Abstract
The study of RNA has continually emphasized the structural and functional versatility of RNA molecules. This versatility has inspired translational and clinical researchers to explore the utility of RNA-based therapeutic agents for a wide variety of medical applications. Several RNA therapeutics, with diverse modes of action, are being evaluated in large late-stage clinical trials, and many more are in early clinical development. Hundreds of patients are enrolled in large trials testing messenger RNAs to combat cancer, small interfering RNAs to treat renal and hepatic disorders, and aptamers to combat ocular and cardiovascular disease. Results from these studies are generating considerable interest among the biomedical community and the public and will be important for the future development of this emerging class of therapeutic agents.
During the past 25 years, the idea of using RNA molecules as therapeutic agents has grown from a concept into a clinical reality (1, 2). Initially, RNA was thought to be a poor choice for a therapeutic agent, given its relatively short half-life in vivo. However, with improvements in stabilization chemistry, and by making informed decisions about where a relatively short-lived molecule may be useful clinically, much of this skepticism has been overcome. RNA molecules possess many properties that potentially make them useful therapeutics. They can fold into complex conformations that allow them to specifically bind proteins, small molecules, or other nucleic acids or even to form catalytic centers (3). Coding mRNAs are the exclusive carriers of genetic instructions from DNA to the ribosome. Many types of noncoding RNA (ncRNA) orchestrate the mobilization and utilization of genetic information through a variety of mechanisms whose discovery has dramatically changed our understanding of RNA biology (3). The four classes of therapeutic RNAs that have received the most attention can be classified by their modes of action: (i) encoding therapeutic proteins or vaccine antigens (mRNAs), (ii) inhibiting pathogenic RNA activity [small interfering RNAs (siRNAs), microRNAs (miRNAs), and antisense RNAs], (iii) modulating protein activity (RNA aptamers), and (iv) reprogramming genetic information [trans-splicing ribozymes and CRISPR guide RNAs (gRNAs)]. Here we briefly review progress in and challenges for the clinical development of such coding and noncoding RNA therapeutics, focusing on those that have reached large phase 3 clinical studies (table S1).
Therapeutic coding RNAs: mRNA-based immunotherapy
Preclinical studies suggest that direct delivery of therapeutic mRNAs into cells ex vivo or in vivo may result in substantial health benefits in a number of clinical settings where transient generation of a therapeutic protein is effective and often preferred. mRNAs that encode tumor antigens have been used to induce tumor-specific immune responses. Such mRNA-based vaccination has largely been performed by either loading dendritic cells (DCs) with mRNA ex vivo and reintroducing these DCs back into the body to translate, present, and elicit an immune response against the encoded tumor antigen (Fig. 1) (4, 5), or by direct injection of tumor antigen–encoding mRNA in vivo for uptake, translation, and presentation by antigen-presenting cells such as DCs (6, 7). In animal studies, such transient antigen presentation has led to long-lasting antitumor immune responses. Phase 1 clinical trials have been carried out in patients with several types of cancer [reviewed in (4, 5)], and an open-label phase 2 clinical study suggests that mRNA-based DC immunotherapy can improve median overall survival rates in renal cancer patients (8). Other phase 2 clinical studies in HIV-infected individuals have demonstrated that introducing mRNAs that encode viral antigens into DCs can induce long-lasting HIV-specific T cell responses (9, 10). Thus, even transient expression of tumor or viral antigens via mRNA transfection can result in prolonged effects. An ongoing phase 3 clinical study is using patient-derived DCs loaded with mRNA amplified from metastatic renal cell carcinoma tumors (Fig. 1 and table S1); this will help determine whether such transient mRNA-based immunotherapy improves long-term survival in patients.
Fig. 1. RNA-based tumor vaccines.
A DC-based vaccine is made using tumor antigen, either RNA isolated from a patient’s tumor biopsy or mRNA encoding a known tumor antigen. Patient peripheral blood mononuclear cells are used to isolate CD14+ monocytes, which in turn are used to generate immature DCs. RNA is transfected into the patient’s immature DCs, followed by DC activation and maturation ex vivo. These RNA-transfected mature DCs are administered to patients to elicit an antitumor immune response.
