblot analysis of total lysates from WT and KO muscle shows no increase in FBXO32 (F-box protein 32, formerly MAFBx, Atrogin-1) and TRIM63 (tripartite motif containing 63 protein, formerly MuRF-1) in KO muscle (n = 3 for each condition). (E) Differences in muscle amino acid levels between WT and KO samples. Metabolome analysis (see Materials and Methods) shows a significant increase in the level of total and individual amino acids in young (top panel; 3-month-old females) and older (bottom panel; 5- to 6-month-old males) KO muscle. (F) Surface sensing of translation (SUnSET) analysis was used to evaluate the rate of protein synthesis in the gastrocnemius muscle of KO and WT mice. The animals were injected intraperitoneally with the aminoacyl-tRNA analog puromycin 30 min prior to sacrifice. The incorporation of puromycin into nascent polypeptides was detected by western blot with anti-puromycin antibody; western blot with anti-GAPDH antibody and Ponceau S staining were used as loading controls. Total intensity of puromycin-labeled polypeptides was quantified (n = 6 for each group). Graphs represent mean ± SE. *p < 0.05; **p < 0.01; ***p < 0.001. Student’s t test.
Taken together, the data point to the activation of protein breakdown in Pompe skeletal muscle. However, consistent with previous studies,28 the two muscle-specific ubiquitin ligases, MAFbx (atrogin-1/FBXO32) and MuRF-1 (TRIM63), commonly upregulated in atrophying muscle in conditions such as diabetes, cancer cachexia, fasting, and denervation,41, 42 were not increased in KO muscle (Figure 1D). Similarly, additional E3 ubiquitin ligases, known to be involved in muscle protein ubiquitylation and breakdown, such as Trim32 and TRAF6,39 were also not upregulated in Pompe muscle (data not shown).
Increased proteasomal degradation in KO muscle is expected to generate excess of amino acids. Indeed, metabolome analysis of muscle samples from 3-month-old KO (n = 7) and WT (n = 5) mice using capillary electrophoresis mass spectrometry (CE-MS) revealed increased levels of total amino acids in KO muscle; the changes were statistically significant for essential (His, Lys, and Thr) and non-essential (Ala, Asp, Gln, and Ser) amino acids (Figure 1E, top). The total level of amino acids continued to rise in older 5- to 6-month-old KO (KO, n = 7; WT, n = 5) mice, and the changes were significant for additional essential and non-essential amino acids (the only exception was His, which was slightly increased in younger but not in older KO) (Figure 1E, bottom). Complete data on 116 annotated metabolites and a set of metabolites that discriminate between the WT and KO are shown in Tables S7 and S7A.
Next, we analyzed the rate of protein synthesis in vivo in KO muscle by surface sensing of translation (SUnSET) method, which relies on the incorporation of puromycin, a tyrosyl-tRNA analog, into nascent peptide chains leading to the termination of their elongation.43 Puromycin was injected intraperitoneally (0.04 μmol/g) for 30 min followed by muscle collection and western blotting with an anti-puromycin antibody. In contrast to myotubes derived from the KO mice,44 a significant increase in anti-puromycin immunoreactivity was detected in KO muscle compared to WT control (Figure 1F). Our previous data on the reduction of cross-sectional area of myofibers—the typical attributes of muscle atrophy—in different muscle groups of KO mice22, 45, 46, 47 suggest that the increased rate of protein synthesis does not fully compensate, but may, at least to some degree, counterbalance increased protein degradation in the KO muscle.
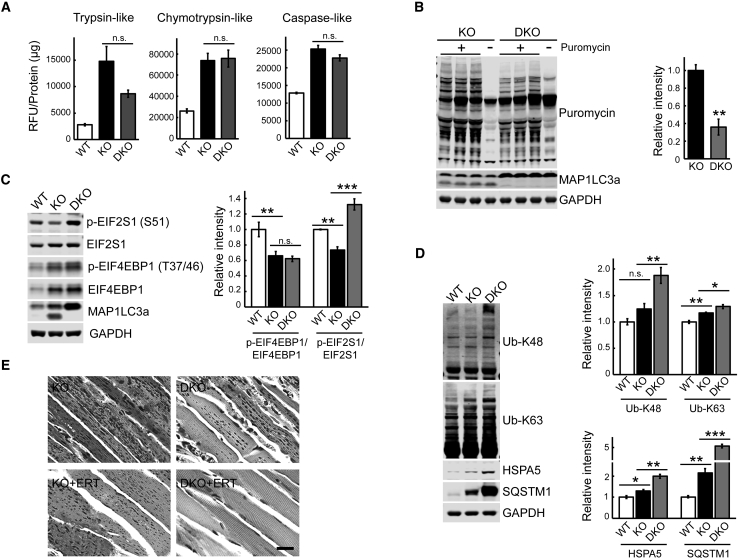
Suppression of autophagy in the diseased muscle, somewhat unexpectedly, did not further increase proteasome activities in DKO muscle compared to KO (Figure 2A). On the other hand, the rate of protein synthesis was lower in DKO compared to KO, as shown by the SUnSET assay (Figure 2B), suggesting that autophagy, although defective in KO, still confers some protection against muscle loss observed in DKO. Consistent with our previous data, KO muscle exhibits a diminished activity of mTORC1, as shown by a significant decrease in p-EIF4EBP1/EIF4EBP1 ratio, and this ratio remains unchanged in DKO (Figure 2C).
Figure 2.
Effects of Suppression of Autophagy on Muscle Proteostasis and the Outcome of ERT
(A) No significant changes in proteasome activity were observed in KO muscle following genetic suppression of autophagy. The activity was measured in proteasome-enriched fractions isolated from muscle WT, KO, and autophagy-deficient KO (DKO) muscle extracts. The results are displayed in relative fluorescence units (RFU)/mg protein. Four-month-old male GFP-LC3:KO were used for the experiments (n = 3 for KO and DKO; n = 2 for WT). (B) The rate of protein synthesis was significantly decreased in muscle from DKO mice. SUnSET analysis was performed as in Figure 1. Western blot with anti-MAP1LC3a antibody was performed to confirm efficient suppression of autophagy in DKO muscle as indicated by the absence of the lower band. n = 3 for each group. (C) Western blot analysis of muscle lysates from WT, KO, and DKO mice with the indicated antibodies. No changes in the mTOR activity are detected in DKO compared to KO samples; both KO and DKO exhibit diminished mTOR activity when compared to WT as shown by the degree of phosphorylation of EIF4EBP1, a downstream target of mTOR. In contrast, the level of p-EIF2S1S51 in the KO muscle was significantly decreased compared to the WT control, whereas it was markedly increased in DKO exceeding the WT level. n = 8 for KO and DKO; n = 4 for WT. (D) Western blot analysis of muscle lysates from WT, KO, and DKO mice with the indicated antibodies. Suppression of autophagy raised both K48-linked and K63-linked Ub conjugates. Note, only K63-linked Ub conjugates are increased in the KO muscle. n = 6 for KO and DKO; n = 3 for WT. ER stress marker HSPA5 and autophagosomal marker SQSTM1 are increased in muscle from KO mice and continue to rise in muscle from DKO mice. n = 4 for each condition. (E) PAS-stained sections (shown in black and white) of gastrocnemius muscle from 7- to 8-month-old untreated (top panels) and ERT-treated (lower panels) KO and DKO mice. Reduction of glycogen storage is observed in muscle from untreated DKO compared to KO mice; near-complete clearance of muscle glycogen is observed in ERT-treated DKO but not in ERT-treated KO mice (see also Table S8). Graphs represent mean ± SE. *p < 0.05; **p < 0.01; ***p < 0.001. Student’s t test.
Considering the reduced mTOR activity in both KO and DKO muscle, we have looked for an mTOR-independent mechanism to account for the enhanced protein synthesis in KO muscle and for the difference in the rate of protein synthesis between the two strains. Eukaryotic initiation factor 2 (EIF2S1) is indispensable for translation initiation; in its active guanosine triphosphate (GTP)-bound state EIF2S1binds to and delivers initiator Met-tRNAi to the 40S ribosomal subunit. Eukaryotic translation initiation factor 2B (EIF2B) serves as a guanosine diphosphate (GDP)/GTP exchange factor for EIF2S1. When EIF2S1 is phosphorylated on Ser51, p-EIF2S1S51 binds to and becomes a competitive inhibitor of EIF2B, thus leading to the repression of translation while stimulating expression of activating transcription factor 4.48, 49, 50, 51 We observed a significant decrease in the level of p-EIF2S1S51 in the KO muscle compared to the WT control (Figures 2C and S1), a finding consistent with the increase in protein synthesis. Several stress-responsive kinases, such as general control non-derepressible 2 (GCN2), phosphorylate EIF2S1 on Ser51.52 Despite our efforts, we were unable to detect GCN2 in muscle lysates by immunoblot. GCN2 is activated by uncharged aminoacyl-tRNAs that accumulate during amino acid starvation, and excess amino acids reverses this process.53, 54, 55 Unlike in KO muscle, phosphorylation of EIF2S1Ser51 was markedly increased in DKO exceeding the WT levels (Figure 2C), which may explain the lower rate of protein synthesis in DKO compared to KO muscle.
Consistent with our previous results,28 the level of the autophagy-specific substrate SQSTM1/p62 was markedly increased in DKO compared to KO (Figure 2D). In addition, the amounts of accumulated K63- and K-48 linked Ub proteins (targeted for autophagy-lysosomal pathway and for proteasomal degradation, respectively56) and glucose-regulated protein GRP78/BiP (HSPA5, the marker of endoplasmic reticulum stress) were also significantly higher in DKO compared to KO muscle (Figure 2D). Importantly, the increase in the amount of K-48 Ub proteins was seen only in DKO but not in KO muscle when compared to the WT levels (Figure 2D). These data again suggest that the partially functional autophagic pathway in KO muscle provides at least some defense against excessive accumulation of potentially toxic substrates and the endoplasmic reticulum (ER) stress.
