Abstract
Elucidating the rules for receptor triggering in cell-cell and cell-matrix contacts requires precise control of ligand positioning in three dimensions. Here, we use the T cell receptor (TCR) as a model and subject T cells to different geometric arrangements of ligands, using a nanofabricated single-molecule array platform. This is comprised of monovalent TCR ligands anchored to lithographically patterned nanoparticle clusters surrounded by mobile adhesion molecules on a supported lipid bilayer (SLB). The TCR ligand could be co-planar with the SLB (2D), excluding the CD45 transmembrane tyrosine phosphatase, or elevated by 10 nm on solid nanopedestals (3D), allowing closer access of CD45 to engaged TCR. The two configurations resulted in different T cell responses, depending on the lateral spacing between the ligands. These results identify the important contributions of lateral and axial components of ligand positioning and create a more complete foundation for receptor engineering for immunotherapy.
T cell receptor (TCR) clustering by surface bound ligands is highly effective in triggering the tyrosine kinase cascade leading to T cell activation1 (Fig. 1a). LCK phosphorylates the immunoreceptor tyrosine-based activation motifs (ITAMs) in the TCR/CD3 complex. Phosphorylated ITAMs then recruit ZAP70, which is phosphorylated by LCK and trans-autophosphorylation2. Active ZAP70 phosphorylates the adapter LAT, leading to the generation of a signalosome rich in phosphotyrosine (pY)3. These events take place in “microclusters” that are ~ 200 nm in size4. The large transmembrane tyrosine phosphatase CD455 both maintains the activity of LCK by removal of the inhibitory C-terminal tyrosine phosphorylation, but continuously dephosphoryates LCK and ZAP70 substrates to maintain a poised basal state of the TCR. The kinetic-segregation (KS) model posits that close membrane apposition generated by TCR-ligand interactions locally excludes CD45 due to the effect of its large extracellular domain6, and triggering ensues when the duration of the TCR-ligand interaction, and thus CD45 exclusion, exceeds the time required for LCK and ZAP70 mediated phosphorylations to progress to a fully active signalosome7. Clustering of TCR by soluble ligands, which cannot exclude CD45 based on size, can trigger signaling but requires spacing between TCR of less than 3.6 nm8. The spatial requirements for TCR ligands on surfaces, the most physiological setting, are less well understood.
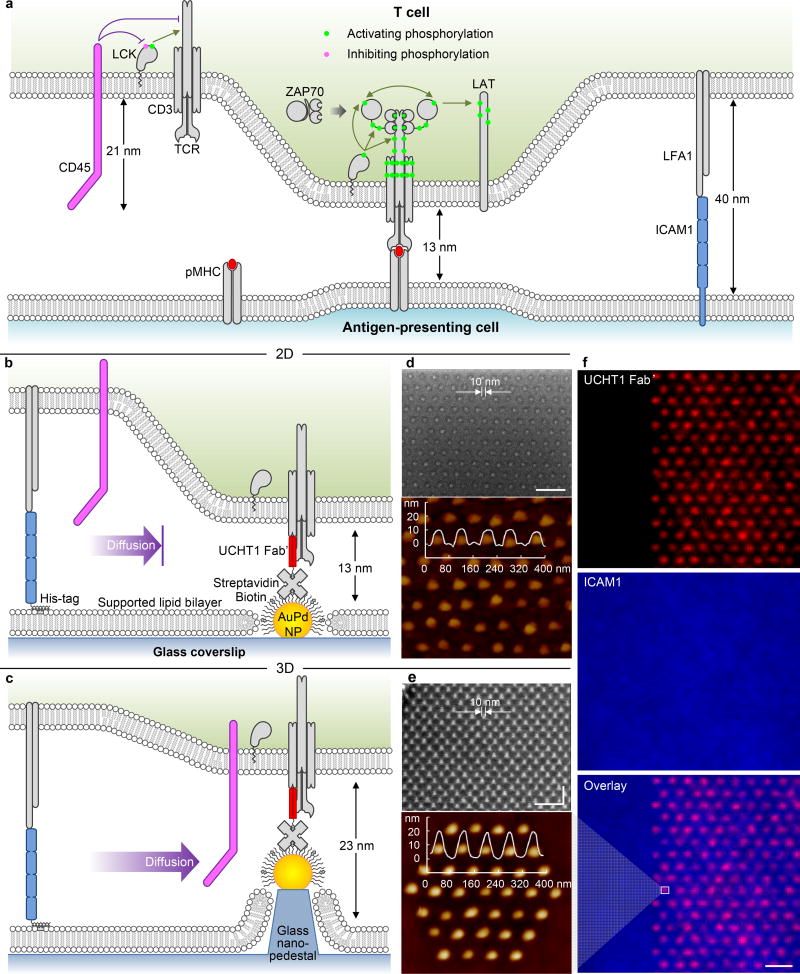
Figure 1. TCR triggering by surface bound ligands.
(a) Triggering by peptide-major histocompatibility complex (pMHC)on an antigen-presenting cell: schematic diagram of the cellular junction, indicating the steps in the TCR signalling cascade, as described in the text. (b–c)Triggering by anti-CD3 antibodies on the nanofabricated platform: schematic diagrams of (b) 2D and (c) 3D surfaces. Scanning electron microscopy (SEM, 40 nm spacing, lateral size ~ 10 nm) and atomic force microscopy (AFM, 80 nm spacing) images of (d) 2D and (e) 3D surfaces (10 nm increment in height). This platform shows precise control on the nanoarray geometry (extended/cluster) and dimension (both in-plane and out-of-plane). (f) Fluorescence images of the platform: UCHT1 Fab’ selectively immobilized on the nanoarrays (NP cluster in e), ICAM1 backfilled on the mobile SLB. Size bar = 100 nm for the SEMs (d,e) and 2 µm for the fluorescence images (f).
Nanofabricated surfaces have previously been used to study the effect of ligand spacing on TCR triggering9, 10, 11. Those studies used extended arrays (meaning much larger than the cell) of gold nanoparticle (NP)-bound TCR ligands surrounded by a non-adhesive background9, 10. A graded increase in TCR triggering with decreasing inter-ligand spacing between 200 nm and 40 nm was observed, but there was no evidence of a spacing threshold10, 11. Those arrays, created by self-assembly, could not decouple ligand spacing from density. Further, the absence of a sufficient adhesion system resulted in a requirement for TCR to mediate both triggering and adhesion, the latter role usually played by abundant adhesion molecules.
