Abstract
Immune checkpoint inhibitors targeting the programmed cell death-1 receptor (PD-1) improve survival in a subset of patients with clear cell renal cell carcinoma (ccRCC). To identify genomic alterations in ccRCC that correlate with response to anti-PD-1 monotherapy, we performed whole exome sequencing of metastatic ccRCC from 35 patients. We found that clinical benefit was associated with loss-of-function mutations in the PBRM1 gene (p=0.012), which encodes a subunit of a SWI/SNF chromatin remodeling complex (the PBAF subtype). We confirmed this finding in an independent validation cohort of 63 ccRCC patients treated with PD-(L)1 blockade therapy alone or in combination with anti-CTLA-4 therapies (p=0.0071). Gene expression analysis of PBAF-deficient ccRCC cell lines and PBRM1-deficient tumors revealed altered transcriptional output in JAK/STAT, hypoxia, and immune signaling pathways. PBRM1 loss in ccRCC may alter global tumor cell expression profiles to influence responsiveness to immune checkpoint therapy.
Immune checkpoint inhibitors such as nivolumab extend the survival of a subset of patients with metastatic ccRCC (1). Whether specific genomic features of ccRCC are associated with clinical benefit is unclear. In contrast to other human tumor types that respond to immunotherapy, such as non-small cell lung cancer (NSCLC), melanoma, and microsatellite-unstable colorectal adenocarcinoma, ccRCC harbors a low burden of somatic mutations (2–5). Melanoma and NSCLC typically harbor 10 to 400 mutations per megabase (Mb) and these genetic variants can generate tumor-specific antigens (neoantigens) that stimulate a strong anti-tumor immune response (1–4). In contrast, ccRCC harbors an average of only 1.1 mutations/Mb (6, 7) yet it ranks highly among tumor types in terms of immune cytolytic activity (8), immune infiltration score, and T cell infiltration score in the tumor microenvironment (9). These observations led us to hypothesize that distinct molecular mechanisms underlie the immunologically active tumor microenvironment and responsiveness to immune checkpoint therapy in patients with ccRCC.
As part of a prospective clinical trial (10), we first analyzed pre-treatment tumors from 35 patients with metastatic ccRCC on a clinical trial of anti-programmed cell death-1 receptor (anti-PD-1) therapy (nivolumab). Whole exome sequencing (WES) from paired tumor/normal tissue was performed to identify genetic correlates of clinical benefit. To validate the findings, we analyzed an independent cohort of 63 patients with metastatic ccRCC treated with therapies blocking PD-1 (e.g., nivolumab) or its ligand PD-L1 (e.g., atezolizumab) (Fig. 1A and table S1A) (11).
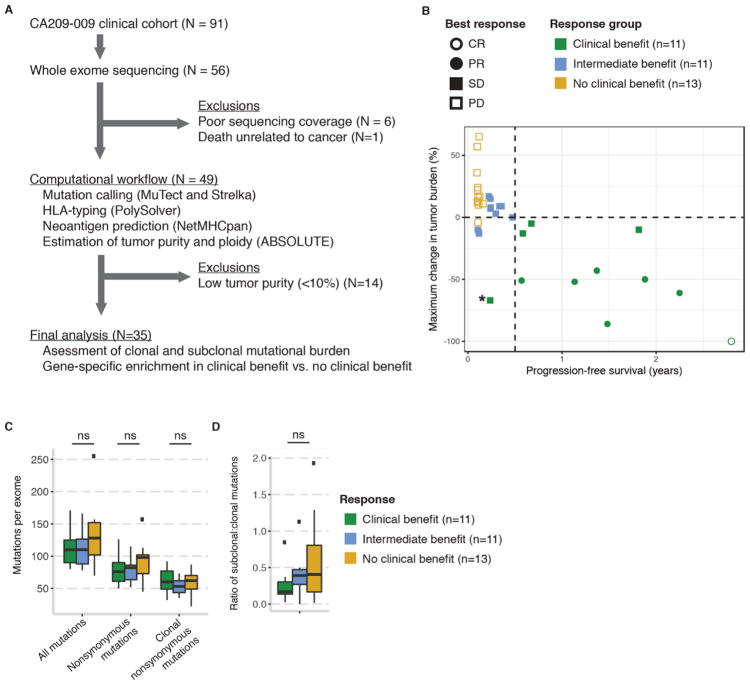
Fig. 1. Cohort consolidation and clinical characteristics of the discovery cohort.
(A) Sample inclusion/exclusion criteria and computational workflow. (B) Clinical stratification by degree of objective change in tumor burden (y-axis) and duration of progression-free survival (x-axis). One patient (RCC_99) not shown due to lack of tumor response data. *Patient RCC_50 was classified as clinical benefit despite PFS<6 months because there was continued tumor shrinkage after an initial period of minor tumor progression (see fig. S2). (C) Mutation burden in the discovery cohort by response group. (D) Ratio of subclonal to clonal mutations, as estimated by ABSOLUTE, by response group. ns = not significant. Abbreviations: CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease.