However, in the setting of GAA deficiency, the benefits of suppression of autophagy in skeletal muscle may outweigh the risk. The removal of the autophagic buildup resulted in a significant increase in the force level (normalized by the fiber cross-sectional area) generated by single DKO fibers: 10.1 ± 0.7 N/cm2 compared to 6.4 ± 0.4 and 12.0 ± 0.8 N/cm2 in KO and WT, respectively.45 Furthermore, we have extended our earlier study on the effect of ERT in young KO mice28 by including older, 7- to 8-month-old DKO; again, biochemical analysis and periodic acid-Schiff (PAS) staining of muscle biopsies showed an efficient clearance of lysosomal glycogen in DKO but not in KO following the same regimen of three intravenous (i.v.) injections of recombinant human GAA (rhGAA) at a dose of 100 mg/kg (Figure 2E; Table S8).
Inhibition of Autophagy by TSC-Knockdown in KO Muscle
We have recently shown that a dramatic reduction in the number of fibers with massive autophagic buildup can be achieved by short hairpin RNA (shRNA)-mediated tuberous sclerosis complex (TSC) knockdown in KO muscle.44 TSC2, a GTPase-activation protein toward Rheb (Ras homolog enriched in brain), is a negative regulator of mTORC1 by keeping Rheb, a direct activator of mTORC1, in its inactive GDP-bound state.57, 58 In addition to the effect on autophagy, activation of mTOR in KO muscle by TSC knockdown led to an increase in muscle mass and reversal of atrophy.44
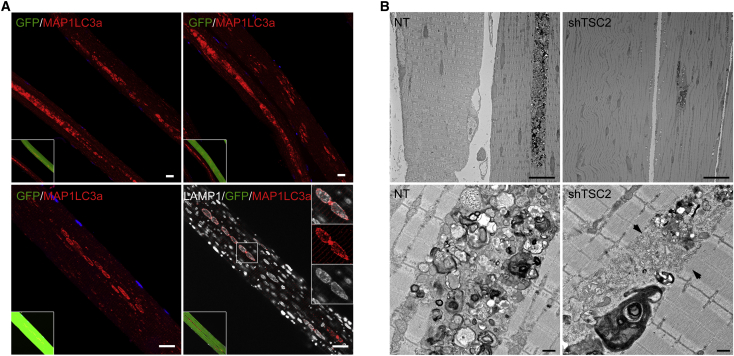
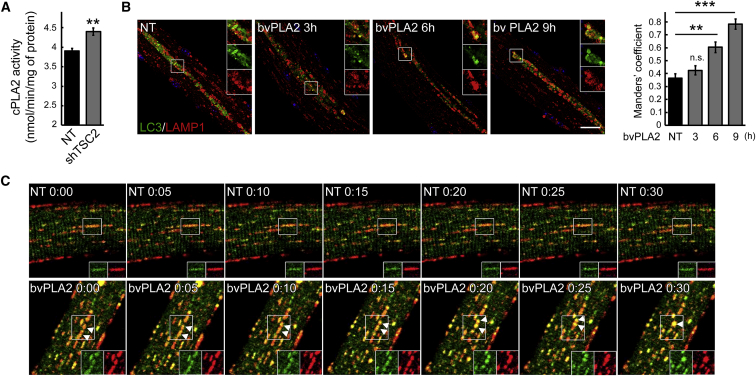
The possibility of affecting autophagic buildup while preventing muscle loss seems particularly attractive for Pompe disease. Consistent with our previous data,44 Adeno-associated virus serotype 1 (AAV1)-mediated shRNA-TSC knockdown increased p-EIF4EBP1/EIF4EBP1 ratio and decreased the MAP1LC3a-II level (Figure S2A). In control experiments using AAV1-null and AAV1-scrambled shRNA, no changes in the EIF4EBP1 or MAP1LC3a levels were observed (Figure S2B). The mechanism of TSC-mediated alleviation of the autophagic buildup in the infected KO fibers remains unclear. mTOR-mediated inhibition of autophagy, no doubt, contributes to the phenomenon. In addition, a closer look at the AAV1-shRNA-TSC-infected KO fibers offered a clue into how the process may advance. Immunostaining of the infected KO fibers with MAP1LC3a showed a striking difference in the shape and distribution of autophagosomes in the infected versus non-infected fibers: clearly discernable large MAP1LC3a-positive vesicular structures aligned in the core of muscle fibers or grouped in clusters can be seen in the infected (Figure 3A, green) but not in the neighboring non-infected fibers (Figure 3A, top panels). Immunostaining of the infected fibers with both LAMP1 and MAP1LC3a showed a near perfect colocalization of the two stains, indicating efficient lysosomal-autophagosomal fusion (lower panels in Figures 3A and S3). This pattern could represent an intermediate stage preceding the full resolution of the autophagic buildup in the diseased muscle. Examination of TSC-knockdown muscle by electron microscopy (EM) confirmed a dramatic reduction in the areas occupied by the autophagic buildup (Figure 3B, top panels). Furthermore, the buildup areas in the infected fibers appear to contain vesicle-free spaces, whereas densely packed vesicles are typically seen in the “overcrowded” autophagic areas of non-infected fibers (lower panels in Figures 3B and S4). These data suggested the possibility of an additional effect of TSC on vesicular fusion in the diseased muscle. TSC has been shown to negatively regulate phospholipase A2 (PLA2),59, 60 a family of enzymes involved in intracellular trafficking, membrane fusion, and lysosome activation.61, 62 mTOR-independent enhanced expression of PLA2 was shown in vitro and in vivo in TSC-deficient cells.60 Indeed, we have detected an increase in PLA2 activity in KO muscle following TSC knockdown (Figure 4A). We then analyzed the effect of PLA2 (from bee venom; 10 μg/mL) on autophagosome-lysosome fusion in live KO fibers. For these experiments, we used flexor digitorum brevis (FDB) muscle derived from GFP-LC3:KO mice; isolated cultured live fibers were incubated with PLA2 for up to 9 hr, followed by fixation and staining with anti-LAMP1 antibody. Figure 4B shows time-dependent increase in the number of LAMP1/MAP1LC3a-positive structures in the buildup area, indicating improved lysosomal-autophagosomal fusion. In an additional series of experiments, FDB muscles from GFP-LC3:KO mice were transfected by electroporation with plasmids containing mCherry-LAMP1; isolated live fibers were then analyzed by time-lapse confocal microscopy in the presence of PLA2. Once again, PLA2 treatment stimulated vesicular fusion as indicated by the gradual appearance of yellow color (Figure 4C; Videos S1 and S2). Thus, PLA2 may become a new target in the strategy to rid the diseased muscle of the autophagic buildup.
Figure 3.
Effect of AAV-Mediated TSC2 Inhibition on Autophagic Buildup in Muscle of the KO Mice
(A) Analysis of muscle biopsies from KO mice following AAV1-GFP-shRNA-TSC2 infection. Immunostaining of single fibers with anti-MAP1LC3a antibody only (red; top and lower left panels) or a combination of anti-MAP1LC3a (red) and anti-LAMP1 (white; lower right panel) antibodies showing distinct MAP1LC3a-positive autophagosomes and extensive co-localization of the two stains in the infected (green) but not in non-infected fibers. The pattern of MAP1LC3a-positive structures in non-infected fibers (GFP-negative; top panels) is strikingly different compared to that in the infected fibers. Scale bars, 20 μm. (B) Electron microscopy confirms a dramatic reduction in the size of the area occupied by autophagic buildup in muscle following TSC knockdown (shTSC2; top right panel). A single continuous area of autophagic accumulation in the gastrocnemius muscle of a 6-month-old untreated KO (NT) mouse illustrates the typical size of such an area (top left panel). 3-month-old KO mice were injected with AAV1- shTSC into the right gastrocnemius muscle; PBS was injected into the contralateral muscle. Animals were sacrificed 5.5 weeks after the procedure. Scale bars, 10 μm. A detailed view of the autophagic accumulation (lower panels) reveals a difference in the content of autophagic areas between the sham-treated (NT) and AAV-shTSC-infected (shTSC2) myofibers; reduced number of vesicles and vesicle-free areas (arrows) are seen in the infected fibers. Scale bar, 0.5 μm.
Figure 4.
A Putative Role for PLA2 in the Resorption of Autophagic Buildup
(A) Cytosolic phospholipase A2 (cPLA2) activity was increased in KO muscle following TSC knockdown. The activity was measured in muscle lysates from untreated (NT) and AAV-shTSC-infected (shTSC2) KO mice (n = 4 for each condition). (B) Bee venom PLA2 (10 μg/mL) stimulated lysosomal-autophagosomal fusion in KO muscle fibers. Cultured live fibers were isolated from FDB muscle derived from GFP-LC3:KO mice; the fibers were incubated with PLA2 for 3, 6, or 9 hr, followed by fixation and staining with anti-LAMP1 antibody. Improved lysosomal-autophagosomal fusion is indicated by a significant time-dependent increase in the number of LAMP1/MAP1LC3a-positive structures. (n = 4 for NT; n = 7 for 3 hr; n = 10 for 5 hr; n = 3 for 9 hr). Scale bar, 20 μm. (C) Time-lapse confocal microscopy of live muscle fibers cultured in the presence of PLA2 showing a gradual increase in the lysosomal-autophagosomal fusion. FDB muscles from GFP-LC3:KO mice were in vivo transfected by electroporation with plasmid containing mCherry-LAMP1. Images were taken on a Carl Zeiss LSM 780 confocal microscope with a 20× objective. Graphs represent mean ± SE. **p < 0.01; ***p < 0.001. Student’s t test.