To test KS and alternative models it is also important to control the axial position (i.e., normal to the plane of the membrane) of the TCR ligands (Fig. 1a). The CD45 extracellular domain stands ~ 20 nm in height6. The intermembrane spacing generated by TCR-pMHC is 13 nm12, which excludes CD4513. Thus, increasing the spacing between apposed membranes by 10 nm should disable the KS mechanism. To date, all experiments testing axial positioning of TCR ligands have involved either the extension of TCR ligand height12, 14 or the truncation of the CD45 extracellular domain15, 16. These flexible or “soft matter” approaches have non-linear effects on the inter-membrane spacing12, 14, 17 and thus lateral spacing and affinity18. These caveats make it difficult to distinguish competing models14, 15. If extension could be accomplished with a rigid, non-tilting structure, this would enable a more precise test of the KS model and a better general understanding of TCR triggering.
The nanofabricated platform applied here (Fig. 1b–f) incorporates lateral and axial control of anti-CD3 position with a mobile adhesion layer. NPs were patterned by electron beam lithography19 (Supplementary Fig. S1), allowing independent control of inter-NP spacing and NP clustering to decouple lateral spacing and the amount of anti-CD3 accessible to the cell (Supplementary Fig. S2). The NPs were functionalized (Supplementary Fig. S3, S4) with monovalent anti-CD3 (UCHT1) Fab’ at an average molecular occupancy of 1 per NP (Supplementary Fig. S5)20. The arrays were backfilled with a supported lipid bilayer (SLB) to which adhesion molecule ICAM1 was tethered to achieve lateral mobility and physiological density21 (Supplementary Fig. S6). We refer to these as 2D surfaces, as the NPs and SLB are co-planar (Fig 1b, d).
Some arrays were subjected to an etching step using the NPs as a mask to create 10 nm nanopedestals tipped with NPs, which we refer to as 3D surfaces (Fig. 1c, e). This created an additional 10 nm axial spacing for TCR ligands between the T cell and the SLB, whilst maintaining control of lateral spacing. The glass pedestal/NP complex is a rigid structure22, 23. This topographic24 feature eliminates the limitations of the soft matter approaches used previously.
On both 2D and 3D surfaces the NPs were ~10 nm in diameter (Fig. 1d, e), and fluorescence signals were equivalent on both (Fig. 1f). This platform allows independent control of lateral and axial positioning of ligands to fully address the geometric requirement for triggering of TCR by monovalent ligands.
Effects of Ligand Spacing and Density on 2D Arrays
Human CD4+ T cells were plated on 2D surfaces at 37°C and were fixed after 5 min in order to investigate early TCR triggering events. The cells were stained for pY as an indicator of TCR signalling strength25 (see Supplementary Fig. S7 for a comparison of pY vs. pZAP70). 2D surfaces were prepared with a range of inter-NP spacings, cluster sizes and inter-cluster spacings that were relevant to the scale of TCR microclusters (~200 nm diameter) and immunological synapses (10 µm diameter) (Exp. I in Supplementary Fig. S2, Fig. 2a). Inter-NP spacing from 40 nm to 120 nm, cluster sizes from 240 nm to 1440 nm diameter, and densities from ~100/µm2 to ~30/µm2 were compared (Fig 2a). The pY signal on all patterns was multifocal, likely reflecting natural patterns of small, F-actin-dependent projections that mediate first contact of T cells with the surface26. The pY signal was strongly colocalized with the UCHT1 Fab’ clusters regardless of the geometry (Fig. 2a, Supplementary Fig. S8a), and it also increased with decreasing inter-NP spacing (Fig. 2b), in line with previous observations10, 11, 20. Similar spatial effects were observed for cell spreading area and the number of adherent cells, which was also generally increased by including the mobile ICAM1 layer (Supplementary Fig. S8b, c).
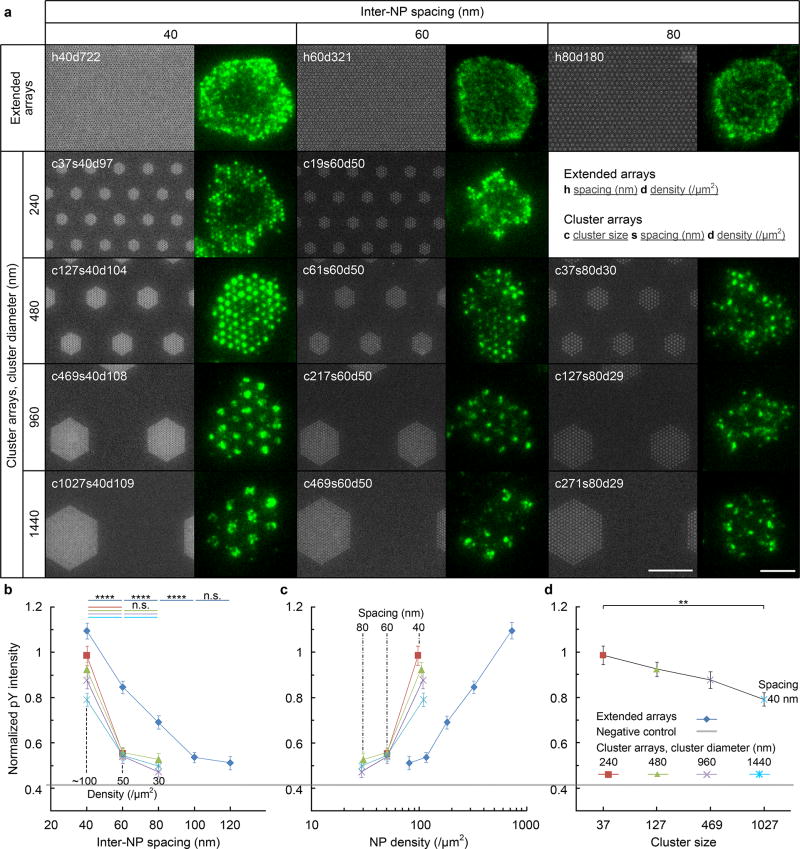
Figure 2. Small clusters of 37 ligands are optimal for TCR triggering.
(a) SEM images of nanoarrays (Ti hard mask shown for better contrast, size bar = 1 µm)and fluorescence images of pY fluorescent pattern for representative T cell (size bar = 5 µm). Nanoarray patterns are named by the geometric arrangement and parameters as noted in the inset. (b–d) pY intensity, normalized to the signal (=1) on a continuous AuPd surface coated with UCHT1 Fab’ for (b) inter-NP spacing, (c) NP density and (d) cluster size. Data are means ± s.e.m., n > 48 cells from 3 independent experiments, ** = p < 0.01, **** = p < 0.0001 (Mann–Whitney test, two-sided). The exact sample sizes and p values are summarized in Supplementary Table S1.