Baseline clinical and demographic features in the discovery cohort have been previously described (10). The subset of patients with complete pre-treatment molecular profiling did not differ substantially in clinical or demographic features from patients whose data did not meet technical quality control standards (fig. S1, A and B, and Supplemental Methods) or from the larger published cohort (10). Given previous evidence suggesting that refined clinical stratifications are necessary to assess clinical benefit from immune checkpoint blockade (12), we defined a composite response endpoint incorporating RECIST (Response Evaluation Criteria In Solid Tumors) (13), radiographic tumor shrinkage, and progression-free survival (PFS) (Fig. 1B and table S1B). Clinical benefit (CB) included patients with complete response (CR) or partial response (PR) by RECIST 1.1 (i.e., tumor shrinkage >30% from baseline) (13) or stable disease (SD) if they had any objective reduction in tumor burden lasting at least 6 months. This modification to include some patients with SD is intended to differentiate those patients with naturally indolent disease (i.e., slow tumor growth not surpassing 20% of baseline tumor size) from those with tumor response to immune checkpoint inhibitors (14). No clinical benefit (NCB) patients experienced progressive disease (PD) by RECIST 1.1 and were discontinued from immunotherapy within three months. All other patients were termed “intermediate benefit” (IB). One patient in the discovery cohort was classified as CB despite PFS < 6 months because there was continued tumor shrinkage (−67% of baseline tumor size) after an initial period of minor tumor progression, and the patient had overall survival exceeding 32 months (fig. S2, A and B). Consistent with prior observations (1), the dose of nivolumab, patient gender, and baseline PD-L1 immunohistochemical staining from metastatic biopsies did not predict patient overall survival (OS) following initiation of anti-PD-1 therapy (p>0.05 for all; log-rank test) (fig. S3).
Mean exome-wide target coverage in the discovery cohort was 128-fold for tumor sequencing and 91-fold for matched germline sequencing (tables S1A and S2A). Overall nonsynonymous mutation burden was moderate in the discovery cohort (median 82 per exome, range 45–157). The tumors of patients with CB and those with NCB showed similar mutation burdens and intratumoral heterogeneity (Fig. 1, C and D, and table S1, C and D). Mutations and copy number alterations affecting antigen presentation machinery and HLA class I alleles were uncommon and were present in tumors of both CB and NCB patients (fig. S4, A and B).
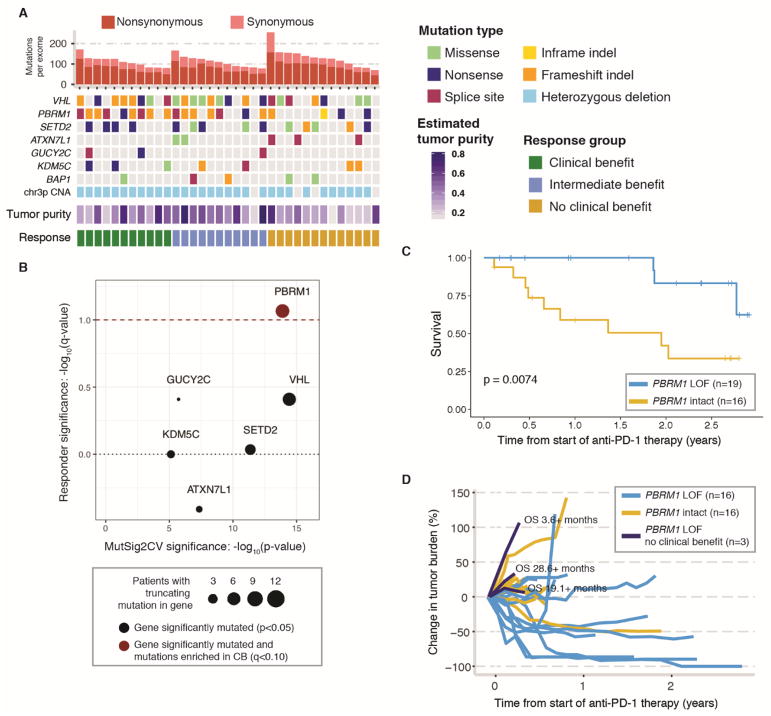
We next focused our analysis on the mutations most likely to be functionally important. We applied MutSig2CV (15) to identify genes recurrently mutated in the discovery cohort. Of these genes, we limited our search to highly deleterious variants, meaning known hotspot or putative truncating (frameshift insertion or deletion, nonsense mutation, or splice-site) mutations. Of the seven recurrently mutated genes (Fig. 2A and table S1E) (6), PBRM1 was the only gene in which truncating, or loss-of-function (LOF) (11), mutations were enriched in tumors from patients in the CB vs. NCB group (9/11 vs. 3/13; Fisher’s exact p=0.012, q=0.086, odds ratio for CB=12.93, 95% C.I. 1.54–190.8) (Fig. 2B and table S1F). In this cohort, all truncating PBRM1 alterations co-occurred with deletion of the non-mutated allele on chromosome 3p (Fig. 2A), resulting in complete LOF of PBRM1, and most of the mutations were predicted to be clonal (present in all tumor cells) (table S1F). Prior large-scale sequencing studies have shown that PBRM1 LOF alterations occur in up to 41% of ccRCC tumors (16) and are commonly clonal events present in all or nearly all tumor cells (17). Patients whose tumors showed biallelic PBRM1 loss had significantly prolonged OS and PFS compared to patients without PBRM1 LOF (log-rank p=0.0074 and p=0.029, respectively) (Fig. 2C and fig. S5), and they experienced sustained reductions in tumor burden (Fig. 2D).
Fig. 2. Analysis of tumor genome features in discovery cohort reveals a correlation between PBRM1 LOF mutations and clinical benefit from anti-PD-1 therapy.