Time-lapse confocal microscopy images were taken every 5 min. There is little (if any) interaction between lysosomes (red) and autophagosomes (green).
The fiber was cultured in the presence of bvPLA2 (10 μg/mL). Time-lapse confocal microscopy images were taken every 5 min. PLA2 stimulates the fusion between autophagosomes and lysosomes.
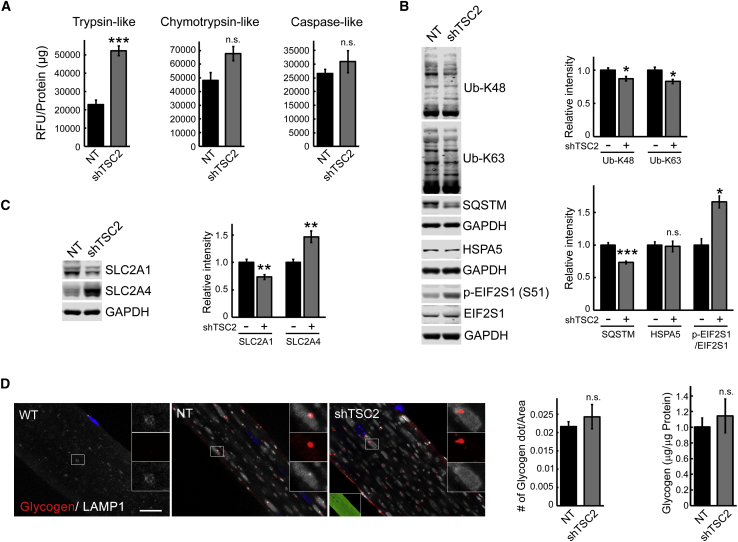
Proteasome activity, in particular, trypsin-like activity, was increased following TSC knockdown (Figure 5A), most likely to supply additional amino acids for mTOR-mediated protein synthesis. Also, EIF2S1-mediated translation was inhibited, as indicated by enhanced phosphorylation of EIF2S1 in TSC knockdown muscle (Figure 5B). Unlike genetic suppression of autophagy, TSC2 knockdown significantly decreased the amount of accumulated K48- and K63-linked Ub proteins and SQSTM1/p62 in KO muscle (Figure 5B); these changes were not seen in control experiments using AAV1-null and AAV1-scrambled shRNA (Figure S2B). In addition, TSC knockdown did not exacerbate ER stress, as shown by the levels of GRP78/BiP (HSPA5)—another advantage compared to suppression of autophagy (Figure 5B).
Figure 5.
Consequences of TSC Knockdown in Muscle of KO Mice
(A) The proteasome activity was measured in proteasome-enriched fractions isolated from sham-treated (NT) left gastrocnemius and AAV1-shRNA-TSC2 (shTSC2)-infected right gastrocnemius muscle from KO mice. An increase in trypsin-like activity is observed following TSC knockdown. The results are displayed in relative fluorescence units (RFU)/mg protein. Six-month-old female mice were used for the experiments (n = 4 for each condition). (B) Immunoblot analysis of total lysates from sham-treated (NT) and AAV1-shRNA-TSC2 (shTSC2)-infected KO muscle with the indicated antibodies. TSC knockdown reduces the amount of accumulated K48- and K63-linked Ub proteins and SQSTM1/p62 (n = 5 for each condition) and increases the level of phosphorylated EIF2S1 (n = 3; p-EIF2S1S51) in KO muscle; no changes are seen in the levels of GRP78/BiP (HSPA5) between the two groups (n = 3). (C) Immunoblot analysis of total lysates from sham-treated (NT) and AAV1-shRNA-TSC2 (shTSC2)-infected KO muscle shows a decrease in SLC2A1 but an increase in SLC2A4 following TSC knockdown (n = 5 for each condition). (D) Immunostaining of single fibers from WT, sham-treated (NT), and AAV1-shRNA-TSC2 (shTSC2)-infected KO muscle with anti-LAMP1 (white) and anti-glycogen (red) antibodies. Intralysosomal glycogen is not detected in the WT fibers. The difference in the amount of lysosomal (left graph) and total (right graph) glycogen between the infected and sham-infected fibers is not significant. Scale bar, 20 μm. Data illustrate the mean ± SE; *p < 0.05, **p < 0.01. n = 3. Student’s t test.
A potential pitfall of TSC knockdown in KO muscle is an increase in glucose uptake and glycogen synthesis, as has been reported in muscle-specific TSC KO mice.63 The abundance of the glucose transporter SLC2A1 was decreased, but the level of SLC2A4 was, indeed, increased in KO muscle following TSC knockdown (Figure 5C). However, the amount of lysosomal glycogen did not appreciably increase in the infected muscle, as indicated by immunostaining of muscle fibers with anti-glycogen antibody (Figure 5D, left graph). Similarly, the levels of total muscle glycogen in the sham-infected and infected muscles were not significantly different (1.01 ± 0.113 μg/μg protein in the control versus 1.14 ± 0.216 μg/μg protein in the infected muscle; n = 4; p = 0.398) (Figure 5D, right graph).
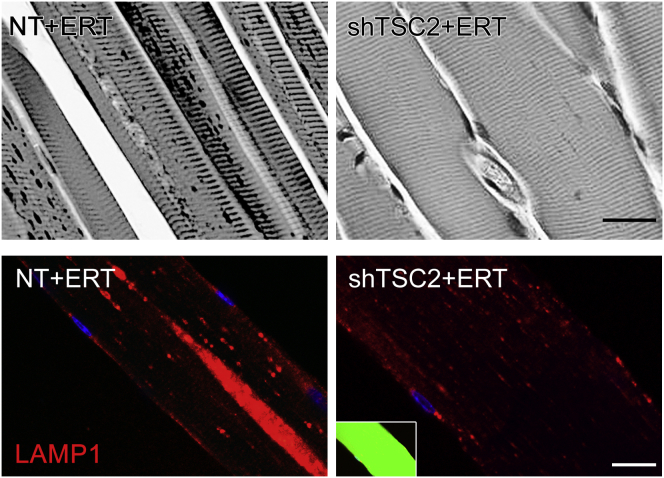
Most importantly, as in DKO, ERT following TSC inhibition resulted in efficient clearance of lysosomal glycogen and normalization of the lysosomal size. AAV1 vector expressing TSC2 shRNA (rAAV1-shRNA-TSC1/2) was introduced to 3- to 4-month-old KO mice (n = 5) by a single intramuscular (i.m.) injection in the three sites of the right gastrocnemius muscle with a total dose of 1 × 1011 genome copies (GC)/muscle. The left muscle, injected with PBS, served as a control. After 4 weeks, animals received four weekly i.v. injections of rhGAA (at a dose of 40 mg/kg); the mice were sacrificed 4–5 days after the last ERT injection. Efficient clearance of glycogen is demonstrated by PAS staining of muscle biopsies and immunostaining of myofibers with LAMP1 antibody (Figure 6). Furthermore, a significant decrease in the lysosomal size was observed in many fibers after a single i.v. injection of rhGAA (100 mg/kg) in combination with shRNA-mediated TSC knockdown (Figure S5).
Figure 6.
Effect of AAV-Mediated TSC2 Inhibition on the Outcome of ERT
Top panels: PAS-stained sections (shown in black and white) of gastrocnemius muscle from 6-month-old ERT-treated KO mice show efficient glycogen clearance in AAV-shTSC-infected (shTSC2 + ERT), but not in sham-infected muscle. 4-month-old KO mice were infected with AAV1-GFP-shRNA TSC2 vector preparation into the right gastrocnemius muscle; PBS was injected into the contralateral muscle; after 4 weeks, animals received four weekly i.v. injections of rhGAA (at a dose of 40 mg/kg); the mice were sacrificed 4–5 days after the last ERT injection. Lower panels: immunostaining of single fibers from ERT-treated KO mice with anti-LAMP1 antibody. The infected fiber (green; right) contains small dot-like lysosomes and no autophagic buildup. Scale bars, 20 μm.
Together, these data indicate that ERT works well when autophagic buildup is removed or diminished and that the manipulation of the TSC-mTOR signaling pathway appears to provide a greater benefit than genetic suppression of autophagy. Furthermore, the study reveals the possibility of reversing the fully formed autophagic buildup.
Discussion
Accumulation of autophagic debris along with the enlargement of glycogen-filled lysosomes in skeletal muscle are the histological hallmarks of Pompe disease.24 Multiple studies indicate that dysfunctional autophagy contributes to muscle atrophy in a wide range of disorders including Pompe disease,26, 29, 30, 31, 32, 33 in which muscle weakness and wasting are prominent features. Another major degradation pathway, the UPS, as well as protein biosynthetic pathways, are no less critical in the pathogenesis of muscle wasting, but few studies have examined them in Pompe disease. This work is an attempt to fill the gap and to investigate how these two interconnected processes, protein synthesis and degradation, are regulated in the diseased muscle. mTOR protein kinase is a key factor involved in the control of muscle mass by coordinating both synthesis and degradation (reviewed in Yoon64).