The normalized pY intensity was highest on 240 nm-diameter clusters with 40 nm inter-NP spacing and 37 NPs (corresponding to a global density of 97 NP/µm2 and designated c37s40d97), which closely resembled the multifocal pattern of pY observed on the extended array with the same inter-NP spacing (designated h40d722) (Fig. 2a) and a similar pY intensity despite the 7-fold lower global NP density (Fig. 2b, c). As the NP cluster size increased, the cells began to form multiple pY foci within each NP cluster, apparently treating them as small extended arrays rather than single ligand clusters (Fig. 2a); this correlated with a relative decrease in normalized pY intensity with increasing cluster size (Fig. 2d). Based on this, clusters of 37 NPs were used as a starting point for further analysis decoupling spacing and density effects.
37-NP cluster arrays with different inter-NP spacings and global densities (Exp. II in Supplementary Fig. S2 and S9) were compared to extended arrays for normalized pY intensity. On 2D cluster and extended arrays, the pY intensity increased with decreasing inter-NP spacing in a graded manner between 150 nm and 40 nm (Fig. 3a). When these same data are replotted against global NP density, it again is apparent that for inter-NP spacing of 40 nm the clusters of 37 are up to 7-fold more efficient than the extended arrays (Fig. 3b). This advantage diminishes as inter-NP spacing increases at the same global density: the clusters of 37 NPs become larger in area and less efficient. Thus, we were able to clearly distinguish effects due to spacing after correcting for global density, confirming that spacing plays a far more dominant role in TCR triggering than does density. Nonetheless, we did not detect a distinct triggering threshold for inter-NP spacing on 2D surfaces.
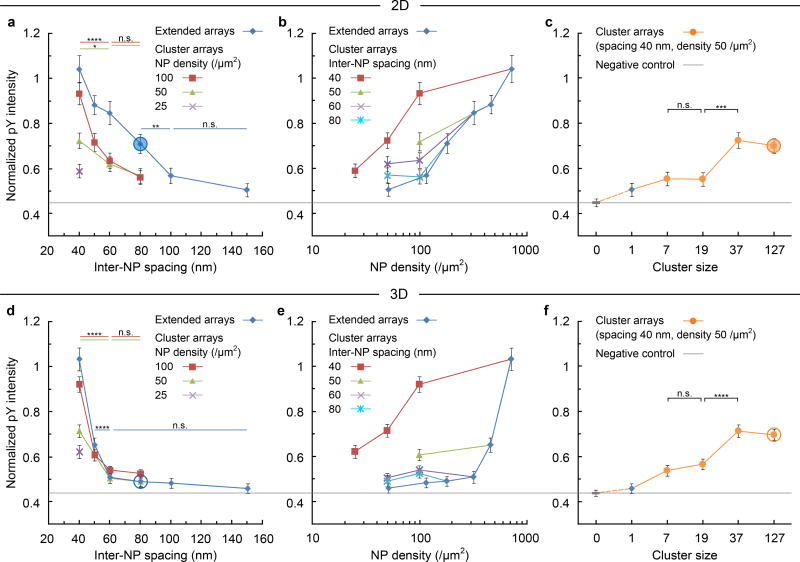
Figure 3. Ligand spacing has distinct effect on TCR triggering on 2D and 3D arrays.
TCR geometric effects on triggering indicated by pY intensity, normalized to the signal (= 1) on a continuous AuPd surface coated with UCHT1 Fab’. 2D surfaces: plots against (a) inter-NP spacing, (b) NP density, (c) cluster size. For a through comparison, an extended array (inter-NP spacing 150 nm, density ~ 50 /µm2) is included as a cluster size of 1; and the SLB passivation background (ICAM1 only) is included as a cluster size of 0, which is also the negative control providing a baseline (the gray line across the plots). 3D surfaces: plots against (d) inter-NP spacing, (e) NP density, and (f) cluster size. The circled points in panels a, c, d and f are referred to in discussion of data from Figs 4 and 6. Data are means ± s.e.m., n > 180 cells from 3 independent experiments, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001 (Mann–Whitney test, two-sided). The exact sample sizes and p values are summarized in Supplementary Table S2. For clarity, the same data sets are either sorted by density in the spacing plots, or sorted by spacing in the density plots. As a result, the correspondence between data points and symbols is not consistent among the plots. Supplementary Fig. S10 provides a consistent version with a detailed symbol library of systematically varied geometries.
Effects of Axial Distance on 3D Arrays
We next investigated the same patterns on 3D arrays with etched pedestals (Fig. 1c). In contrast to the 2D arrays, on the 3D arrays a robust pY signal was observed only for spacing < 50 nm (Fig. 3d). All the curves collapse to a uniform trend with the same threshold, a marked difference relative to the 2D surfaces (Fig. 3a). Thus, on the 3D surfaces, close packing of engaged TCR at inter-TCR spacing of < 50 nm is required for robust signalling with little response at inter-TCR spacing > 60 nm. When plotted against global density, the advantage of the optimal cluster arrays over the extended arrays was even more apparent than for the 2D surfaces (Fig. 3e). It is notable that for arrays with 40 nm spacing, the 2D and 3D systems perform similarly, whereas the 2D surface outperforms the 3D surface for all other geometries (Fig. 3b vs. 3e). Interestingly, when cluster size was further decreased whilst maintaining an inter-NP spacing of 40 nm (and a global density of 50 NP/µm2), the pY intensity displayed a similar decrease on 2D and 3D arrays (Fig. 3c, f). Small clusters (c ≤ 7) with low density (≤ 7 NP/µm2) gave low pY signals averaged over the entire cell, but pY foci were detected around many clusters of as few as 3 NPs (Supplementary Fig. S11).