(A) Mutations in the discovery cohort. Patients are ordered by response category, with tumor mutation burden in decreasing order within each response category. Shown are the genes that were recurrently mutated at a significant frequency, as assessed by MutSig2CV analysis (table S1E). CNA = copy number alteration. (B) Enrichment of truncating mutations in tumors from patients in the CB vs. NCB groups. Red dashed line denotes q<0.1 (Fisher’s exact test). Mutations in genes above the black dotted line are enriched in tumors of patients with CB from anti-PD-1 therapy and mutations in genes below the line are enriched in tumors of patients with NCB. (C) Kaplan-Meier curve comparing overall survival of patients treated with anti-PD-1 therapy whose tumors did or did not harbor LOF mutations in PBRM1. See also fig. S5 for Kaplan-Meier curve comparing progression-free survival of these patients. (D) Spider plot showing objective decrease in tumor burden in PBRM1-LOF (blue) vs. PBRM1-intact (yellow) tumors. Three patients with early progression on anti-PD-1 therapy and truncating mutations in PBRM1 (dark blue) had long and/or censored OS.
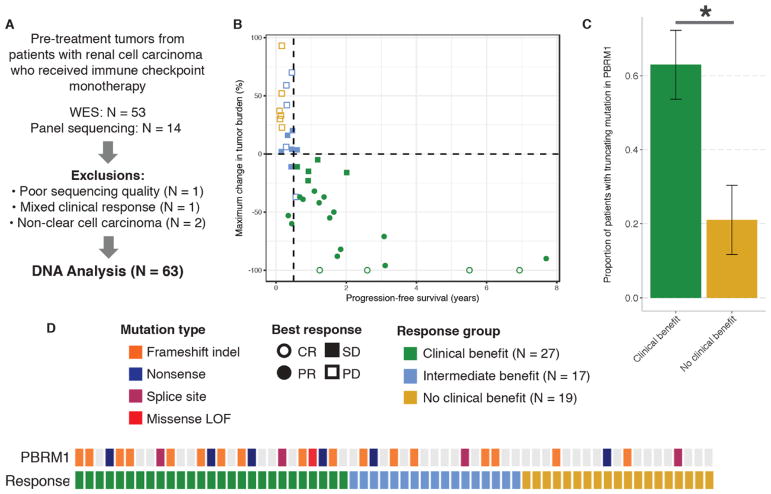
To evaluate the reproducibility of this finding, we then examined matched pre-treatment tumor and germline genomic data from an additional 63 patients treated with anti-PD-(L)1 therapy, either alone or in combination with anti-CTLA-4 therapy. Of these 63 patients, PBRM1 mutation status was derived from WES in 49 and panel sequencing in 14 patients (Fig. 3, A and B, and table S2, A and B) (11). Tumors from CB patients were more likely to harbor truncating alterations in PBRM1 (17/27 vs. 4/19, Fisher’s exact p=0.0071, odds ratio for CB=6.10, 95% C.I. 1.42–32.64) (Fig. 3, C and D, and table S2C). Although we could not assess copy number alterations in all samples in the validation cohort, the PBRM1 LOF mutations likely represented biallelic loss, as chromosome 3p deletions are nearly ubiquitous in ccRCC (6). Notably, one of the four NCB patients whose tumor showed a PBRM1 LOF mutation also had an alteration in B2M, which codes for a protein important in antigen presentation. This provides a potential explanation for the patient’s lack of clinical benefit from immune checkpoint blockade therapy despite having a truncating PBRM1 mutation.
Fig. 3. PBRM1 LOF mutations correlate with clinical benefit in a validation cohort of ccRCC patients treated with immune checkpoint inhibitors.
(A) Selection of the validation cohort. (B) Clinical outcomes in the validation cohort. Ten patients without post-treatment re-staging scans (eight with clinical PD, two with SD, and one with PR) as well as 14 patients with targeted panel sequencing are not shown. (C) Proportion of tumors harboring PBRM1 LOF mutations in patients in the CB vs. NCB groups. Error bars are S.E. *Fisher’s exact p<0.05. (D) Truncating alterations in PBRM1 and response to anti-PD-(L)1 therapies by sample. Colored boxes indicate samples with truncating mutations in PBRM1 while gray denotes samples without PBRM1 truncating mutations. Missense LOF denotes a missense mutation detected by targeted sequencing that was confirmed to be LOF by PBRM1 immunohistochemistry (see Supplemental Methods).
While primary analyses excluded patients with intermediate benefit (IB) due to the unclear effect of immune checkpoint blockade therapy on patient outcomes in this group, the observed trend between PBRM1 mutation status and clinical benefit persisted with the inclusion of these patients as an intermediate phenotype. In both the discovery and validation cohorts, patients in the IB group had intermediate rates of PBRM1 LOF (82%, 64%, 23% for CB, IB, NCB in the discovery cohort and 63%, 41%, 21% for CB, IB, NCB in the validation cohort; Fisher-Freeman-Halton Exact p = 0.017 and 0.017). Additionally, while no difference in clinical benefit was observed between treatment-naive and previously-treated patients in the discovery cohort (fig. S2), the progression-free survival benefit conferred by PBRM1 LOF was more prominent in tumors from previously-treated patients compared to those from patients receiving anti-PD-1 therapy as their first cancer therapy (p=0.009) (fig. S6 and tables S1 and S2).