We have recently demonstrated a diminished activity of mTORC1 in muscle fibers from KO mice.44 This is consistent with reports of reduced mTORC1 activation in GAA-deficient C2C12 myoblasts65 and in patients’ fibroblasts and myocytes derived from induced pluripotent stem cells (iPSCs).66, 67 The molecular link between mTOR activity and autophagosome formation is well defined: activation of mTOR by nutrients and growth factors stimulates protein synthesis and inhibits autophagy and, conversely, inhibition of mTOR by nutrient or serum withdrawal stimulates protein breakdown by inducing autophagy.68, 69 In contrast, the relationship between this serine-threonine protein kinase and the ubiquitin proteasome system is less clear. A recent study indicates that inhibition of mTOR activates both autophagic and proteasomal degradation pathways in different cell types.70 Indeed, we found increased proteasome content and activity in Pompe skeletal muscle. Activation of calpain 2 in the diseased muscle further supports the enhanced proteasomal degradation. Calpains, calcium-dependent cysteine proteases, are believed to initiate limited proteolysis of myofibrillar proteins, followed by their degradation by the UPS.37 The catabolic response in Pompe skeletal muscle is also supported by the reported increase in the levels of urinary 3-methylhistidine (3-MH), an indirect indicator of muscle proteolysis, in KO mice.65
Enhanced protein degradation through the ubiquitin-proteasome pathway is largely responsible for loss of muscle mass in a wide range of systemic disorders including diabetes, renal failure, immobilization, and cancer. Two muscle-specific E3 ubiquitin ligases, Atrogin1/MAFbx and MuRF-1 (muscle RING finger-1 protein) are dramatically upregulated and activated in these conditions.41, 42, 71 Both ubiquitin ligases as well as other autophagy-related genes Bnip3, Beclin1, and p62 were upregulated in muscle biopsies from three Pompe patients with the most severe infantile form of the disease.26 However, Atrogin1 and MuRF-1 were not increased in KO muscle, suggesting that a lesser extent of muscle damage in the KO may explain the difference. As in Pompe disease, upregulation and activation of the proteasome system in skeletal muscle have been reported in other muscle dystrophies, including Duchenne Muscular Dystrophy (DMD) and congenital muscular dystrophy with laminin alpha2 chain deficiency (also known as MDC1A).72, 73, 74 A ubiquitin-ligase, TRIM32, but not Atrogin-1 or MuRF-1, was selectively induced in muscle biopsies from patients with dystrophin deficiency.75 Again, this ubiquitin-ligase remained unchanged in KO muscle. An induction of Atrogin-1, but not MuRF-1, was reported in muscle atrophy associated with Danon disease,76 an X-linked dominant deficiency of lysosome-associated membrane protein 2 (LAMP2).77 These data suggest that the mechanisms regulating excessive protein breakdown in myopathies and muscle dystrophies are disease-specific and different from those in systemic muscle-wasting conditions when the initial insult is external. Interestingly, muscle biopsies from both young Pompe patients and KO mice bear a strong resemblance to aging skeletal muscle,78 in which no changes in the Atrogin-1 and MuRF-1 were observed.79
The proteasome-dependent release of amino acids may stimulate protein synthesis by activating mTORC1 to counterbalance increased protein degradation as was shown in denervation-induced muscle atrophy.80 Higher amino acid content was recently reported in Pompe disease iPSC-derived cardiomyocytes.81 We, too, detected an excess of amino acids and an increase in the rate of protein synthesis in KO muscle, but this increase was not through activation of mTOR. Yet another scenario was described in a mouse model of hindlimb immobilization: a reduction in the rate of protein synthesis and muscle mass was associated with an increase in mTOR signaling,82 again suggesting that there is no common mechanism regulating protein synthesis in different muscle wasting conditions.
Excessive protein breakdown accompanied by depressed protein synthesis may lead to rapid muscle loss, as observed in muscle atrophying in response to various catabolic stimuli (reviewed in Sandri;30 Cohen et al.83). Therefore, increased protein synthesis in KO muscle can be considered a compensatory (although not sufficient to prevent atrophy) mechanism to counteract the increased proteolysis. Of note, we have previously reported a decrease in the rate of protein synthesis in cultured muscle cells derived from the KO mice,44 indicating that caution should be used when extrapolating the in vitro data. On the other hand, our results in the KO mimic early (and somehow forgotten) data in Pompe patients: a rapid fall in circulating amino acids after protein intake in a child with Pompe disease, as well as isotope turnover study in an adult patient, suggested an increase in both protein synthesis and degradation with a shift of protein balance toward degradation.84, 85 These observations, although limited, formed the basis of the high protein and exercise therapy for Pompe patients.84, 86
An mTOR-independent increase in the global rates of protein synthesis in KO muscle was associated with decreased phosphorylation of the α subunit of the EIF2S1, a key regulatory mechanism for translational control.87 Several stress-responsive kinases, including GCN2, phosphorylate EIF2S1 on an evolutionarily conserved serine residue (Ser51); this phosphorylation leads to a block in general translation while promoting translation of ATF4 that upregulates the expression of a selective set of genes to mitigate stress condition49, 50 (reviewed in Baird and Wek51 and Castilho et al.88). GCN2 is activated under amino acid starvation by the uncharged aminoacyl-tRNAs,89 but its effect on EIF2S1 phosphorylation may vary in different cell types.90 Several studies showed that excess of amino acids reverses this process. When amino acids are abundant, GCN2 remains inactive, leaving Ser51 on EIF2S1 mostly in a non-phosphorylated state.91 A reduction in EIF2S1 phosphorylation was observed in vitro (in human tumor fibrosarcoma cells) following addition of glutamine54 and in vivo (in neonatal pigs) after long-term parenteral leucine infusion.55 Systemic infusion of amino acids to healthy individuals resulted in a significant dephosphorylation of EIF2S1 in untreated and dexamethasone-treated subjects.53
Thus, the reduced mTOR activity in Pompe muscle is associated with an EIF2S1-mediated increase in the rate of protein synthesis that partially compensates for the massive proteolysis. Considering activation of the proteasome system in Pompe disease and other muscle-wasting conditions, it seems intuitive that proteasome inhibitors may have beneficial effect. However, the data on the subject are controversial: some studies showed beneficial effects of proteasome inhibitors, such as Velcade (bortezomib or PS-341), MLN273 (PS-273), or MG-132, in muscle dystrophy models,72, 73, 74 whereas others fail to demonstrate a positive outcome.92 In general, there is a growing consensus that chronic proteasome inhibition to combat atrophy may be deleterious, leading to potentially harmful effects on cell metabolism and protein quality75 (reviewed in Cohen et al.83 and Sandri et al.93).
The approaches in this study were designed to address the underlying pathological and metabolic changes in Pompe skeletal muscle with the goal to improve the effect of ERT: (1) elimination of the autophagic buildup by genetic suppression of autophagy and (2) inhibition of autophagy by TSC-mediated activation of mTOR. TSC2 interacts with and inhibits activity of Rheb, which acts upstream of mTOR to stimulate its activity.58 Both approaches made the diseased muscle amenable to ERT as shown by complete or near-complete reversal of lysosomal pathology. Of course, these strategies are no substitute for gene therapy, which is being actively pursued for treatment of Pompe disease. Extensive preclinical studies using AAV-mediated delivery of human GAA have led to the first clinical trial as well as to the upcoming planned trials.94, 95, 96 However, it is by no means clear if the gene therapy would effectively reverse muscle atrophy and the accumulation of autophagic debris. The experiments in this study are proof of principle aimed to identify new targets to address these aspects of the disease pathogenesis.
We have previously tested ERT in two DKO strains, in which Atg5 or Atg7 gene was inactivated in the KO muscle (Atg5 DKO and Atg7 DKO, respectively). These previous experiments were performed in young 2.5-month-old mice. Both strains responded well to ERT but, unexpectedly, unlike Atg7 DKO, Atg5 DKO mice were clinically more affected and had a shortened lifespan compared to KO mice.28 Since then, we have clarified the discrepancy and showed that the adverse effect of Atg5 inactivation in Pompe mice was unrelated to suppression of autophagy.97 Therefore, in this study, only Atg7 DKO mice were used to evaluate the effect of ERT in old animals that are difficult to treat, and again, we observed a significant decrease in the amount of accumulated glycogen and its near-complete clearance following ERT. Furthermore, the removal of autophagic buildup (also referred to as non-contractile inclusions98) improved the level of myofiber force despite atrophy. Of note, positive outcomes, such as enhanced β-oxidation and energy expenditure, as well as protection from diet-induced obesity and insulin resistance, have been reported in muscle-specific Atg7 KO mice.99
The flipside of this approach involves an increase in the levels of K48- and K63- linked Ub conjugates, an increase in the amount of accumulated autophagic substrate SQSTM/p62 and ER stress marker GRP78/HSPA5, and increased phosphorylation of EIF2S1 leading to suppression of translation. These findings are consistent with the results obtained in muscle-specific Atg7 KO mice on a WT background.35, 36
The second approach provided a better outcome: TSC2-mediated activation of mTOR resulted in an increase in muscle mass, inhibition of autophagy, and a decrease in K48- and K63-linked Ub conjugates and SQSTM/p62, without exacerbation of ER stress. As with genetic suppression of autophagy, TSC knockdown inhibited EIF2S1-mediated translation, consistent with the data in muscle-specific TSC KO mice.63 A potential problem with long-term chronic mTOR hyperactivation is an increase in muscle glycogen content as shown in muscle-specific TSC KO mice.63
Perhaps most important, the study demonstrates that the fully developed autophagic buildup can be reversed in the diseased myofibers. A combination of mTOR-mediated inhibition of autophagy and the upregulation of PLA2 following TSC knockdown may explain the phenomenon. A combination of TSC-mediated activation of mTOR with ERT has the potential to address multiple aspects of the disease pathology.