While this initial ex vivo strategy of using mRNA-based immunotherapy has been moving through clinical development, several improvements have been established to enhance mRNA-induced immunity. Electroporation has been used to more effectively load DCs with mRNA ex vivo (11). To further enhance the immune response, mRNAs encoding CD40L, CD70, and a constitutively active form of TLR4 have been delivered in conjunction with tumor antigens to generate optimized human DCs for use as cancer vaccines (12). Also, preconditioning the skin site before intradermal injection of a DC-loaded mRNA vaccine leads to increased DC migration to lymph nodes and improved survival in patients with glioblastoma (13).
Ex vivo DC vaccine strategies require complicated personalized cell therapy. Direct injection in vivo of mRNA encoding tumor antigens has the potential to develop into an off-the-shelf therapeutic agent. Direct injection of naked or protamine-protected mRNA leads to the induction of T and B cell responses (14), and intradermal administration of mRNA promotes the induction of antitumor immunity (15). However, such unmodified mRNA is expected to have a very short half-life in vivo. To optimize mRNA pharmacokinetics and limit unwanted inflammatory responses, investigators have modified elements of mRNA structure such as the 5′ and 3′ untranslated region; 5′ cap; coding region, poly(A) tail, and nucleotide chemistry [reviewed in-depth in (16)]. These optimized mRNAs are now being evaluated in ongoing clinical studies to determine whether they can effectively replace the more complicated and costly cell-based approaches.
In addition to active immunization, mRNA can be used to transiently generate T cells (or natural killer cells) that express tumor antigen–specific T cell receptors or chimeric antigen receptors for therapeutic applications (17, 18). Cell therapy using T cells electroporated with mRNA encoding mesothelin chimeric antigen receptors is in clinical trials in patients with pancreatic cancer and other solid malignancies (19). Lastly, in addition to being used as cancer vaccines, tumor mRNA–loaded DCs have been used to expand autologous T cells in vitro for adoptive cell therapy. Thus, coding RNA is central to a number of potential applications in immunotherapy settings where even transient production of a therapeutic agent can elicit a prolonged therapeutic effect.
Therapeutic proteins generated by mRNA
mRNA can be used to generate a variety of therapeutic proteins for applications that include protein replacement therapy, regenerative medicine, and therapeutic genome editing. Methods have been developed to reduce innate immune activation caused by the mRNA itself and to increase mRNA translation by incorporating modified nucleotides (16), because many therapeutic protein replacement applications will probably require repeated administration.
mRNAs can be used to reprogram cells. Electroporation into B cells of a cocktail of four different mRNAs encoding immune stimulatory proteins produces reprogrammed cells that are similar to natural DCs and as effective at stimulating T cells (20). A cocktail of mRNAs that encode four transcription factors was shown to efficiently reprogram cells to pluripotency, even though these mRNAs are expected to have a relatively short half-life in cells (21). mRNA-based approaches will probably be evaluated in the clinic in the near future, given that using mRNA for regenerative medicine applications poses less of a safety concern than DNA-based reprogramming. Another potential application of mRNA-based therapeutics is to transiently generate ZFNs (zinc-finger nucleases), TALENs (transcription activator-like effector nucleases), and Cas9 for therapeutic genome editing. For example, genetically modified cynomolgus monkeys have been generated by injecting Cas9 mRNA and CRISPR gRNA into one-cell embryos (22). Given ongoing concerns about off-target effects, mRNA-based transient expression of such nucleases is being actively pursued as a strategy to reduce risk. A number of clinical studies slated to begin in 2016 or 2017 will explore these new applications. Thus, though coding RNAs only transiently induce protein production, an increasing number of potential clinical applications appear to be well suited for these relatively short-lived therapeutic agents.