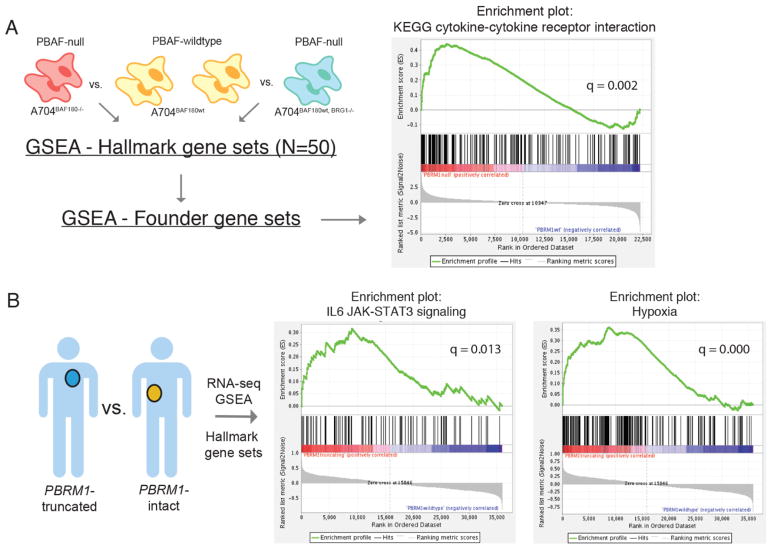
The PBRM1 gene codes for BAF180, a subunit of the PBAF subtype of the SWI/SNF chromatin remodeling complex. The PBAF complex suppresses the hypoxia transcriptional signature in VHL−/− ccRCC (18, 19) but its effects on tumor-immune interactions have not been thoroughly studied. To explore the potential impact of this complex on the immunophenotype of ccRCC, we analyzed previously reported whole transcriptome sequencing (RNA-seq) data from A704 ccRCC cell lines with perturbations in the PBAF complex (19). Loss of BAF180 or the related PBAF subunit BRG1, encoded by the gene SMARCA4, prevent formation of the intact PBAF complex (19). We performed gene expression analyses of BAF180-null (A704BAF180−/−) cell lines vs. PBAF-wildtype (A704BAF180wt) cell lines, as well as BRG1-null (A704BAF180wt, BRG1−/−) cell lines vs. PBAF-wildtype (A704BAF180wt) cell lines (Fig. 4A). Differential gene expression analysis showed substantial overlaps (~50%) between the top 100 genes differentially expressed in A704BAF180−/− vs. A704BAF180wt and A704BAF180wt, BRG1−/− vs. A704BAF180wt (table S4). This reflects the fact that BAF180 is essential to the PBAF but not the BAF complex, while BRG1 is a required subunit of both. Thus, the BAF180-null and BRG1-null cell lines have some shared characteristics but are also biologically and phenotypically distinct.
Fig. 4. PBRM1 mutational status in ccRCC influences immune gene expression.
(A) GSEA was performed on PBAF-deficient (A704BAF180−/− and A704BAF180wt, BRG1−/−) vs. PBAF-proficient (A704BAF180wt) kidney cancer cell lines using both Hallmark and corresponding Founder gene sets. GSEA enrichment plot shown for the KEGG cytokine-cytokine receptor interaction gene set in A704BAF180−/− vs. A704BAF180wt (PBRM1 null vs. wildtype). Enrichment plot is similar for A704BAF180wt, BRG1−/− vs. A704BAF180wt (BRG1 null vs. wildtype); see table S4. (B) GSEA was also performed on RNA-seq from pre-treatment tumors in the discovery and validation cohorts of this study (n = 18 PBRM1-LOF vs. n = 14 PBRM1-intact) using the Hallmark gene sets. Enrichment plots show increased expression of the hypoxia and IL6/JAK-STAT3 gene sets in the PBRM1-LOF tumors.
Gene set enrichment analysis (GSEA) on 50 “hallmark” gene sets representing major biological processes (20) revealed five gene sets whose expression was significantly enriched in cell lines that were PBAF-deficient. These included genes linked to IL6/JAK-STAT3 signaling, TNF-α signaling via NF-κB, and IL2/STAT5 signaling (Fig. 4A and table S5, A and B). As expected, the hallmark hypoxia gene set was up-regulated in A704BAF180−/− vs. A704BAF180wt cell lines (family-wise error rate - FWER q=0.071) (table S5A) (19). Across the more refined “founder” gene sets describing these five significantly enriched hallmark gene sets, the most strongly enriched gene set in PBAF-deficient cell lines was the KEGG cytokine-cytokine receptor interaction gene set (FWER q=0.0020 for A704BAF180−/− vs. A704BAF180wt and q=0.023 for A704BAF180wt, BRG1−/− vs. A704BAF180wt) (Fig. 4A and table S5, C to L). This gene set includes both immune-stimulatory (e.g., IL12, CCL21) and immune-inhibitory (e.g., IL10) genes, but Gene Ontology term analysis (11) showed that the genes most strongly enriched in PBAF-deficient cell lines were immune-stimulatory (table S6). Previously reported GSEA analysis of untreated ccRCC from The Cancer Genome Atlas (TCGA) and a murine model of PBRM1 loss also show amplified transcriptional outputs of HIF1 and STAT3, involved in hypoxia response and JAK-STAT signaling respectively, in PBRM1-mutant vs. PBRM1-wildtype states (18). GSEA analysis of RNA-seq from pre-treatment tumors in the discovery and validation cohorts of this study (n = 18 PBRM1-LOF vs. n = 14 PBRM1-intact) confirmed increased expression of the hypoxia and IL6/JAK-STAT3 gene sets in the PBRM1-LOF tumors (Fig. 4B and tables S7, A and B, and S8). Given JAK-STAT3 pathway gene involvement in the interferon gamma (IFN-γ-) signaling pathway and IFN-γ-dependent cancer immunostimulation (21), differential expression of these genes may impact PBRM1-LOF patients’ response to anti-PD-(L)1 therapy.