Materials and Methods
Antibodies
The following primary antibodies were used: anti-Calpain2 (2539), anti-phospho-EIF2αS51 (3398), anti-EIF2α (9722), anti-ATF4 (11815), anti-EIF2B5 (3595), anti-phospho-4EBP1S65 (13443), anti-phospho-4EBP1T37/46 (9459), anti-4EBP1 (9644), anti-phospho-S6S235/236 (4858), anti-S6 (2217), and anti-cPLA2 (2832) were from Cell Signaling Technology; anti-GLUT1 (ab115730), anti-RFP (ab62341), anti-FBXO32 (ab168372), anti-Ub-K48 (ab140601), anti-phospho-EIF2B5S539 (ab4775), anti-SQSTM1 (ab56416), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ab9484) were from Abcam; anti-HSPA5 (SPA-826), anti-PSMC1 (PW8305), and anti-PSMA5 (PW8125) from Enzo Life Sciences; anti-Puromycin (MABE343) and anti-Ub-K63 (05-1308) from Millipore; anti-GLUT4 (GT41-A) from Alpha Diagnostic International; anti-PSMA2 (sc-67339) from Santa Cruz Biotechnology; anti-TRIM63 (NBP1-31207) from Novus Biologicals; anti-LC3B (L7543) from Sigma; anti-mouse LAMP-1 (553792) from BD Biosciences.
Animal Models, Treatments, and Muscle Tissue Processing and Analysis
Three previously described mouse models of Pompe disease were used for the experiments: GAA−/− mice (referred to as KO),100 transgenic GAA−/− mice expressing GFP-MAP1LC3a (referred to as GFP-LC3:KO),101 and muscle-specific autophagy-deficient GAA−/− mice (MLCcre:Atg7F/F:GAA−/−; referred to as DKO).28 Gender- and age-matched WT and GFP-LC3:WT mice were used as controls. The white part of gastrocnemius muscle and FDB muscle were used for the experiments. Force measurements of single muscle fibers was performed as described.45
PLA2 Treatment of FDB Muscle
Endotoxin-free plasmid containing full-length rat LAMP1 gene (mCherry-LAMP1) (GENEWIZ, NJ) was electroporated into FDB muscle of 3- to 4-month-old GFP-LC3:KO mice. Electric-pulse-mediated gene transfer was performed as described.102 The animals were sacrificed 7 days after the procedure. Isolation and culturing of single live fibers on Matrigel-coated plates were done as previously described.101 The fibers were allowed to settle overnight and incubated with PLA2 (10 μg/mL) (Sigma, P9279) for 3 hr. The fibers were then imaged in the presence of PLA2 on a Carl Zeiss LSM 780 confocal microscope (Göttingen, Germany) every 5 min with a 20× objective. Alternatively, non-electroporated live-cultured FDB fibers were isolated from GFP-LC3:KO mice and incubated with PLA2 for 3, 6, and 9 hr. The fibers were then fixed in 2% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 30 min at room temperature, immunostained with anti-LAMP1 antibody, and analyzed by confocal microscopy as described.101, 103 The extent of MAP1LC3a/LAMP1 co-localization was evaluated by calculating the Mander’s coefficients in ImageJ software (1.51p version) using the JACoP plugin.
Intramuscular Injection of AAV1- shRNA TSC2
Two preparations of AAV1- shRNA TSC2 were used for the experiments: one with GFP tag (AAV1-GFP-shRNA-TSC244), and the other with red fluorescent protein (RFP) tag (AAV1-RFP-shRNA TSC2; Vector Biolabs, PA). The latter was used for injections into GFP-LC3:KO mice. Two- four-month-old KO or GFP-LC3:KO mice were injected with a total dose of 1011 GC of AAV1 vector preparation into three sites of the right gastrocnemius (three injections of 25 μL each) using a Hamilton syringe. Equal volumes of PBS were injected into the contralateral muscle. Animals were sacrificed 4 to 6 weeks after the procedure. The white part of gastrocnemius muscle was used for analysis. Part of the sample was used to test the efficiency of the gene knockdown, and the remaining muscle was used for western blotting, enzyme assays, isolation and immunostaining of single muscle fibers, or EM. In a series of control experiments, 3-month-old KOs were injected with empty vector (AAV1 null) or AAV1 containing scrambled shRNA (AAV1-sc shRNA) as described above. A group of mice injected with AAV1-GFP-shRNA-TSC2 was also treated with the recombinant human GAA (Myozyme, alglucosidase alfa, Genzyme, a Sanofi company). The enzyme was provided under a cooperative research and development agreement (CRADA) between the NIH and Genzyme.
ERT
Myozyme was administered intravenously at a dose of 40 mg/kg twice every week into KO mice; the therapy started 3–4 weeks after intramuscular injection of AAV1-shRNA TSC2. ERT was also used to treat 7- to 8-month-old KO (n = 6) and DKO (n = 6) mice; these mice received three biweekly injections at a dose of 100 mg/kg. Diphenhydramine was injected intraperitoneally at a dose of 5 mg/kg 15 min prior to the second and subsequent doses of ERT to prevent anaphylaxis. The mice were sacrificed 1 week after the last Myozyme injection. Isolated gastrocnemius muscle was fixed in 3% glutaraldehyde/0.2 M sodium cacodylate buffer and stained with PAS reagent according to standard protocol. Part of the muscle sample was used for immunostaining of single muscle fibers.
Western Blot Analysis
For western blot analysis, whole muscle tissues were homogenized in radioimmunprecipitation assay (RIPA) buffer (PBS containing 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, and a protease and phosphatase inhibitor cocktail [Cell Signaling Technology, 5872]). Samples were centrifuged at 16,000 × g at 4°C for 15 min. Protein concentrations of the supernatants were measured using the Bio-Rad Protein Assay (Bio-Rad). Equal amounts of protein were run on SDS-PAGE gels (Invitrogen, Carlsbad, CA) followed by transfer onto nitrocellulose membranes (Invitrogen, Carlsbad, CA). Membranes were blocked in 1:1 PBS/Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE), incubated with primary antibodies overnight at 4°C, washed, incubated with secondary antibodies, and washed again. Blots were scanned on an infrared imager (LI-COR Biosciences).
Enzyme Activity Assays
Proteasome-enriched fraction of gastrocnemius muscle was isolated as described104 with some modifications. In brief, muscles were homogenized in ice-cold buffer containing 50 mM Tris-HCl, 5 mM MgCl2, and 250 mM sucrose (pH 7.5), followed by sequential centrifugation at 16,000 × g for 15 min, 100,000 × g for 1 hr, and 150,000 × g for 5 hr. The pellet from the final centrifugation containing the proteasome was resuspended in ice-cold buffer containing 40 mM Tris (pH 7.2), 50 mM NaCl, 2 mM β-mercaptoethanol, 5 mM MgCl2, and 10% glycerol. Protein concentration was determined by Bradford assay using bovine serum albumin as a standard. Proteasome activity was measured using the Proteasome Activity Fluorometric Assay Kit II (UBPBio, Aurora, CO, USA; #J4120) per the manufacturer’s instructions. Fluorogenic peptide substrates, Z-LLE-AMC, Suc-LLVY-AMC, and Boc-LRR-AMC, were used for caspase-, chymotrypsin-, and trypsin-like activities, respectively. To evaluate calpain activity, whole-muscle tissue was homogenized in RIPA buffer (without protease and phosphatase inhibitors), centrifuged at 16,000 × g for 15 min at 4°C, and the activity was measured using InnoZyme Calpain 1/2 Activity Assay kit (Millipore, CBA054) per the manufacturers’ protocol. To evaluate PLA2 activity, muscle tissue was homogenized in buffer containing 50 mM HEPES and 1 mM EDTA (pH 7.4), centrifuged at 10,000 × g for 15 min at 4°C, and the activity was measured using cytosolic Phospholipase A2 Assay kit (Abcam, ab133090) per the manufacturers’ protocol.
Measuring the Rate of Protein Synthesis, Metabolome Analysis, Immunostaining for Glycogen, and EM
Protein synthesis in WT, KO, and DKO mice was evaluated using SUnSET method as described.43 In brief, puromycin was injected intraperitoneally at a dose of 0.040 μmol/g. Thirty minutes after injection, muscle tissue was isolated and the amount of puromycin incorporated into nascent peptides was then evaluated by western blot using antibody against puromycin.
Gastrocnemius muscle (white part) from 3-month-old female (WT, n = 5; KO, n = 7) and 5- to 6-month-old male (WT, n = 5; KO, n = 7) mice was extracted and frozen by the isopentane-freezing process. Metabolome analysis was performed by Human Metabolome Technologies (HMT, Tsuruoka, Japan). In brief, the samples were homogenized, filtered, centrifuged, and subjected to metabolomics analysis using CE-MS system (Agilent Technologies, Santa Clara, CA, USA) in the cation and anion analysis modes. Metabolite peaks were quantified and annotated based on an HMT metabolite database (http://humanmetabolome.com/).
Immunostaining of single fixed muscle fibers for glycogen was performed according to a recently described protocol.105 The number of intra-lysosomal glycogen-positive particles in AAV1-shRNA TSC2 treated and untreated fibers was evaluated using ImageJ.
For EM, muscles were fixed in 4% formaldehyde/2% glutaraldehyde in 0.1 M cacodylate buffer and processed as described,103 except that osmication was done in the presence of 1.5% potassium ferrocyanide (“reduced osmium”) for improved glycogen staining.