The three translational applications of therapeutic ncRNAs that have received the most clinical attention to date are (i) inhibition of mRNA translation, (ii) protein modulation, and (iii) genetic reprogramming. As shown in table S1, 10 ncRNA-based therapeutics have already entered large randomized phase 3 clinical studies.
ncRNA-mediated inhibition of mRNA translation
Antisense RNA binding to and regulating the expression of a complementary mRNA is a well-characterized phenomenon in prokaryotes, and expression of antisense RNA in mammalian cells has been used to inhibit a wide variety of target mRNAs. These observations have led to the development of several antisense oligonucleotide–based therapeutic agents that have been making their way through clinical trials. Because many of these antisense oligonucleotides contain DNA, we do not discuss them here in detail; a recent review considers the clinical development of this class of therapeutic agents (23).
The discovery of RNA interference (RNAi) (24) greatly increased interest in the potential utility of ncRNAs that can inhibit therapeutically relevant mRNAs through posttranscriptional gene silencing. Gene silencing can be induced by synthetic siRNAs (25) or miRNA mimics (1) or by expression of short hairpin RNAs (shRNAs) or primary miRNAs that are processed into therapeutic siRNAs or miRNAs. After loading into the RNA-induced silencing complex (RISC), targeted RNAs have their translation repressed (miRNA) or are cleaved and degraded (siRNA). In addition, once associated with silencing complexes, RNAi can induce the silencing of multiple target RNAs. Moreover, related RNA complexes have recently been shown to induce transcriptional gene silencing, which may elicit an even longer-lasting effect in cells (26). More than 20 clinical trials are underway evaluating the potential utility of RNAi-based therapeutics for a range of human diseases. RNAi-based therapies that have incorporated modified nucleotides to limit innate immune system activation seem to be well tolerated in patients (26), and six RNAi-based therapeutic agents have progressed into phase 3 clinical trials (table S1). Of the six, five use synthetic siRNAs and the sixth uses an expressed bifunctional shRNA in cancer cells. The shRNA approach involves two shRNAs targeted to furin mRNA (27) and is designed to augment immune responses against patient-derived tumor cells that are transfected with the shRNA expression cassette, lethally irradiated, and intradermally re-administered to the patient. Currently, this ex vivo shRNA-based cancer immunotherapy approach is being evaluated in a phase 3 clinical study to determine whether such transient gene silencing can improve long-term survival in cancer patients (table S1). Several other RNAi-based therapeutic agents are being evaluated for treatment of a variety of cancers, and several that do not involve ex vivo cell therapy are progressing through early clinical development [reviewed in (26)].
Two major pharmacokinetic challenges exist for developing synthetic oligonucleotide therapeutics: their limited oral bioavailability and the rapid rate at which short nuclease-resistant RNAs are cleared from circulation. All five of the current synthetic siRNA phase 3 clinical trials either use local delivery of siRNA to the eye or target delivery of siRNA to the liver or kidney, organs that are involved in oligonucleotide clearance after intravenous systemic administration (table S1). The siRNA bevasiranib was designed to silence vascular endothelial growth factor (VEGF) mRNA after direct intraocular administration in patients with age-related macular degeneration. Despite encouraging phase 2 results, this first-generation siRNA-based therapy did not reach the intended clinical endpoint in a phase 3 trial and was discontinued; this is probably because the chemistry used was not particularly potent, and because other VEGF inhibitors (aptamer and antibody) made it to market first and proved superior (28). Subsequent early-phase clinical studies with siRNAs targeting different mRNAs in the eye have been more promising (26). The ocular siRNA QPI-1007 targets Cas2 (29) to reduce retinal ganglion apoptosis in patients with optic neuropathy (table S1). This chemically modified siRNA drug has been granted an orphan drug designation by the U.S. Food and Drug Administration (FDA) and is moving into a phase 3 clinical trial. A large clinical trial is under way of an intravenously administered siRNA (QPI-1002) that targets p53 mRNA, localizes to the kidney, and is designed to transiently limit p53-induced apoptosis and thereby mitigate programmed cell death and kidney injury after transplantation (table S1) (30). Thus, siRNA appears to be particularly well suited for local or transient therapeutic applications.