In addition to assessing tumor-intrinsic gene expression with GSEA, we further characterized the quality of the tumor-immune microenvironment in PBRM1-LOF vs. PBRM1-intact ccRCC in three independent cohorts: TCGA (6), an independent cohort of untreated ccRCC tumors (Sato) (22), and patient tumors from this study (table S8). In all three cohorts, tumors harboring LOF mutations in PBRM1 showed lower expression of immune inhibitory ligands (e.g., CD276 and BTLA) (23) than those without PBRM1 mutations. This finding was somewhat unexpected, as high PD-L1 staining is associated with increased responsiveness to anti-PD-1 and anti-PD-L1 agents in other cancer types (24, 25). However, the magnitudes of these differences were small and potentially confounded by differing degrees of tumor-stromal admixture (fig. S7, A to C) (9). We also examined LOF mutations in VHL, the most commonly-mutated gene in the TCGA ccRCC cohort. VHL mutation status did not correlate with immune-related gene expression (fig. S8), suggesting that observed differences in immune gene expression in the context of PBRM1 LOF may be specific to the PBRM1 gene.
In summary, we have shown that patients with metastatic ccRCC harboring truncating mutations in PBRM1 experienced increased clinical benefit from immune checkpoint therapy. This may be due to distinct immune-related gene expression profiles in PBRM1-mutant or PBAF-deficient tumor cells compared to their PBAF-intact counterparts, as shown by RNA-seq analyses in this study, though further in vivo studies will be needed to further explore these findings. Given the high prevalence of PBRM1 LOF in ccRCC and of SWI/SNF alterations across all cancer types (more than 20%) (26), this finding has important implications as a molecular tool for considering immunotherapy-responsiveness in ccRCC and across cancer types.
In vivo studies of mice harboring tumor clones with inactivation of PBRM1 – or the related essential PBAF complex components ARID2 or BRD7 – show that cells with PBAF loss are more sensitive to T-cell-mediated cytotoxicity compared to their PBAF-intact counterparts (27). This finding lends a mechanistic basis to the results observed here, and helps explain the conflicting results regarding PBRM1 mutation status as a prognostic variable in ccRCC (in the absence of immunotherapy) in prior studies (28–36). PBRM1 also previously has been linked to longer PFS with VEGF-targeted therapies (37). The observed interaction between PBRM1 status, prior treatment (largely with VEGF inhibitors), and response to immune checkpoint therapy in this study argues for further investigation of patient outcomes from sequential and combination treatment regimens that include anti-PD-(L)1. The relationship between PBRM1 LOF and clinical benefit from anti-PD-(L)1 therapies in ccRCC, as well as the immunological significance of PBAF loss in other cancer types, merit further preclinical and prospective clinical validation.
Supplementary Material
Table S1A: Whole exome sequencing metrics and inclusions/exclusions for patients in the discovery cohort
Table S1B: Clinical characteristics of patients receiving anti-PD1 therapy (nivolumab) in discovery cohort (N=35)
Table S1C. All called mutations in the discovery cohort (N=35)
Table S1D. Summary of mutational burden in the discovery cohort (N=35)
Table S1E. MutSig2CV results in discovery cohort (N=35)
Table S1F: Truncating PBRM1 alterations in patients discovery cohort passing whole exome quality control (N=35)
Table S2A: Sequencing Metrics and Inclusion/Exclusion Criteria for Whole Exome Sequencing in Validation Cohort (N=67)
Table S2B: Clinical information for immune checkpoint-treated patients in validation cohort (N=63)
Table S2C: Truncating PBRM1 alterations in validation cohort (N=63)
Table S3: SWI/SNF genes
Table S4. Intersection of top 100 positively differentially expressed genes in PBRM1null and BRG1null, and top 100 negatively differentially expressed genes in PBRM1 null and BRG1 null, both with respect to wild type using EdgeR
Table S5A. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on Hallmark gene sets
Table S5B. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on Hallmark gene sets
Table S5C. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on Cholesterol Homeostasis Founder gene sets
Table S5D. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on Cholesterol homeostasis founder gene sets
Table S5E. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on IL6_JAK_STAT Founder gene sets
Table S5F. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on IL6_JAK_STAT founder gene sets
Table S5G. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on E2F Founder gene sets
Table S5H. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines onE2F founder gene sets
Table S5I. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on TNFA Founder gene sets
Table S5J. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on TNFA founder gene sets
Table S5K. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on IL2 Founder gene sets
Table S5L. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on IL2 founder gene sets