Glycogen Measurement and Light Microscopy
Glycogen levels in skeletal muscle were evaluated by measuring the amount of glucose released after treatment of tissue extracts with glucoamylase as described.106 Alternatively, Glycogen Assay Kit (ab65620, Abcam) was used for glycogen measurements according to the manufacturer’s instructions. Muscle biopsies were fixed in 3% glutaraldehyde (EM grade, Electron Microscopy Sciences, Hatfield, PA) in 0.2 M sodium cacodylate buffer for 4 hr at 4°C, washed in 0.1 M sodium cacodylate buffer and stored at 4°C in the same buffer. Samples were then imbedded in paraffin, sectioned, and stained with PAS by standard procedures.
Statistical Analysis
Statistical significance was determined by two-tailed Student’s t test; error bars represent SE. Benjamini-Hochberg procedure was used to calculate the adjusted p values (q values) to decrease the number of RNaseq false positives. *p < 0.05 was considered statistically significant. ** indicates p values < 0.01 and *** indicates p values < 0.001.
Animal care and experiments were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. We have routinely used white part of gastrocnemius muscle for all the experiments unless indicated otherwise.
Author Contributions
J.L. performed experiments, analyzed and interpreted the data, and participated in preparation of the manuscript; B.S. analyzed and interpreted the data; R.P. analyzed and interpreted the data and participated in writing of the manuscript; N.R. designed, interpreted, and analyzed data, and wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We would like to thank Dr. Lishu Li and Dr. Sengen Xu for their assistance. We are also grateful to Dr. Alexander V. Skurat (Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis) for the generous gift of ant-glycogen antibody. We would like to thank Dr. Hossein Zare for help with statistical analysis. We would also like to thank Dr. Kunio Nagashima for help with the EM. This research was supported in part by the Intramural Research Program of the NHLBI of the NIH. J.L. was supported in part by a CRADA between the NIH and Genzyme Corporation and from the Acid Maltase Deficiency Association.
Footnotes
Supplemental Information includes five figures, eight tables, and two videos and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.04.025.
Contributor Information
Rosa Puertollano, Email: puertolr@nhlbi.hih.gov.
Nina Raben, Email: rabenn@mail.nih.gov.
Supplemental Information
References
- 1.DeRuisseau L.R., Fuller D.D., Qiu K., DeRuisseau K.C., Donnelly W.H., Jr., Mah C., Reier P.J., Byrne B.J. Neural deficits contribute to respiratory insufficiency in Pompe disease. Proc. Natl. Acad. Sci. USA. 2009;106:9419–9424. doi: 10.1073/pnas.0902534106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filosto M., Todeschini A., Cotelli M.S., Vielmi V., Rinaldi F., Rota S., Scarpelli M., Padovani A. Non-muscle involvement in late-onset glycogenosis II. Acta Myol. 2013;32:91–94. [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J., Desai A.K., Kazi Z.B., Corey K., Austin S., Hobson-Webb L.D., Case L.E., Jones H.N., Kishnani P.S. The emerging phenotype of late-onset Pompe disease: A systematic literature review. Mol. Genet. Metab. 2017;120:163–172. doi: 10.1016/j.ymgme.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Falk D.J., Todd A.G., Lee S., Soustek M.S., ElMallah M.K., Fuller D.D., Notterpek L., Byrne B.J. Peripheral nerve and neuromuscular junction pathology in Pompe disease. Hum. Mol. Genet. 2015;24:625–636. doi: 10.1093/hmg/ddu476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntosh P.T., Hobson-Webb L.D., Kazi Z.B., Prater S.N., Banugaria S.G., Austin S., Wang R., Enterline D.S., Frush D.P., Kishnani P.S. Neuroimaging findings in infantile Pompe patients treated with enzyme replacement therapy. Mol. Genet. Metab. 2018;123:85–91. doi: 10.1016/j.ymgme.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroos M., Hoogeveen-Westerveld M., van der Ploeg A., Reuser A.J. The genotype-phenotype correlation in Pompe disease. Am. J. Med. Genet. C. Semin. Med. Genet. 2012;160C:59–68. doi: 10.1002/ajmg.c.31318. [DOI] [PubMed] [Google Scholar]
- 7.van der Ploeg A.T., Reuser A.J. Pompe’s disease. Lancet. 2008;372:1342–1353. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 8.Kishnani P.S., Corzo D., Nicolino M., Byrne B., Mandel H., Hwu W.L., Leslie N., Levine J., Spencer C., McDonald M. Recombinant human acid [alpha]-glucosidase: major clinical benefits in infantile-onset Pompe disease. Neurology. 2007;68:99–109. doi: 10.1212/01.wnl.0000251268.41188.04. [DOI] [PubMed] [Google Scholar]
- 9.Strothotte S., Strigl-Pill N., Grunert B., Kornblum C., Eger K., Wessig C., Deschauer M., Breunig F., Glocker F.X., Vielhaber S. Enzyme replacement therapy with alglucosidase alfa in 44 patients with late-onset glycogen storage disease type 2: 12-month results of an observational clinical trial. J. Neurol. 2010;257:91–97. doi: 10.1007/s00415-009-5275-3. [DOI] [PubMed] [Google Scholar]
- 10.Schoser B., Hill V., Raben N. Therapeutic approaches in glycogen storage disease type II/Pompe Disease. Neurotherapeutics. 2008;5:569–578. doi: 10.1016/j.nurt.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoser B., Stewart A., Kanters S., Hamed A., Jansen J., Chan K., Karamouzian M., Toscano A. Survival and long-term outcomes in late-onset Pompe disease following alglucosidase alfa treatment: a systematic review and meta-analysis. J. Neurol. 2017;264:621–630. doi: 10.1007/s00415-016-8219-8. [DOI] [PubMed] [Google Scholar]
- 12.van der Ploeg A.T., Kruijshaar M.E., Toscano A., Laforêt P., Angelini C., Lachmann R.H., Pascual Pascual S.I., Roberts M., Rösler K., Stulnig T., European Pompe Consortium European consensus for starting and stopping enzyme replacement therapy in adult patients with Pompe disease: a 10-year experience. Eur. J. Neurol. 2017;24 doi: 10.1111/ene.13285. 768–e31. [DOI] [PubMed] [Google Scholar]
- 13.Chakrapani A., Vellodi A., Robinson P., Jones S., Wraith J.E. Treatment of infantile Pompe disease with alglucosidase alpha: the UK experience. J. Inherit. Metab. Dis. 2010;33:747–750. doi: 10.1007/s10545-010-9206-3. [DOI] [PubMed] [Google Scholar]
- 14.Prater S.N., Banugaria S.G., DeArmey S.M., Botha E.G., Stege E.M., Case L.E., Jones H.N., Phornphutkul C., Wang R.Y., Young S.P., Kishnani P.S. The emerging phenotype of long-term survivors with infantile Pompe disease. Genet. Med. 2012;14:800–810. doi: 10.1038/gim.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prater S.N., Patel T.T., Buckley A.F., Mandel H., Vlodavski E., Banugaria S.G., Feeney E.J., Raben N., Kishnani P.S. Skeletal muscle pathology of infantile Pompe disease during long-term enzyme replacement therapy. Orphanet J. Rare Dis. 2013;8:90–101. doi: 10.1186/1750-1172-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Case L.E., Beckemeyer A.A., Kishnani P.S. Infantile Pompe disease on ERT: update on clinical presentation, musculoskeletal management, and exercise considerations. Am. J. Med. Genet. C. Semin. Med. Genet. 2012;160C:69–79. doi: 10.1002/ajmg.c.31321. [DOI] [PubMed] [Google Scholar]
- 17.Parini R., De Lorenzo P., Dardis A., Burlina A., Cassio A., Cavarzere P., Concolino D., Della Casa R., Deodato F., Donati M.A. Long term clinical history of an Italian cohort of infantile onset Pompe disease treated with enzyme replacement therapy. Orphanet J. Rare Dis. 2018;13:32–44. doi: 10.1186/s13023-018-0771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien Y.H., Hwu W.L., Lee N.C. Pompe disease: early diagnosis and early treatment make a difference. Pediatr. Neonatol. 2013;54:219–227. doi: 10.1016/j.pedneo.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Peng S.S., Hwu W.L., Lee N.C., Tsai F.J., Tsai W.H., Chien Y.H. Slow, progressive myopathy in neonatally treated patients with infantile-onset Pompe disease: a muscle magnetic resonance imaging study. Orphanet J. Rare Dis. 2016;11:63. doi: 10.1186/s13023-016-0446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim J.A., Li L., Raben N. Pompe disease: from pathophysiology to therapy and back again. Front. Aging Neurosci. 2014;6:177. doi: 10.3389/fnagi.2014.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukuda T., Ahearn M., Roberts A., Mattaliano R.J., Zaal K., Ralston E., Plotz P.H., Raben N. Autophagy and mistargeting of therapeutic enzyme in skeletal muscle in Pompe disease. Mol. Ther. 2006;14:831–839. doi: 10.1016/j.ymthe.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman A.P., Puertollano R., Raben N., Slaugenhaupt S., Walkley S.U., Ballabio A. Autophagy in lysosomal storage disorders. Autophagy. 2012;8:719–730. doi: 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim J.A., Kakhlon O., Li L., Myerowitz R., Raben N. Pompe disease: shared and unshared features of lysosomal storage disorders. Rare Dis. 2015;3:e1068978. doi: 10.1080/21675511.2015.1068978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shea L., Raben N. Autophagy in skeletal muscle: implications for Pompe disease. Int. J. Clin. Pharmacol. Ther. 2009;47(Suppl 1):S42–S47. doi: 10.5414/cpp47042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nascimbeni A.C., Fanin M., Masiero E., Angelini C., Sandri M. The role of autophagy in the pathogenesis of glycogen storage disease type II (GSDII) Cell Death Differ. 2012;19:1698–1708. doi: 10.1038/cdd.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nascimbeni A.C., Fanin M., Tasca E., Angelini C., Sandri M. Impaired autophagy affects acid α-glucosidase processing and enzyme replacement therapy efficacy in late-onset glycogen storage disease type II. Neuropathol. Appl. Neurobiol. 2015;41:672–675. doi: 10.1111/nan.12214. [DOI] [PubMed] [Google Scholar]