Advances in siRNA formulation allow efficient delivery to the liver (26). The utility of siRNA-mediated silencing of transthyretin (TTR) mRNA in the liver of patients with familial TTR amyloidosis (31, 32) is being studied in two separate phase 3 clinical trials. The first trial uses a siRNA formulated with lipid nanoparticle technology (ALN-TTR02) and intravenous administration to attempt to improve neuropathy in patients. This is the therapeutic siRNA that is closest to potential approval. In contrast, the second trial uses a TTR-targeted siRNA (ALN-TTRSC) that is formulated with N-acetylgalactosamine (GalNAc) to facilitate uptake by and release into the cytoplasm of hepatocytes, as well as to allow for subcutaneous administration (Fig. 2 and table S1). Many other receptor-targeting agents are being evaluated preclinically to determine whether siRNA can be selectively and effectively delivered to the cytoplasm of other cell types in other tissues.
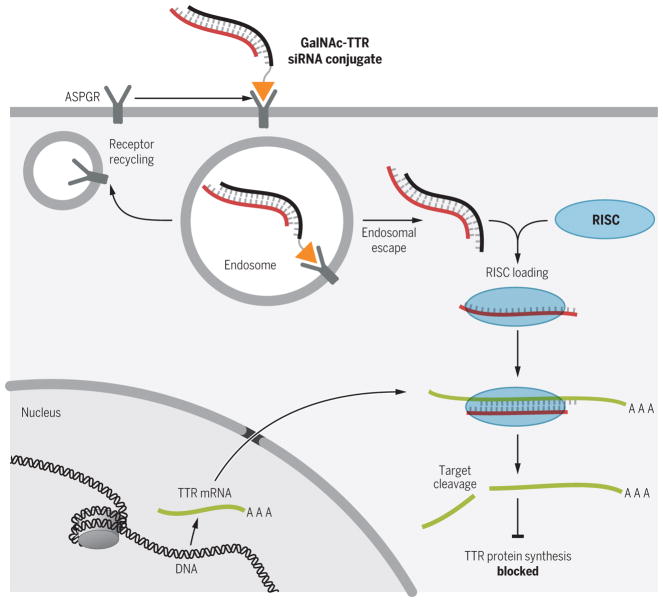
Fig. 2. siRNA-mediated treatment of TTR amyloidosis.
siRNA-mediated inhibition of mutant TTR synthesis in the liver is used to limit TTR protein generation and subsequent protein misfolding, oligomerization, and systemic pathology. GalNAc (orange triangle) is appended to the siRNA (ALN-TTRSC, black line) to facilitate delivery to hepatocytes through the asialoglycoprotein receptor (ASGPR) and release from the endosomal compartment into the cytoplasm, where the targeting strand (red line) is taken up by the RISC complex. The RISC complex then targets the TTR mRNA (green line; AAA, poly(A) tail) for destruction, preventing synthesis of the TTR protein.
ncRNAs as protein antagonists
Certain viruses express short, structured ncRNAs (aptamers) that bind and antagonize cellular proteins involved in antiviral responses. That these aptamers can have therapeutic applications has been demonstrated by the expression of the short, stem-loop HIV-derived TAR ncRNA in CD4+ T cells, where it bound and repressed the viral protein Tat and thereby inhibited HIV replication (33). RNA aptamers can be generated against novel targets in vitro by using a combinatorial chemistry method termed SELEX (systematic evolution of ligands by exponential enrichment); well over 1000 proteins can now be targeted by high-affinity and -specificity aptamers, including many proteins of therapeutic interest (34). SELEX allows the incorporation of modified nucleotides so that RNA aptamers can be generated that are highly nuclease-resistant and therefore suitable for animal and clinical studies. Because nuclease-resistant RNA aptamers can target extracellular proteins and do not have to cross the cell membrane to elicit a therapeutic effect, their clinical development has focused largely on targeting soluble extracellular proteins or extracellular domains of cell-surface receptors. Several aptamers have now been evaluated in the clinic (1), with four moving into large phase 3 clinical studies (table S1).