Local Signalling Response to Ligand Spacing and Topology
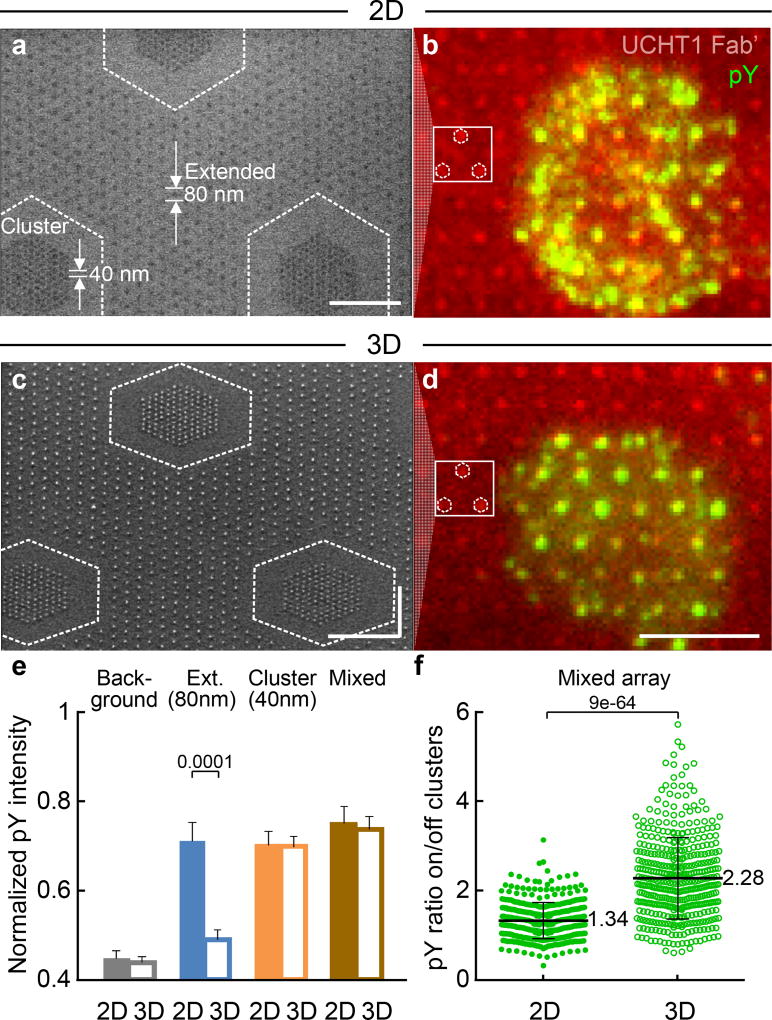
To test whether or not T cells sense ligand geometry locally, T cells were plated on 2D or 3D mixed nanoarrays comprising clusters of 127 NPs with an inter-NP spacing of 40 nm and a global density of 50 NP/µm2 embedded in an extended array with an inter-NP spacing of 80 nm (c127s40d50/h80d180) (Fig. 4a–d). The global density of the mixed array was kept the same as the extended array (the average Fab’ intensity on the mixed array was virtually identical to that of the 80 nm extended array, Supplementary Fig. S4b). On the 2D surfaces, both the cluster array (circled in Fig. 3c) and extended array (circled in Fig. 3a) displayed elevated pY intensity. Consistent with the responses to the individual surfaces, T cells showed no obvious preference for cluster or extended arrays and formed pY foci on both (Fig. 4a, b). In contrast, on the 3D surfaces, only the cluster array (circled in Fig. 3f), but not the extended array (circled in Fig. 3d) showed increased pY intensity. Although the overall pY signal per cell was similar to that on the 2D surfaces (Fig. 4e), the pY foci on the 3D surfaces are seen mostly on the clusters with 40 nm spacing, but not on the extended arrays with 80 nm spacing (Fig. 4c, d). As a result, the T cells displayed a clear preference of pY colocalization for the 40 nm clusters over the surrounding 80 nm extended array (Fig. 4f). These results provide direct evidence for a non-linear role played by spacing – both in and out of the plane of the membrane – in local TCR triggering.
Figure 4. T cells sense ligand position locally.
A mixed array of 40 nm clusters embedded in an 80 nm extended array (c127s40d50/h80d180) for 2D and 3D surfaces. (a) and (c) SEM images (size bar = 500 nm). (b) and (d) Epifluorescence images (size bar = 5 µm). Locations of 40 nm clusters are indicated in SEMs, with corresponding clusters indicated in fluorescence image to aid the eye. On the 2D surfaces, the pY is seen in (b) to be distributed over a cell, both on and off the clusters, with no obvious preference for the 80 nm or 40 nm spaced NPs. On the 3D surfaces, increased pY colocalization with the 40 nm clusters is clearly evident in (d), with much lower intensity off clusters as compared to (b). The experiment was repeated 3 times independently with similar results. (e) Normalized pY intensity on mixed arrays, compared with data from Fig. 3. Data are means ± s.e.m, n >180 cells from 3 independent experiments (Mann–Whitney test, two-sided),as summarized in Supplementary Table S2. (f) pY intensity ratio on/off clusters. Data are means ± s.d., each point representing a cluster in cell, n = 11 cells from 2 independent experiments(Mann–Whitney test, two-sided).Whilst the overall pY intensity for the cells is similar for the 2D and 3D mixed arrays, the difference of pY distribution between 2D and 3D surfaces highlights that a T cell can sense the ligand position and respond locally, even when the spacing is not uniform over the entire cell.
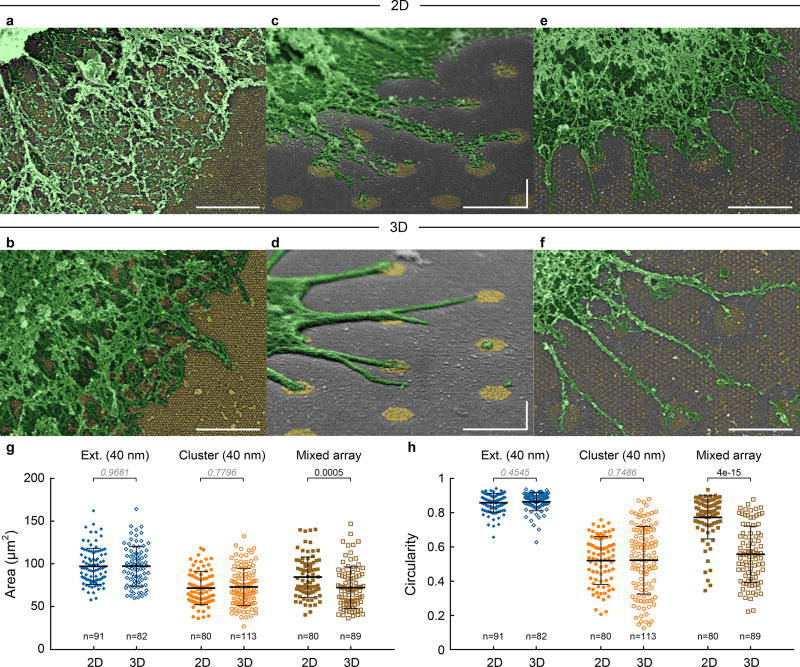
In addition to TCR triggering, T cells spread differently on extended and cluster arrays: they formed lamellipodia on homogeneous extended arrays and individual, long filopodia on clusters (Fig. 5a–d. Filopodia were observed on clusters with as few as only 3 NPs in Supplementary Fig. S11). On a singular geometry, there was no significant difference between 2D and 3D surfaces (Fig. 5g, h). On the mixed arrays, contacts formed on both cluster and extended arrays on 2D surfaces (Fig. 5e), whereas on 3D surfaces, a significant preference for clusters over the surrounding extended array was observed (Fig. 5f), in line with the pY distribution (Fig. 4f). This difference resulted in smaller area and lower circularity (more filopodia) on the 3D mixed array than the 2D (Fig. 5g, h).