- 28.Raben N., Schreiner C., Baum R., Takikita S., Xu S., Xie T., Myerowitz R., Komatsu M., Van der Meulen J.H., Nagaraju K. Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder—murine Pompe disease. Autophagy. 2010;6:1078–1089. doi: 10.4161/auto.6.8.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandri M. Autophagy in health and disease. 3. Involvement of autophagy in muscle atrophy. Am. J. Physiol. Cell Physiol. 2010;298:C1291–C1297. doi: 10.1152/ajpcell.00531.2009. [DOI] [PubMed] [Google Scholar]
- 30.Sandri M. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int. J. Biochem. Cell Biol. 2013;45:2121–2129. doi: 10.1016/j.biocel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raben N., Roberts A., Plotz P.H. Role of autophagy in the pathogenesis of Pompe disease. Acta Myol. 2007;26:45–48. [PMC free article] [PubMed] [Google Scholar]
- 32.Raben N., Takikita S., Pittis M.G., Bembi B., Marie S.K.N., Roberts A., Page L., Kishnani P.S., Schoser B.G., Chien Y.H. Deconstructing Pompe disease by analyzing single muscle fibers: to see a world in a grain of sand... Autophagy. 2007;3:546–552. doi: 10.4161/auto.4591. [DOI] [PubMed] [Google Scholar]
- 33.Nascimbeni A.C., Fanin M., Masiero E., Angelini C., Sandri M. Impaired autophagy contributes to muscle atrophy in glycogen storage disease type II patients. Autophagy. 2012;8:1697–1700. doi: 10.4161/auto.21691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J.J., Quijano C., Chen E., Liu H., Cao L., Fergusson M.M., Rovira I.I., Gutkind S., Daniels M.P., Komatsu M., Finkel T. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany N.Y.) 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Masiero E., Sandri M. Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy. 2010;6:307–309. doi: 10.4161/auto.6.2.11137. [DOI] [PubMed] [Google Scholar]
- 37.Powers S.K., Kavazis A.N., DeRuisseau K.C. Mechanisms of disuse muscle atrophy: role of oxidative stress. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R337–R344. doi: 10.1152/ajpregu.00469.2004. [DOI] [PubMed] [Google Scholar]
- 38.Glass D.J. Skeletal muscle hypertrophy and atrophy signaling pathways. Int. J. Biochem. Cell Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 39.Bonaldo P., Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 2013;6:25–39. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim J.A., Li L., Kakhlon O., Myerowitz R., Raben N. Defects in calcium homeostasis and mitochondria can be reversed in Pompe disease. Autophagy. 2015;11:385–402. doi: 10.1080/15548627.2015.1009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomes M.D., Lecker S.H., Jagoe R.T., Navon A., Goldberg A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lecker S.H., Jagoe R.T., Gilbert A., Gomes M., Baracos V., Bailey J., Price S.R., Mitch W.E., Goldberg A.L. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 43.Goodman C.A., Mabrey D.M., Frey J.W., Miu M.H., Schmidt E.K., Pierre P., Hornberger T.A. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25:1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim J.A., Li L., Shirihai O.S., Trudeau K.M., Puertollano R., Raben N. Modulation of mTOR signaling as a strategy for the treatment of Pompe disease. EMBO Mol. Med. 2017;9:353–370. doi: 10.15252/emmm.201606547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu S., Galperin M., Melvin G., Horowits R., Raben N., Plotz P., Yu L. Impaired organization and function of myofilaments in single muscle fibers from a mouse model of Pompe disease. J. Appl. Physiol. 2010;108:1383–1388. doi: 10.1152/japplphysiol.01253.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N., Ralston E., Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takikita S., Schreiner C., Baum R., Xie T., Ralston E., Plotz P.H., Raben N. Fiber type conversion by PGC-1α activates lysosomal and autophagosomal biogenesis in both unaffected and Pompe skeletal muscle. PLoS ONE. 2010;5:e15239. doi: 10.1371/journal.pone.0015239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimball S.R., Jefferson L.S. Role of amino acids in the translational control of protein synthesis in mammals. Semin. Cell Dev. Biol. 2005;16:21–27. doi: 10.1016/j.semcdb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 50.Kilberg M.S., Shan J., Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baird T.D., Wek R.C. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berlanga J.J., Santoyo J., De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur. J. Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z., Li G., Kimball S.R., Jahn L.A., Barrett E.J. Glucocorticoids modulate amino acid-induced translation initiation in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2004;287:E275–E281. doi: 10.1152/ajpendo.00457.2003. [DOI] [PubMed] [Google Scholar]
- 54.Ye J., Kumanova M., Hart L.S., Sloane K., Zhang H., De Panis D.N., Bobrovnikova-Marjon E., Diehl J.A., Ron D., Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson F.A., Suryawan A., Gazzaneo M.C., Orellana R.A., Nguyen H.V., Davis T.A. Stimulation of muscle protein synthesis by prolonged parenteral infusion of leucine is dependent on amino acid availability in neonatal pigs. J. Nutr. 2010;140:264–270. doi: 10.3945/jn.109.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nathan J.A., Kim H.T., Ting L., Gygi S.P., Goldberg A.L. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013;32:552–565. doi: 10.1038/emboj.2012.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J., Manning B.D. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inoki K., Li Y., Xu T., Guan K.L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Priolo C., Ricoult S.J., Khabibullin D., Filippakis H., Yu J., Manning B.D., Clish C., Henske E.P. Tuberous sclerosis complex 2 loss increases lysophosphatidylcholine synthesis in lymphangioleiomyomatosis. Am. J. Respir. Cell Mol. Biol. 2015;53:33–41. doi: 10.1165/rcmb.2014-0379RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li C., Zhang E., Sun Y., Lee P.S., Zhan Y., Guo Y., Osorio J.C., Rosas I.O., Xu K.F., Kwiatkowski D.J., Yu J.J. Rapamycin-insensitive up-regulation of adipocyte phospholipase A2 in tuberous sclerosis and lymphangioleiomyomatosis. PLoS ONE. 2014;9:e104809. doi: 10.1371/journal.pone.0104809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown W.J., Chambers K., Doody A. Phospholipase A2 (PLA2) enzymes in membrane trafficking: mediators of membrane shape and function. Traffic. 2003;4:214–221. doi: 10.1034/j.1600-0854.2003.00078.x. [DOI] [PubMed] [Google Scholar]
- 62.Burlando B., Marchi B., Panfoli I., Viarengo A. Essential role of Ca2+ -dependent phospholipase A2 in estradiol-induced lysosome activation. Am. J. Physiol. Cell Physiol. 2002;283:C1461–C1468. doi: 10.1152/ajpcell.00429.2001. [DOI] [PubMed] [Google Scholar]
- 63.Guridi M., Tintignac L.A., Lin S., Kupr B., Castets P., Rüegg M.A. Activation of mTORC1 in skeletal muscle regulates whole-body metabolism through FGF21. Sci. Signal. 2015;8:ra113. doi: 10.1126/scisignal.aab3715. [DOI] [PubMed] [Google Scholar]
- 64.Yoon M.S. mTOR as a key regulator in maintaining skeletal muscle mass. Front. Physiol. 2017;8:788. doi: 10.3389/fphys.2017.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shemesh A., Wang Y., Yang Y., Yang G.S., Johnson D.E., Backer J.M., Pessin J.E., Zong H. Suppression of mTORC1 activation in acid-α-glucosidase-deficient cells and mice is ameliorated by leucine supplementation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014;307:R1251–R1259. doi: 10.1152/ajpregu.00212.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nishiyama Y., Shimada Y., Yokoi T., Kobayashi H., Higuchi T., Eto Y., Ida H., Ohashi T. Akt inactivation induces endoplasmic reticulum stress-independent autophagy in fibroblasts from patients with Pompe disease. Mol. Genet. Metab. 2012;107:490–495. doi: 10.1016/j.ymgme.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 67.Yoshida T., Awaya T., Jonouchi T., Kimura R., Kimura S., Era T., Heike T., Sakurai H. A skeletal muscle model of infantile-onset Pompe disease with patient-specific iPS cells. Sci. Rep. 2017;7:13473. doi: 10.1038/s41598-017-14063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sabatini D.M. Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. USA. 2017;114:11818–11825. doi: 10.1073/pnas.1716173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng Y., Yao Z., Klionsky D.J. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015;25:354–363. doi: 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao J., Zhai B., Gygi S.P., Goldberg A.L. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. USA. 2015;112:15790–15797. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jagoe R.T., Goldberg A.L. What do we really know about the ubiquitin-proteasome pathway in muscle atrophy? Curr. Opin. Clin. Nutr. Metab. Care. 2001;4:183–190. doi: 10.1097/00075197-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 72.Bonuccelli G., Sotgia F., Capozza F., Gazzerro E., Minetti C., Lisanti M.P. Localized treatment with a novel FDA-approved proteasome inhibitor blocks the degradation of dystrophin and dystrophin-associated proteins in mdx mice. Cell Cycle. 2007;6:1242–1248. doi: 10.4161/cc.6.10.4182. [DOI] [PubMed] [Google Scholar]