Table S6. Enriched GO terms for KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION genes in BAF180-mutant GSEA enriched vs. BAF180-mutant GSEA depleted
Table S7A. GSEA results for gene sets enriched in pre-treatment patient tumors with truncating mutations in PBRM1
Table S7B. GSEA results for gene sets enriched in pre-treatment patient tumors wildtype at PBRM1
Table S8. Gene expression for all patient samples (TPM)
Acknowledgments
This project was supported by the Bristol-Myers Squibb II-ON consortium, the American Association for Cancer Research KureIt Grant for Kidney Cancer Research (EMV, TKC), and the Cancer Immunologic Data Commons (NIH U24CA224316). DM is a Howard Hughes Medical Institute Medical Research Fellow. TKC is supported in part by the Dana-Farber/Harvard Cancer Center Kidney SPORE, the Kohlberg chair at Harvard Medical School and the Trust Family, Michael Brigham, and Loker Pinard Funds for Kidney Cancer Research at the Dana-Farber Cancer Institute. THH is supported by the Gerstner Family Career Development Award, the National Cancer Institute (K12CA90628), and the Department of Defense (W81XWH-17-1-0546). Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. This study makes use of data generated by the Department of Pathology and Tumor Biology, Kyoto University (Sato cohort). WGK is a paid consultant for Agios, Fibrogen, Nextech Ventures, Peloton Therapeutics, Tracon, Third Rock Ventures, and serves on the Lilly Pharmaceuticals Board of Directors. EMV is a paid consultant for Third Rock Ventures, Genome Medical Inc., and Tango Therapeutics and receives research support from Bristol-Myers Squibb and Novartis. TKC is a paid advisor for AstraZeneca, Bayer, Bristol-Myers Squibb, Cerulean, Foundation Medicine, Genentech, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, Prometheus Labs, Roche, and Eisai. TKC receives institutional research funding from AstraZeneca, Bristol-Myers Squibb, Exelixis, Genentech, GSK, Merck, Novartis, Peloton, Pfizer, Roche, Tracon, and Eisai (for clinical trials). S.S. is a paid consultant for Merck and Bristol-Myers Squibb. R.J.M is a paid consultant for Pfizer, Genentech/Roche, Novartis, Exelixis, and Eisai. T. H. is a paid consultant for Pfizer, Exelixis, and Roche. F.S. H. is a paid consultant for Bristol-Myers Squibb, Merck, Genentech, Novartis, Amgen, and EMD Serono. M.D.H. is a paid consultant for Bristol-Myers Squibb, Merck, Genentech/Roche, AstraZeneca, Mirati, Janssen, and Novartis. E.M.V., T.K.C. and D.M. are inventors on patent application submitted by Dana-Farber Cancer Institute that covers PBRM1 mutational status in tumors and response to immunotherapy. The sequencing data are deposited in dbGap (accession number phs001493.v1.p1). The cell line transcriptome data are deposited in GEO (accession number PRJNA371283). DM, CAM, MB, MEA, and EMV performed genomic analyses. WL and WG performed the cell line experiments and generated the cell line genomic data. MG, TKC, DC, CH, MWR, MV, and RJM gathered the discovery cohort clinical and biological materials. SS contributed to immunohistochemistry. SMW, DJM, DB, MHV, AS, MDH, THH, and CN collected the biological materials and clinical annotations for the validation cohort. AT contributed to project management. DM, CAM, EMV, and TKC prepared the initial draft of the manuscript. FSH, WGK, DC, CH, MWR, AS, MHV, RJM, TKC and EMV supervised the study.
Footnotes
www.sciencemag.org/cgi/content/full/science.aan5951/DC1
Materials and Methods
REFERENCES AND NOTES
- 1.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, Castellano D, Choueiri TK, Gurney H, Donskov F, Bono P, Wagstaff J, Gauler TC, Ueda T, Tomita Y, Schutz FA, Kollmannsberger C, Larkin J, Ravaud A, Simon JS, Xu LA, Waxman IM, Sharma P. CheckMate 025 Investigators, Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA., Jr PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creighton CJ, et al. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Velasco G, Miao D, Voss MH, Hakimi AA, Hsieh JJ, Tannir NM, Tamboli P, Appleman LJ, Rathmell WK, Van Allen EM, Choueiri TK. Tumor mutational load and immune parameters across metastatic renal cell carcinoma risk groups. Cancer Immunol Res. 2016;4:820–822. doi: 10.1158/2326-6066.CIR-16-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Şenbabaoğlu Y, Gejman RS, Winer AG, Liu M, Van Allen EM, de Velasco G, Miao D, Ostrovnaya I, Drill E, Luna A, Weinhold N, Lee W, Manley BJ, Khalil DN, Kaffenberger SD, Chen Y, Danilova L, Voss MH, Coleman JA, Russo P, Reuter VE, Chan TA, Cheng EH, Scheinberg DA, Li MO, Choueiri TK, Hsieh JJ, Sander C, Hakimi AA. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17:231. doi: 10.1186/s13059-016-1092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choueiri TK, Fishman MN, Escudier B, McDermott DF, Drake CG, Kluger H, Stadler WM, Perez-Gracia JL, McNeel DG, Curti B, Harrison MR, Plimack ER, Appleman L, Fong L, Albiges L, Cohen L, Young TC, Chasalow SD, Ross-Macdonald P, Srivastava S, Jure-Kunkel M, Kurland JF, Simon JS, Sznol M. Immunomodulatory activity of nivolumab in metastatic renal cell carcinoma. Clin Cancer Res. 2016;22:5461–5471. doi: 10.1158/1078-0432.CCR-15-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supplementary materials.