Figure 5. Ligand position affects T cell spreading.
SEM images of lamellipodia on (a) 2D and (b) 3D extended arrays (40 nm spacing, h40d722). Individual filopodia on (c) 2D and (d) 3D cluster arrays (40 nm spacing, c127s40). (e) 2D and (f) 3D mixed arrays (c127s40d50/h80d180). Pseudo-colour, size bar = 1 µm. The experiment was repeated 3 times independently with similar results. (g) Cell spreading area and (h) circularity. Data are means ± s.d., each point representing a cell, n > 80 cells from 2 independent experiments (Mann–Whitney test, two-sided).The difference is not significant if p-value > 0.05 (italic).
Ligand Spacing and Kinetic Segregation
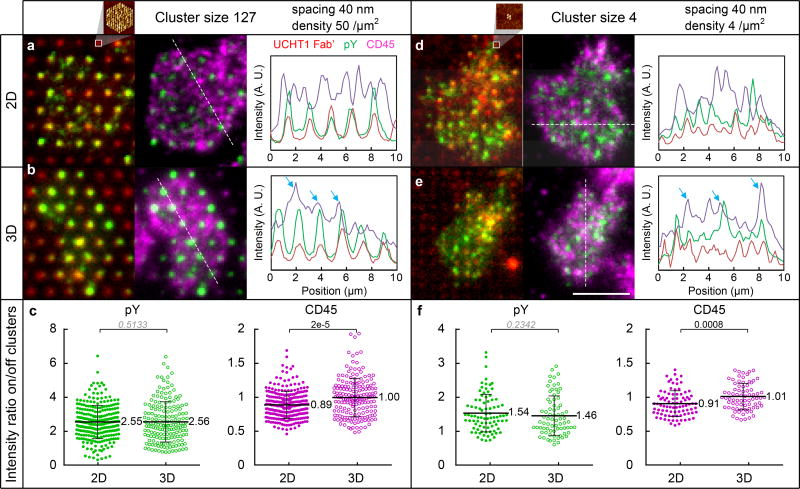
The emergence of a distinct lateral spacing threshold with the 10 nm increase in the separation between the T cell and substrate evokes a possible role of CD45 and the KS model7. We found that CD45 exclusion on clusters (inter-NP spacing 60 nm) is weaker on 3D than 2D surfaces, which explains the pY signal drop on 3D cluster arrays (Supplementary Fig. S12), in agreement with the KS model. On the other hand, for inter-NP spacings < 50 nm, the pY signal on 3D surfaces was comparable to that on 2D surfaces. Figure 6a, b shows the CD45 distribution for large clusters of 127 NPs with inter-NP spacing of 40 nm and density of 50 NP/µm2 (c127s40d50, inset AFM image). The pY foci and UCHT1 Fab’ clusters were also detected (Fig. 6a, b, with correlated line profiles of pY and Fab’). On 2D surfaces,R0 was largely depleted from Fab’/pY spots (Fig. 6a, with anti-correlated line profiles of CD45 and Fab’/pY), whilst on 3D surfaces, some of the bright CD45 signals overlapped with Fab’/pY spots (Fig. 6b, white spots in fluorescence image and correlated peaks in line profiles, highlighted by arrows), indicating that CD45 penetrated the 3D clusters. The CD45 intensity ratio on the clusters relative to the surrounding area was 0.89 on the 2D surfaces, indicating CD45 exclusion from the clusters, compared with a ratio of 1.00 on the 3D surfaces, implying weaker exclusion. The pY intensity ratios were 2.55 and 2.56 respectively, indicating that TCR triggering is similar on 3D and 2D surfaces (Fig. 6c). Indeed, the overall pY intensity per cell was similar (the points circled in Fig. 3c, f are compared in Fig. 4e). Clusters as small as 4 NPs with a density of 4 NP/µm2 (c4s40d4, Fig. 6d inset AFM image) displayed pY colocalization on UCHT1 Fab’ for both 2D and 3D surfaces (intensity ratio > 1), whereas a constant anti-correlation between CD45 and Fab’/pY was found for 2D (intensity ratio < 1) but not 3D surfaces (intensity ratio ~ 1) (Fig. 6d–f, CD45 colocalized with Fab’/pY also highlighted by arrows). Thus, greater relative exclusion of CD45 on 2D than 3D surfaces provides a potential mechanism for the emergence of a sharp 50 nm lateral spacing threshold for TCR triggering on the 3D, but not 2D surfaces.
Figure 6. Axial positioning of ligand controls CD45 exclusion.
CD45 exclusion and pY colocalization on NP clusters (inter-NP spacing 40 nm). Large clusters c127s40d50 (cluster size 127, density 50 NP/µm2): Fluorescence images with line profiles on (a) 2D and (b) 3D surfaces, (c) pY and CD45 intensity ratio of the cluster over surrounding area. Small clusters c4s40d4 (cluster size 4, density 4 NP/µm2): Fluorescence images with line profiles on (d) 2D and (e) 3D surfaces, (f) pY and CD45 intensity ratio. The experiment was repeated 3 times independently with similar results. The insets of a, d, are zoom-in AFM images of NP clusters. Size bar in fluorescence images: 5 µm. Data in c, f, are means ± s.d., each point representing a cluster in cell, n =8 cells from 2 independent experiments for each data set (Mann–Whitney test, two-sided).The difference is not significant if p-value > 0.05 (italic).