Ocular diseases present a natural fit for the clinical development of therapeutic aptamers, because local delivery to the eye (2) would theoretically eliminate pharmacokinetic challenges associated with systemic delivery of short oligonucleotides. Aptamers have been created against several proteins that are thought to be important therapeutic targets for ocular indications, including VEGF, platelet-derived growth factor (PDGF), and complement component 5 (C5) (2); aptamers targeting each of these are in or have completed phase 3 clinical trials. Patients suffering from neovascular age-related macular degeneration and treated with the aptamer that targets VEGF (Macugen) demonstrated a 45% improvement in limiting disease progression compared with the standard of care (2). Macugen received FDA approval for ophthalmic use in 2004 and reached sales of $185 million in the United States in 2005. Protein-based inhibitors of VEGF in the eye have since largely replaced Macugen in the market because of their improved efficacy (2). Nonetheless, Macugen demonstrated that an RNA therapeutic was marketable and addressed a then-unmet medical need. Two additional aptamers targeting PDGF and C5 in combination with VEGF protein inhibitors (e.g., Lucentis) are now progressing through phase 3 clinical studies (table S1) after encouraging phase 2 studies, which indicate that such aptamer-based combination therapy further improves outcomes for patients with age-related macular degeneration (2).
A drug-antidote approach has been used to develop a rapid-onset and rapidly reversible RNA aptamer–based anticoagulant agent for use in acute care and clinical settings requiring short-term yet potent anticoagulation, such as percutaneous coronary intervention in cardiovascular disease patients. The aptamer RB006, which was developed for systemic delivery [formulated with a 40-kDa polyethylene glycol (PEG)], targets coagulation factor IXa (35), and the matched RNA oligonucleotide (RB007) is the “antidote” that forms base pairs with the aptamer (Fig. 3). This drug-antidote approach highlights a major advantage of aptamers over antibody-based drugs: It is easy to generate antidotes to aptamers to provide physicians with additional control over aptamer-based medications. Unfortunately, after treating only 25% of an anticipated 6600 patients, the phase 3 trial was halted because of serious allergic reactions in a very small number of recipients that received the PEGylated aptamer (36). These rare allergic reactions are associated with the presence of high amounts of preexisting antibodies to PEG in allergic individuals (37). However, it is clear that the factor IXa RNA aptamer can serve as a potent rapid-onset anticoagulant, controlling coagulation and limiting thrombosis as effectively as the standard of care in this common clinical setting (Fig. 3). The study also demonstrated that RNA aptamers are rapidly controllable therapeutic agents with the potential to improve safety and efficacy and that alternative formulations to PEG should be considered to improve aptamer pharmacokinetic properties. The collective clinical experience with the ocular aptamers in phase 3 clinical studies and the anticoagulant activity achieved with the factor IXa aptamer in over 1600 cardiovascular disease patients bode well for the future development of RNA aptamers as a class of therapeutic agents. The major challenge for the widespread commercialization of RNA aptamer–based therapeutics remains the need to distinguish them from monoclonal antibody competitors, which naturally have longer-lasting pharmacokinetic properties after systemic administration.
Fig. 3. The factor IXa aptamer anticoagulation system.