- 12.Wolchok JD, Hoos A, O’Day S, Weber JS, Hamid O, Lebbé C, Maio M, Binder M, Bohnsack O, Nichol G, Humphrey R, Hodi FS. Guidelines for the evaluation of immune therapy activity in solid tumors: Immune-related response criteria. Clin Cancer Res. 2009;15:7412–7420. doi: 10.1158/1078-0432.CCR-09-1624. [DOI] [PubMed] [Google Scholar]
- 13.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 14.Gofrit ON, Yutkin V, Zorn KC, Duvdevani M, Landau EH, Hidas G, Pode D. The growth rate of “clinically significant” renal cancer. Springerplus. 2015;4:580. doi: 10.1186/s40064-015-1385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortés ML, Auclair D, Saksena G, Voet D, Noble M, DiCara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau DA, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll SA, Mora J, Crompton B, Onofrio R, Parkin M, Winckler W, Ardlie K, Gabriel SB, Roberts CWM, Biegel JA, Stegmaier K, Bass AJ, Garraway LA, Meyerson M, Golub TR, Gordenin DA, Sunyaev S, Lander ES, Getz G. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin M-L, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LFA, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, Martinez P, Phillimore B, Begum S, Rabinowitz A, Spencer-Dene B, Gulati S, Bates PA, Stamp G, Pickering L, Gore M, Nicol DL, Hazell S, Futreal PA, Stewart A, Swanton C. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nargund AM, Pham CG, Dong Y, Wang PI, Osmangeyoglu HU, Xie Y, Aras O, Han S, Oyama T, Takeda S, Ray CE, Dong Z, Berge M, Hakimi AA, Monette S, Lekaye CL, Koutcher JA, Leslie CS, Creighton CJ, Weinhold N, Lee W, Tickoo SK, Wang Z, Cheng EH, Hsieh JJ. The SWI/SNF protein PBRM1 restrains VHL-loss-driven clear cell renal cell carcinoma. Cell Reports. 2017;18:2893–2906. doi: 10.1016/j.celrep.2017.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao W, Li W, Xiao T, Liu XS, Kaelin WG., Jr Inactivation of the PBRM1 tumor suppressor gene amplifies the HIF-response in VHL−/− clear cell renal carcinoma. Proc Natl Acad Sci USA. 2017;114:1027–1032. doi: 10.1073/pnas.1619726114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, Ogawa S. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 23.Ramsay AG. Immune checkpoint blockade immunotherapy to activate anti-tumour T-cell immunity. Br J Haematol. 2013;162:313–325. doi: 10.1111/bjh.12380. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, Srinivas S, Retz MM, Grivas P, Joseph RW, Galsky MD, Fleming MT, Petrylak DP, Perez-Gracia JL, Burris HA, Castellano D, Canil C, Bellmunt J, Bajorin D, Nickles D, Bourgon R, Frampton GM, Cui N, Mariathasan S, Abidoye O, Fine GD, Dreicer R. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma A, Tsoucas D, Qiu X, Lim K, Rao P, Long HW, Yuan G-C, Doench J, Brown M, Liu S, Wucherpfennig KW. A major chromatin regulator determines resistance of tumor cells to T cell–mediated killing. Science. 2018 doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beuselinck B, Job S, Becht E, Karadimou A, Verkarre V, Couchy G, Giraldo N, Rioux-Leclercq N, Molinié V, Sibony M, Elaidi R, Teghom C, Patard JJ, Méjean A, Fridman WH, Sautès-Fridman C, de Reyniès A, Oudard S, Zucman-Rossi J. Molecular subtypes of clear cell renal cell carcinoma are associated with sunitinib response in the metastatic setting. Clin Cancer Res. 2015;21:1329–1339. doi: 10.1158/1078-0432.CCR-14-1128. [DOI] [PubMed] [Google Scholar]
- 29.Fay AP, de Velasco G, Ho TH, Van Allen EM, Murray B, Albiges L, Signoretti S, Hakimi AA, Stanton ML, Bellmunt J, McDermott DF, Atkins MB, Garraway LA, Kwiatkowski DJ, Choueiri TK. Whole-exome sequencing in two extreme phenotypes of response to VEGF-targeted therapies in patients with metastatic clear cell renal cell carcinoma. J Natl Compr Canc Netw. 2016;14:820–824. doi: 10.6004/jnccn.2016.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen Y-B, Gonen M, Liu H, Takeda S, Voss MH, Tickoo SK, Reuter VE, Russo P, Cheng EH, Sander C, Motzer RJ, Hsieh JJ ccRCC Cancer Genome Atlas (KIRC TCGA) Research Network investigators. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19:3259–3267. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh JJ, Chen D, Wang PI, Marker M, Redzematovic A, Chen YB, Selcuklu SD, Weinhold N, Bouvier N, Huberman KH, Bhanot U, Chevinsky MS, Patel P, Pinciroli P, Won HH, You D, Viale A, Lee W, Hakimi AA, Berger MF, Socci ND, Cheng EH, Knox J, Voss MH, Voi M, Motzer RJ. Genomic biomarkers of a randomized trial comparing first-line everolimus and sunitinib in patients with metastatic renal cell carcinoma. Eur Urol. 2017;71:405–414. doi: 10.1016/j.eururo.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapur P, Peña-Llopis S, Christie A, Zhrebker L, Pavía-Jiménez A, Rathmell WK, Xie XJ, Brugarolas J. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: A retrospective analysis with independent validation. Lancet Oncol. 2013;14:159–167. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwiatkowski DJ, Choueiri TK, Fay AP, Rini BI, Thorner AR, de Velasco G, Tyburczy ME, Hamieh L, Albiges L, Agarwal N, Ho TH, Song J, Pignon J-C, Barrios PM, Michaelson MD, Van Allen E, Krajewski KM, Porta C, Pal S, Bellmunt J, McDermott DF, Heng DYC, Gray KP, Signoretti S. Mutations in TSC1, TSC2, and MTOR are associated with response to rapalogs in patients with metastatic renal cell carcinoma. Clin Cancer Res. 2016;22:2445–2452. doi: 10.1158/1078-0432.CCR-15-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nam SJ, Lee C, Park JH, Moon KC. Decreased PBRM1 expression predicts unfavorable prognosis in patients with clear cell renal cell carcinoma. Urol Oncol. 2015;33:340.e9–340.e16. doi: 10.1016/j.urolonc.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Pawlowski R, Mühl SM, Sulser T, Krek W, Moch H, Schraml P. Loss of PBRM1 expression is associated with renal cell carcinoma progression. Int J Cancer. 2013;132:E11–E17. doi: 10.1002/ijc.27822. [DOI] [PubMed] [Google Scholar]