Further insight into the triggering threshold below an inter-NP spacing ~ 50 nm can be gained by considering what is known about the T cell activation mechanism (Supplementary Fig. S13). The cytoplasmic domains of the CD3 complex include 10 ITAMs, which consist of paired tyrosine motifs that recruit ZAP70 when phosphorylated. The cytoplasmic domain of CD3ζ is initially bound to the inner leaflet of the plasma membrane by interaction with anionic phospholipids, and the two tyrosines are buried in the hydrophobic core of the membrane27. Initial rearrangements triggered by TCR engagement reduce the negative charge leading to greater exposure of the ITAMs to kinases and adaptors27, 28, 29. FRET studies on the B cell antigen receptor also support a greater distance between the C-termini of the Igα and Igβ subunits during triggering30. When fully engaged in signalling, the maximal reach of the ZAP70 linked to ITAMs is 12 nm (6 per complex), 24 nm (2 per complex) or 36 nm (2 per complex), based on the primary sequence (Supplementary Fig. S13a), with a weighted average ~ 19.2 nm (Supplementary Fig. S13b). Whilst the average hydrodynamic radius of random coil peptides is much smaller than these extended values, the sequences are likely substantially extended when 1–3 ZAP70 molecules are bound. Since a key activation step of ZAP70 involves trans-auto-phosphorylation2, an increased potential for activation is achieved when TCR cytoplasmic domains reach a close-packed state at a distance of ~40 nm (Supplementary Fig. S13c). LAT, the target of ZAP70, has a maximal reach from its transmembrane domain of ~70 nm, and the average locus of sites is up to ~ 36 nm from the membrane. The pair-correlation function of LAT clustering decays to 0 at a distance of 200 nm, but is half its maximum value at ~ 40 nm3. The ability of ZAP70 to trans auto-phosphorylate sites in LAT involves electrostatic attraction that may further enhance inter-digitation of the cytoplasmic domains of TCR and LAT once they are within ~ 40 nm31. CD45 exclusion on 3D surfaces was not detected within the limits of our measurement (Fig. 6c, f, S12); however, we cannot rule out local CD45 rearrangement in nanoscale clusters. Local exclusion of CD45 may take place even in the absence of KS as phosphorylated LAT can form a protein “phase” with other TCR signalosome components that can locally exclude the CD45 phosphatase domain32. Further, in the context of B cell antigen receptor cross-linking by antibodies, lipid phase separation generates nanoscale exclusion of CD4533. These effects, if active here, may reasonably have a strong dependence on lateral receptor packing and may contribute to the sharp signalling threshold on 3D surfaces. (We note that in addition to segregation of CD45, we also observed central clearing of ICAM1 on only 2D surfaces. This is discussed in the Supplementary Information and illustrated in Supplementary Fig. S14.)
Conclusions
We have developed a novel single-molecule platform that combines the advantages of precise spatial control of biological ligands and an adhesion system that can be recruited or excluded as needed (SLB platform34, 35). It enables full control of ligand geometry, both in-plane and out-of-plane, and we have applied it to study the role of ligand geometry in TCR triggering. The 3D surfaces provide a rigid, non-tilting structure to fix intermembrane spacing without incurring entropic penalties associated with long-flexible spacers inherent to engineering molecular dimensions using proteins. This allowed us to further refine the KS model and its role in T cell triggering. It should be pointed out that the KS model has been applied to nearly all types of immune cells that express CD45 and even seems to operate in stromal cells when CD45 expression is forced6. Our results support roles for both clustering and KS in TCR triggering while fixing conformational and mechanical parameters. As predicted by the KS model, axial spacing plays a critical role, as T cells respond in a graded fashion to ligands with more relaxed spacing on 2D surfaces where CD45 exclusion is evident. A role for close packing of engaged TCR in triggering is also revealed, as strict clustering at < 50 nm inter-NP spacing was needed on 3D surfaces, where CD45 was only weakly excluded. Clustering was also important on both 2D and 3D surfaces, as ≥ 3 UCHT1 Fab’ within a cluster supported pY signals. Thus, as expected, the UCHT1 Fab’, which lacks the ability to recruit CD4, does require multiple local engagements to function even when CD45 can be excluded. Similar minimal clustering requirement of 3 – 4 have been reported previously for TCRs and chimeric antigen receptors (CARs)36, 37.
Being able to control both lateral and axial spacing has revealed a sharp lateral spacing threshold when the axial spacing is increased by 10 nm. This finding has potential utility in T cell engineering for immunotherapy applications, particularly for CARs, where the axial spacing between membranes may be engineered38. There is a complex relationship between the size of CARs, the position of the epitope in their ligands and induction of killing39. Some observations are consistent with the KS model in that interactions that lead to intermembrane distances close enough to exclude CD45 are optimal for killing by CAR expressing CD8+ T cells40. Many solid tumors overexpress antigens that are also expressed at lower levels on normal tissue cells38. Our results suggest that CARs with sharper thresholds and greater selectivity for high antigen levels could be engineered by imposing > 23 nm spacing between membranes. A possible approach to this challenge could be the use of DNA or protein “origami” in which 3D nanostructures combining clustering and greater resistance to bending might be constructed41, 42. Our results clearly demonstrate that axial positioning that allows entry of CD45 does not preclude TCR triggering, but generates a more distinct threshold for inter-receptor distance needed for triggering. The solid-state platform we have developed isolates axial positioning of surface ligands as a parameter for further investigation in many signalling contexts.
Methods
Nanoarray fabrication and functionalization
AuPd NP arrays were fabricated on glass coverslips (24 × 40 mm, no. 1.5, VWR) using electron beam lithography (NanoBeam nB4, Supplementary Fig. S1). Aquasave polymer was used as the conductive layer, which can be simply removed by DI water after lithography. Cold development (MIBK/IPA 3:1 at 4 °C) with sonication was used to ensure high contrast that yields high-resolution patterns. The lift-off process was facilitated by either single resist layer (PMMA 495K) with hard mask using angle evaporation19 (Ti hard masks are shown in Fig. 2), or resist bilayer (PMMA 35K and 495K) (AuPd film before lift-off and NPs after are shown in Supplementary Fig. S9). 3D samples underwent an additional dry etch process (CHF3, 40 sccm, 60 W) with the 2D nanoarrays as a mask, recessing the glass substrate and forming nano-pedestals (Fig. 1d, e).