An anticoagulant aptamer (RB006, blue and green) targeting blood coagulation factor IXa is used to control blood coagulation during percutaneous coronary intervention in patients with acute coronary syndrome (blue bases are the region of aptamer that is recognized by the antidote). Substantial anticoagulation is required to limit clotting during this procedure, in which a catheter is inserted into a peripheral artery and guided to the heart to administer a contrast agent that will indicate sites of blockage in a coronary angiography. If blockage is found in the arteries of a patient’s heart (red circle), angioplasty is often performed to restore blood flow (green circle). To limit bleeding after treatment, the antidote oligonucleotide (RB007, pink) is administered to rapidly unfold the aptamer and restore normal blood hemostasis.
ncRNA-mediated genetic reprogramming
The observation that ncRNAs can serve as guides to target endonucleases—not simply to cleave and destroy genetic instructions but rather to reprogram them—has received considerable attention. ncRNAs created by trans-splicing ribozymes and CRISPR gRNAs can, in principle, be used to reengineer the transcriptome or genome for therapeutic applications (38, 39). Though this RNA-based genetic reprogramming technology is not nearly as clinically advanced as other RNA therapeutics, the first therapeutic ncRNA designed to reprogram genetic instructions entered a phase 1 clinical trial in 2015. After encouraging pre-clinical animal studies, a trans-splicing ribozyme designed to recognize telomerase reverse transcriptase mRNA and reprogram it to encode herpes simplex virus–thymidine kinase only in cancer cells has now been tested in five cancer patients with hepatic metastases (40). In that study, the trans-splicing ribozyme was expressed after in vivo gene transfer, and the safety and potential efficacy of this ncRNA-mediated transcriptome engineering was assessed. Numerous RNA-guided endonuclease-based therapies are currently in preclinical development. If it can be adequately developed during the next decade, ncRNA-mediated genetic engineering may prove useful for treating a variety of inherited diseases (including genetic disorders) through genome engineering and acquired diseases (such as cancer) through transcriptome engineering, where transient reprogramming is expected to be sufficiently effective and also safer than permanently reprogramming the genome.
Conclusion
The field of RNA therapeutics is currently undergoing a major expansion and a continued renaissance. Eleven RNA-based therapeutics have now reached late-stage clinical development, with one on the market and several others close behind (table S1). The continued discovery of activities performed by RNAs, taken together with the increasing recognition that transient induction of a therapeutic effect can have long-lasting health benefits, promises to keep the clinical development pipeline full of therapeutic RNAs well into the future.
Supplementary Material
Acknowledgments
We would like to thank R. Becker, T. Povsic, S. Nimjee, and D. Boczkowski for useful discussions. This work was supported in part by grants from the NIH (to B.A.S.) and the Department of Defense (to S.N. and B.A.S.). B.A.S. and Duke University have filed U.S. patent applications (numbers 7,300,922 and 7,312,325) that relate to anticoagulant aptamers and antidote oligonucleotides. S.N. is a co-inventor on the patent describing the use of DCs transfected with tumor antigen–encoding RNA that has been licensed by Argos Therapeutics (Durham, NC) through Duke University. S.N. has no financial interest in and is not compensated by Argos Therapeutics.
Footnotes
REFERENCES AND NOTES
- 1.Burnett JC, Rossi JJ. Chem Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drolet DW, Green LS, Gold L, Janjic N. Nucleic Acid Ther. 2016;26:127–146. doi: 10.1089/nat.2015.0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cech TR, Steitz JA. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Benteyn D, Heirman C, Bonehill A, Thielemans K, Breckpot K. Expert Rev Vaccines. 2015;14:161–176. doi: 10.1586/14760584.2014.957684. [DOI] [PubMed] [Google Scholar]