- 36.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnström H, Glimelius B, Sjöblom T, Edqvist PH, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 37.Carlo MI, Manley B, Patil S, Woo KM, Coskey DT, Redzematovic A, Arcila M, Ladanyi M, Lee W, Chen YB, Lee CH, Feldman DR, Hakimi AA, Motzer RJ, Hsieh JJ, Voss MH. Genomic alterations and outcomes with VEGF-targeted therapy in patients with clear cell renal cell carcinoma. Kidney Cancer. 2017;1:49–56. doi: 10.3233/KCA-160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, Jane-Valbuena J, Friedrich DC, Kryukov G, Carter SL, McKenna A, Sivachenko A, Rosenberg M, Kiezun A, Voet D, Lawrence M, Lichtenstein LT, Gentry JG, Huang FW, Fostel J, Farlow D, Barbie D, Gandhi L, Lander ES, Gray SW, Joffe S, Janne P, Garber J, MacConaill L, Lindeman N, Rollins B, Kantoff P, Fisher SA, Gabriel S, Getz G, Garraway LA. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, Berlin AM, Blumenstiel B, Cibulskis K, Friedrich D, Johnson R, Juhn F, Reilly B, Shammas R, Stalker J, Sykes SM, Thompson J, Walsh J, Zimmer A, Zwirko Z, Gabriel S, Nicol R, Nusbaum C. A scalable fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 42.Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: Estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 45.Bell D, et al. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, Fostel JL, Friedrich DC, Perrin D, Dionne D, Kim S, Gabriel SB, Lander ES, Fisher S, Getz G. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stachler MD, Taylor-Weiner A, Peng S, McKenna A, Agoston AT, Odze RD, Davison JM, Nason KS, Loda M, Leshchiner I, Stewart C, Stojanov P, Seepo S, Lawrence MS, Ferrer-Torres D, Lin J, Chang AC, Gabriel SB, Lander ES, Beer DG, Getz G, Carter SL, Bass AJ. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet. 2015;47:1047–1055. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 50.Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, Ligon KL, Tabernero J, Seoane J, Martinez-Saez E, Curry WT, Dunn IF, Paek SH, Park S-H, McKenna A, Chevalier A, Rosenberg M, Barker FG, II, Gill CM, Van Hummelen P, Thorner AR, Johnson BE, Hoang MP, Choueiri TK, Signoretti S, Sougnez C, Rabin MS, Lin NU, Winer EP, Stemmer-Rachamimov A, Meyerson M, Garraway L, Gabriel S, Lander ES, Beroukhim R, Batchelor TT, Baselga J, Louis DN, Getz G, Hahn WC. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, Beroukhim R, Pellman D, Levine DA, Lander ES, Meyerson M, Getz G. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J, Lawrence MS, Getz G, Bader GD, Ding L, Lopez-Bigas N. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, Hruban C, Guthrie VB, Rodgers K, Naidoo J, Kang H, Sharfman W, Georgiades C, Verde F, Illei P, Li QK, Gabrielson E, Brock MV, Zahnow CA, Baylin SB, Scharpf RB, Brahmer JR, Karchin R, Pardoll DM, Velculescu VE. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1A: Whole exome sequencing metrics and inclusions/exclusions for patients in the discovery cohort
Table S1B: Clinical characteristics of patients receiving anti-PD1 therapy (nivolumab) in discovery cohort (N=35)
Table S1C. All called mutations in the discovery cohort (N=35)
Table S1D. Summary of mutational burden in the discovery cohort (N=35)
Table S1E. MutSig2CV results in discovery cohort (N=35)
Table S1F: Truncating PBRM1 alterations in patients discovery cohort passing whole exome quality control (N=35)
Table S2A: Sequencing Metrics and Inclusion/Exclusion Criteria for Whole Exome Sequencing in Validation Cohort (N=67)
Table S2B: Clinical information for immune checkpoint-treated patients in validation cohort (N=63)
Table S2C: Truncating PBRM1 alterations in validation cohort (N=63)
Table S3: SWI/SNF genes
Table S4. Intersection of top 100 positively differentially expressed genes in PBRM1null and BRG1null, and top 100 negatively differentially expressed genes in PBRM1 null and BRG1 null, both with respect to wild type using EdgeR
Table S5A. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on Hallmark gene sets
Table S5B. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on Hallmark gene sets
Table S5C. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on Cholesterol Homeostasis Founder gene sets
Table S5D. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on Cholesterol homeostasis founder gene sets
Table S5E. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on IL6_JAK_STAT Founder gene sets
Table S5F. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on IL6_JAK_STAT founder gene sets
Table S5G. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on E2F Founder gene sets
Table S5H. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines onE2F founder gene sets
Table S5I. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on TNFA Founder gene sets
Table S5J. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on TNFA founder gene sets
Table S5K. GSEA for BAF180-null vs. BAF180-wildtype A704 cell lines on IL2 Founder gene sets
Table S5L. GSEA for BAF180-wildtype, BRG1-null vs. BAF180-wildtype, BRG1-wildtype A704 cell lines on IL2 founder gene sets
Table S6. Enriched GO terms for KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION genes in BAF180-mutant GSEA enriched vs. BAF180-mutant GSEA depleted
Table S7A. GSEA results for gene sets enriched in pre-treatment patient tumors with truncating mutations in PBRM1
Table S7B. GSEA results for gene sets enriched in pre-treatment patient tumors wildtype at PBRM1
Table S8. Gene expression for all patient samples (TPM)