The fabricated nanoarray samples were cleaned in 1.5 hr-aged piranha solution (3:1 H2SO4:H2O2) for 3 min, and in gentle oxygen plasma at 18 W for 5 min, which not only removes organic residuals from fabrication, but also makes the glass surface hydrophilic, both are necessary for the SLB formation. The cleaned samples were incubated in a freshly prepared 1 mM mixture of HS-C11-EG6-Biotin and HS-C11-EG3-OH (ProChimia Surfaces) solution in anhydrous ethanol for ~ 18 h (overnight). A self-assembled monolayer formed on each NP, where the mole fraction of biotin-alkylthiol can be easily adjusted for a given NP size, to ensure single-molecule occupancy20. The thiolated samples were rinsed with ethanol, blown dry, mounted on a chamber and immediately filled with small unilamellar vesicles solution (12.5% DGS-NTA(Ni), 87.5 % DOPC, Avanti Lipids). We used channel slides with self-adhesive underside (Ibidi sticky-Slide VI 0.4). The coverslip was glued and sealed with pattern regions underneath the channel and facing up. After 5 min, SLB formed on the surface and was washed with a flow cell buffer of HBS (HEPES buffered saline43) with 1% HSA (human serum albumin). The defects on SLB were blocked with 5% casein solution for 1 h, and excess casein was washed out with HBS-HSA. The channel was incubated in a buffer with 5 µg/mL streptavidin for 30 min, then a mixed solution of 4 µg/mL biotinylated UCHT1 Fab’-Alexa 568 and 0.625 µg/mL His-tagged ICAM1-Alexa 405 for another 30 min. In this way, UCHT1 Fab’ is immobilized on the nanoarray, and ICAM1 is added to the mobile SLB in the background (Fig. 1f, Supplementary Fig. S3). UCHT1 was used in preference over pMHC as the former ligates only the TCR, whereas MHC molecules also ligate co-receptors, which could have an independent response to changes in ligand spacing. This approach also keeps constant any contributions due to mechanically induced conformational changes. More details on fabrication and functionalization were described in Refs.20, 44.
T cell assay and microscopy
Leukapheresis products (non-clinical and de-identified) were obtained from the New York Blood Center (New York, NY), which are exempt from institutional review board (IRB) review. CD4+ T cells were purified by negative selection (RosetteSep)45. Total human CD4+ T-cells (106 cells per mL) were purified by negative selection, plated on the nanoarrays and incubated for 5 min (This is the optimal time for maximal early TCR signaling) at 37 °C and 0% CO2. Cells were fixed in warm PHEMO buffer (10 mM EGTA, 2 mM MgCl2, 60 mM Pipes, 25 mM HEPES, pH 7.2) with 2% paraformaldehyde for 10 min and permeabilized (0.1% Triton in PBS) for 3 min. Cells were then stained with the pY binding monoclonal antibody Alexa 488-PY20 (Biolegend) and rabbit anti-human CD45 H-230 recognizing C-terminal amino acids 1075–1304 so as to avoid potential differences in extracellular access on 2D and 3D surfaces, which was detected with anti-rabbit F(ab’)2 secondary antibody labeled with Alexa647 (Jackson ImmunoResearch). The “average pY intensity” over the entire cell contact surface was measured to determine the cellular response. Imaging was done using an inverted total interference reflection fluorescence microscope (100×, 1.49NA, Nikon Eclipse Ti) with a back illuminated EM-CCD camera (AndorDU897). For SEM, the fixed cells were processed by critical point drying and sputter coating of 10 nm AuPd.
Fluorescence recovery after photobleaching (FRAP) was used to measure the SLB (ICAM1–405) continuity and mobility. A small patch of the SLB was bleached with intense light through closed aperture, and then a time series was immediately acquired with low intensity light through open aperture. For molecular occupancy analysis, a continuous movie was taken for a small region of interest (ROI) on a 1 µm-spaced nanoarray until the florescence signal (Fab’-Alexa 568) was fully bleached.
Data analysis
The FIJI software package was used for image processing and data analysis. As an indicator of TCR signaling strength, the average pY intensity was measured for the ROI of each cell (Fig. 2, 3). Our fabrication technique based on electron beam lithography enables the integration and therefore direct comparison of various patterns on a same sample. For the comparison between samples, the data was normalized by the positive control on continuous AuPd surfaces (maximal Fab’ density). And the negative control on SLB passivation background with 200 /µm2 ICAM1 and minimal Fab’ density (non-specific adsorption only) provided a baseline. For cell spreading quantification (Fig. 5), area and circularity were measured for the ROI of each cell in SEM images (magnification 1000 ×).
For the analysis of pY colocalization and CD45 exclusion on NP clusters, an intensity ratio of the cluster over surrounding area was measured (Figs. 4f, 6c, f). The ROI selection is described in Supplementary Fig. S15. These are probably slightly underestimated as a result of light microscope resolution limits. For the spatial effect on pY colocalization (Supplementary Fig. S8a), clusters with the same hexagon layout (cluster area, inter-cluster gap) were compared by the Mander’s coefficient measured with the “Colocalization Threshold” plug-in.
For FRAP analysis, the intensity in the center of bleached region was normalized by that of surrounding area, and plotted against time. The FRAP curves confirmed the SLB integrity in the presence of the nanoarrays and revealed a dependence of ICAM1 mobility on nanoarray geometry (Supplementary Fig. S6). There was an average of 1 UCHT1-Fab’ per NP by resolving and counting the fluorescence bleaching steps, on both 2D and 3D surfaces (Supplementary Fig. S5). This indicates that the NPs penetrate the SLB, rather than being covered by it. The sidewalls of the pedestals did not increase non-specific binding, as they were likely covered with lipid membrane and/or blocked by casein. Moreover, the Fab’ intensity scaled linearly with NP density (Supplementary Fig. S4), which further suggests that the NPs were closely surrounded by the SLB with no significant edge effects (defective coverage around NPs). This is important because defects in the SLB, especially around the NPs, would result in nonspecific adsorption of UCHT1 Fab’ and could alter cellular response.
Supplementary Material
Acknowledgments
The authors thank S. Curado for coordination and S. Davis for insightful comments. This work was supported primarily by the National Science Foundation under award no. CMMI-1300590, by the National Institutes of Health Common Fund Nanomedicine program grant PN2 EY016586, R37 AI043542 and P01A1080192. and Wellcome Trust and Kennedy Trust for Rheumatology Research PRF 100262Z/12/Z. The Columbia Nano Initiative provided cleanroom and processing facilities. We thank M. Cammer of NYULMC OCR for microscopy and analysis support.