- 73.Gazzerro E., Assereto S., Bonetto A., Sotgia F., Scarfì S., Pistorio A., Bonuccelli G., Cilli M., Bruno C., Zara F. Therapeutic potential of proteasome inhibition in Duchenne and Becker muscular dystrophies. Am. J. Pathol. 2010;176:1863–1877. doi: 10.2353/ajpath.2010.090468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carmignac V., Quéré R., Durbeej M. Proteasome inhibition improves the muscle of laminin α2 chain-deficient mice. Hum. Mol. Genet. 2011;20:541–552. doi: 10.1093/hmg/ddq499. [DOI] [PubMed] [Google Scholar]
- 75.Assereto S., Piccirillo R., Baratto S., Scudieri P., Fiorillo C., Massacesi M., Traverso M., Galietta L.J., Bruno C., Minetti C. The ubiquitin ligase tripartite-motif-protein 32 is induced in Duchenne muscular dystrophy. Lab. Invest. 2016;96:862–871. doi: 10.1038/labinvest.2016.63. [DOI] [PubMed] [Google Scholar]
- 76.Nascimbeni A.C., Fanin M., Angelini C., Sandri M. Autophagy dysregulation in Danon disease. Cell Death Dis. 2017;8:e2565. doi: 10.1038/cddis.2016.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T., Mora M., Riggs J.E., Oh S.J., Koga Y. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 78.Feeney E.J., Austin S., Chien Y.H., Mandel H., Schoser B., Prater S., Hwu W.L., Ralston E., Kishnani P.S., Raben N. The value of muscle biopsies in Pompe disease: identifying lipofuscin inclusions in juvenile- and adult-onset patients. Acta Neuropathol. Commun. 2014;2:2–17. doi: 10.1186/2051-5960-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sakuma K., Yamaguchi A. Recent advances in pharmacological, hormonal, and nutritional intervention for sarcopenia. Pflugers Arch. 2018;470:449–460. doi: 10.1007/s00424-017-2077-9. [DOI] [PubMed] [Google Scholar]
- 80.Quy P.N., Kuma A., Pierre P., Mizushima N. Proteasome-dependent activation of mammalian target of rapamycin complex 1 (mTORC1) is essential for autophagy suppression and muscle remodeling following denervation. J. Biol. Chem. 2013;288:1125–1134. doi: 10.1074/jbc.M112.399949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sato Y., Kobayashi H., Higuchi T., Shimada Y., Ida H., Ohashi T. Metabolomic profiling of Pompe disease-induced pluripotent stem cell-derived cardiomyocytes reveals that oxidative stress is associated with cardiac and skeletal muscle pathology. Stem Cells Transl. Med. 2017;6:31–39. doi: 10.5966/sctm.2015-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.You J.S., Anderson G.B., Dooley M.S., Hornberger T.A. The role of mTOR signaling in the regulation of protein synthesis and muscle mass during immobilization in mice. Dis. Model. Mech. 2015;8:1059–1069. doi: 10.1242/dmm.019414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cohen S., Nathan J.A., Goldberg A.L. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 2015;14:58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 84.Slonim A.E., Coleman R.A., McElligot M.A., Najjar J., Hirschhorn K., Labadie G.U., Mrak R., Evans O.B., Shipp E., Presson R. Improvement of muscle function in acid maltase deficiency by high-protein therapy. Neurology. 1983;33:34–38. doi: 10.1212/wnl.33.1.34. [DOI] [PubMed] [Google Scholar]
- 85.Umpleby A.M., Wiles C.M., Trend P.S., Scobie I.N., Macleod A.F., Spencer G.T., Sonksen P.H. Protein turnover in acid maltase deficiency before and after treatment with a high protein diet. J. Neurol. Neurosurg. Psychiatry. 1987;50:587–592. doi: 10.1136/jnnp.50.5.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Slonim A.E., Bulone L., Goldberg T., Minikes J., Slonim E., Galanko J., Martiniuk F. Modification of the natural history of adult-onset acid maltase deficiency by nutrition and exercise therapy. Muscle Nerve. 2007;35:70–77. doi: 10.1002/mus.20665. [DOI] [PubMed] [Google Scholar]
- 87.Wek R.C., Jiang H.Y., Anthony T.G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 88.Castilho B.A., Shanmugam R., Silva R.C., Ramesh R., Himme B.M., Sattlegger E. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim. Biophys. Acta. 2014;1843:1948–1968. doi: 10.1016/j.bbamcr.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 89.Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A.G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 90.Wang X., Proud C.G. A novel mechanism for the control of translation initiation by amino acids, mediated by phosphorylation of eukaryotic initiation factor 2B. Mol. Cell. Biol. 2008;28:1429–1442. doi: 10.1128/MCB.01512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dey M., Trieselmann B., Locke E.G., Lu J., Cao C., Dar A.C., Krishnamoorthy T., Dong J., Sicheri F., Dever T.E. PKR and GCN2 kinases and guanine nucleotide exchange factor eukaryotic translation initiation factor 2B (eIF2B) recognize overlapping surfaces on eIF2alpha. Mol. Cell. Biol. 2005;25:3063–3075. doi: 10.1128/MCB.25.8.3063-3075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Selsby J., Morris C., Morris L., Sweeney L. A proteasome inhibitor fails to attenuate dystrophic pathology in mdx mice. PLoS Curr. 2012;4:d8930. doi: 10.1371/4f84a944d8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sandri M., Coletto L., Grumati P., Bonaldo P. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J. Cell Sci. 2013;126:5325–5333. doi: 10.1242/jcs.114041. [DOI] [PubMed] [Google Scholar]
- 94.Byrne B.J., Falk D.J., Pacak C.A., Nayak S., Herzog R.W., Elder M.E., Collins S.W., Conlon T.J., Clement N., Cleaver B.D. Pompe disease gene therapy. Hum. Mol. Genet. 2011;20(R1):R61–R68. doi: 10.1093/hmg/ddr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doerfler P.A., Nayak S., Corti M., Morel L., Herzog R.W., Byrne B.J. Targeted approaches to induce immune tolerance for Pompe disease therapy. Mol. Ther. Methods Clin. Dev. 2016;3:15053. doi: 10.1038/mtm.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bond J.E., Kishnani P.S., Koeberl D.D. Immunomodulatory, liver depot gene therapy for Pompe disease. Cell. Immunol. 2017;17 doi: 10.1016/j.cellimm.2017.12.011. 30238–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lim J.A., Zare H., Puertollano R., Raben N. Atg5flox-derived autophagy-deficient model of Pompe disease: does it tell the whole story? Mol. Ther. Methods Clin. Dev. 2017;7:11–14. doi: 10.1016/j.omtm.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hesselink R.P., Wagenmakers A.J., Drost M.R., Van der Vusse G.J. Lysosomal dysfunction in muscle with special reference to glycogen storage disease type II. Biochim. Biophys. Acta. 2003;1637:164–170. doi: 10.1016/s0925-4439(02)00229-6. [DOI] [PubMed] [Google Scholar]
- 99.Kim K.H., Jeong Y.T., Oh H., Kim S.H., Cho J.M., Kim Y.N., Kim S.S., Kim D.H., Hur K.Y., Kim H.K. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 2013;19:83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- 100.Raben N., Nagaraju K., Lee E., Kessler P., Byrne B., Lee L., LaMarca M., King C., Ward J., Sauer B., Plotz P. Targeted disruption of the acid alpha-glucosidase gene in mice causes an illness with critical features of both infantile and adult human glycogen storage disease type II. J. Biol. Chem. 1998;273:19086–19092. doi: 10.1074/jbc.273.30.19086. [DOI] [PubMed] [Google Scholar]
- 101.Spampanato C., Feeney E., Li L., Cardone M., Lim J.A., Annunziata F., Zare H., Polishchuk R., Puertollano R., Parenti G. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 2013;5:691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DiFranco M., Quinonez M., Capote J., Vergara J. DNA transfection of mammalian skeletal muscles using in vivo electroporation. J. Vis. Exp. 2009;19:1520. doi: 10.3791/1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Raben N., Shea L., Hill V., Plotz P. Monitoring autophagy in lysosomal storage disorders. Methods Enzymol. 2009;453:417–449. doi: 10.1016/S0076-6879(08)04021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hobler S.C., Williams A., Fischer D., Wang J.J., Sun X., Fischer J.E., Monaco J.J., Hasselgren P.O. Activity and expression of the 20S proteasome are increased in skeletal muscle during sepsis. Am. J. Physiol. 1999;277:R434–R440. doi: 10.1152/ajpregu.1999.277.2.R434. [DOI] [PubMed] [Google Scholar]
- 105.Skurat A.V., Segvich D.M., DePaoli-Roach A.A., Roach P.J. Novel method for detection of glycogen in cells. Glycobiology. 2017;27:416–424. doi: 10.1093/glycob/cwx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kikuchi T., Yang H.W., Pennybacker M., Ichihara N., Mizutani M., Van Hove J.L., Chen Y.T. Clinical and metabolic correction of pompe disease by enzyme therapy in acid maltase-deficient quail. J. Clin. Invest. 1998;101:827–833. doi: 10.1172/JCI1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse confocal microscopy images were taken every 5 min. There is little (if any) interaction between lysosomes (red) and autophagosomes (green).
The fiber was cultured in the presence of bvPLA2 (10 μg/mL). Time-lapse confocal microscopy images were taken every 5 min. PLA2 stimulates the fusion between autophagosomes and lysosomes.