- 5.Nair S, Boczkowski D, Pruitt S, Urban J. In: Cancer Vaccines: From Research to Clinical Practice. 1. Bot A, Obrocea M, Marincola FM, editors. CRC Press; 2011. pp. 217–231. [Google Scholar]
- 6.Bringmann A, Held SA, Heine A, Brossart P. J Biomed Biotechnol. 2010;2010:623687. doi: 10.1155/2010/623687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Lint S, et al. Expert Rev Vaccines. 2015;14:235–251. doi: 10.1586/14760584.2015.957685. [DOI] [PubMed] [Google Scholar]
- 8.Amin A, et al. J Immunother Cancer. 2015;3:14. doi: 10.1186/s40425-015-0055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allard SD, et al. Clin Immunol. 2012;142:252–268. doi: 10.1016/j.clim.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Van Gulck E, et al. AIDS. 2012;26:F1–F12. doi: 10.1097/QAD.0b013e32834f33e8. [DOI] [PubMed] [Google Scholar]
- 11.Van Tendeloo VF, et al. Blood. 2001;98:49–56. doi: 10.1182/blood.v98.1.49. [DOI] [PubMed] [Google Scholar]
- 12.Van Nuffel AM, et al. Cancer Immunol Immunother. 2012;61:1033–1043. doi: 10.1007/s00262-011-1176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell DA, et al. Nature. 2015;519:366–369. doi: 10.1038/nature14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoerr I, Obst R, Rammensee HG, Jung G. Eur J Immunol. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 15.Granstein RD, Ding W, Ozawa H. J Invest Dermatol. 2000;114:632–636. doi: 10.1046/j.1523-1747.2000.00929.x. [DOI] [PubMed] [Google Scholar]
- 16.Sahin U, Karikó K, Türeci Ö. Nat Rev Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, et al. Cancer Res. 2010;70:9053–9061. doi: 10.1158/0008-5472.CAN-10-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon SH, et al. Cancer Gene Ther. 2009;16:489–497. doi: 10.1038/cgt.2008.98. [DOI] [PubMed] [Google Scholar]
- 19.Beatty GL, et al. Cancer Immunol Res. 2014;2:112–120. doi: 10.1158/2326-6066.CIR-13-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, Dollins CM, Boczkowski D, Sullenger BA, Nair S. Immunology. 2008;125:229–240. doi: 10.1111/j.1365-2567.2008.02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren L, et al. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu Y, et al. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 23.McClorey G, Wood MJ. Curr Opin Pharmacol. 2015;24:52–58. doi: 10.1016/j.coph.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Fire A, et al. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 25.Elbashir SM, et al. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 26.Bobbin ML, Rossi JJ. Annu Rev Pharmacol Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- 27.Nemunaitis J, et al. Oncology. 2014;87:21–29. doi: 10.1159/000360993. [DOI] [PubMed] [Google Scholar]
- 28.Garba AO, Mousa SA. Ophthalmol Eye Dis. 2010;2:75–83. doi: 10.4137/OED.S4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vigneswara V, et al. Brain. 2014;137:1656–1675. doi: 10.1093/brain/awu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramos AM, et al. Expert Opin Drug Discov. 2015;10:541–556. doi: 10.1517/17460441.2015.1033394. [DOI] [PubMed] [Google Scholar]
- 31.Suhr OB, et al. Orphanet J Rare Dis. 2015;10:109. doi: 10.1186/s13023-015-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanna M. Curr Heart Fail Rep. 2014;11:50–57. doi: 10.1007/s11897-013-0182-4. [DOI] [PubMed] [Google Scholar]
- 33.Sullenger BA, Gallardo HF, Ungers GE, Gilboa E. Cell. 1990;63:601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 34.Mehan MR, et al. Adv Exp Med Biol. 2013;735:283–300. doi: 10.1007/978-1-4614-4118-2_20. [DOI] [PubMed] [Google Scholar]
- 35.Rusconi CP, et al. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 36.Lincoff AM, et al. Lancet. 2016;387:349–356. doi: 10.1016/S0140-6736(15)00515-2. [DOI] [PubMed] [Google Scholar]
- 37.Ganson NJ, et al. J Allergy Clin Immunol. 2016;137:1610–1613.e7. doi: 10.1016/j.jaci.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan N, Howrey RP, Lee SW, Smith CA, Sullenger BA. Science. 1998;280:1593–1596. doi: 10.1126/science.280.5369.1593. [DOI] [PubMed] [Google Scholar]
- 39.Doudna JA, Charpentier E. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 40.Kim YH, et al. Theranostics. 2016;6:357–368. doi: 10.7150/thno.13621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.