Footnotes
Author contributions
H. C. carried out nanofabrication. H. C. and D. D. contributed equally to development of the bilayer backfill methodology. H. C. and J. M. contributed equally to the data acquisition and analysis. V. M. provided purified ICAM1. S. J. W., M. L. D. and M. P. S. designed the experiments. H. C., J. M., M. L. D. and S. J.W. interpreted data and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding authors upon reasonable request.
Additional information
Supplementary information is available in the online version of the paper.
References
- 1.Iwashima M, Irving BA, van Oers NS, Chan AC, Weiss A. Sequential interactions of the TCR with two distinct cytoplasmic tyrosine kinases. Science. 1994;263(5150):1136–1139. doi: 10.1126/science.7509083. [DOI] [PubMed] [Google Scholar]
- 2.Liaunardy-Jopeace A, Murton BL, Mahesh M, Chin JW, James JR. Encoding optical control in LCK kinase to quantitatively investigate its activity in live cells. Nature structural & molecular biology. 2017 doi: 10.1038/nsmb.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherman E, Barr V, Manley S, Patterson G, Balagopalan L, et al. Functional Nanoscale Organization of Signaling Molecules Downstream of the T Cell Antigen Receptor. Immunity. 2011;35(5):705–720. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pageon SV, Tabarin T, Yamamoto Y, Ma Y, Bridgeman JS, et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. U S A. 2016;113(37):E5454–5463. doi: 10.1073/pnas.1607436113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui E, Vale RD. In vitro membrane reconstitution of the T-cell receptor proximal signaling network. Nature structural & molecular biology. 2014;21(2):133–142. doi: 10.1038/nsmb.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang VT, Fernandes RA, Ganzinger KA, Lee SF, Siebold C, et al. Initiation of T cell signaling by CD45 segregation at 'close contacts'. Nat. Immunol. 2016;17(5):574–582. doi: 10.1038/ni.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 2006;7(8):803–809. doi: 10.1038/ni1369. [DOI] [PubMed] [Google Scholar]
- 8.Cochran JR, Cameron TO, Stone JD, Lubetsky JB, Stern LJ. Receptor proximity, not intermolecular orientation, is critical for triggering T-cell activation. J. Biol. Chem. 2001;276(30):28068–28074. doi: 10.1074/jbc.M103280200. [DOI] [PubMed] [Google Scholar]
- 9.Deeg J, Axmann M, Matic J, Liapis A, Depoil D, et al. T cell activation is determined by the number of presented antigens. Nano Lett. 2013;13(11):5619–5626. doi: 10.1021/nl403266t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matic J, Deeg J, Scheffold A, Goldstein I, Spatz JP. Fine Tuning and Efficient T Cell Activation with Stimulatory aCD3 Nanoarrays. Nano Lett. 2013 doi: 10.1021/nl4022623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delcassian D, Depoil D, Rudnicka D, Liu M, Davis DM, et al. Nanoscale ligand spacing influences receptor triggering in T cells and NK cells. Nano Lett. 2013;13(11):5608–5614. doi: 10.1021/nl403252x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature. 2005;436(7050):578–582. doi: 10.1038/nature03843. [DOI] [PubMed] [Google Scholar]
- 13.Carbone CB, Kern N, Fernandes RA, Hui EF, Su XL, et al. In vitro reconstitution of T cell receptor-mediated segregation of the CD45 phosphatase. Proc. Natl. Acad. Sci. USA. 2017;114(44):E9338–E9345. doi: 10.1073/pnas.1710358114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen BM, Al-Aghbar MA, Lee CH, Chang TC, Su YC, et al. The Affinity of Elongated Membrane-Tethered Ligands Determines Potency of T Cell Receptor Triggering. Front. Immunol. 2017;8:793. doi: 10.3389/fimmu.2017.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irles C, Symons A, Michel F, Bakker TR, van der Merwe PA, et al. CD45 ectodomain controls interaction with GEMs and Lck activity for optimal TCR signaling. Nat. Immunol. 2003;4(2):189–197. doi: 10.1038/ni877. [DOI] [PubMed] [Google Scholar]
- 16.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487(7405):64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmid EM, Bakalar MH, Choudhuri K, Weichsel J, Ann HS, et al. Size-dependent protein segregation at membrane interfaces. Nat. Phys. 2016;12(7):704-+. doi: 10.1038/nphys3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, Vendome J, Shapiro L, Ben-Shaul A, Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475(7357):510–513. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schvartzman M, Wind SJ. Robust pattern transfer of nanoimprinted features for sub-5-nm fabrication. Nano Lett. 2009;9(10):3629–3634. doi: 10.1021/nl9018512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai H, Wolfenson H, Depoil D, Dustin ML, Sheetz MP, et al. Molecular Occupancy of Nanodot Arrays. ACS Nano. 2016 doi: 10.1021/acsnano.5b07425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310(5751):1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 22.Roman G, Martin M, Joachim PS. Block copolymer micelle nanolithography. Nanotechnology. 2003;14(10):1153. [Google Scholar]
- 23.Schoen I, Hu W, Klotzsch E, Vogel V. Probing cellular traction forces by micropillar arrays: contribution of substrate warping to pillar deflection. Nano Lett. 2010;10(5):1823–1830. doi: 10.1021/nl100533c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettinger CJ, Langer R, Borenstein JT. Engineering Substrate Topography at the Micro- and Nanoscale to Control Cell Function. Angew. Chem. Int. Edit. 2009;48(30):5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vardhana S, Choudhuri K, Varma R, Dustin ML. Essential Role of Ubiquitin and TSG101 Protein in Formation and Function of the Central Supramolecular Activation Cluster. Immunity. 2010;32(4):531–540. doi: 10.1016/j.immuni.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science. 2017;356(6338) doi: 10.1126/science.aal3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi X, Bi Y, Yang W, Guo X, Jiang Y, et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature. 2013;493(7430):111–115. doi: 10.1038/nature11699. [DOI] [PubMed] [Google Scholar]
- 28.Gagnon E, Schubert DA, Gordo S, Chu HH, Wucherpfennig KW. Local changes in lipid environment of TCR microclusters regulate membrane binding by the CD3ε cytoplasmic domain. The Journal of Experimental Medicine. 2012;209(13):2423–2439. doi: 10.1084/jem.20120790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minguet S, Swamy M, Alarcón B, Luescher IF, Schamel WWA. Full Activation of the T Cell Receptor Requires Both Clustering and Conformational Changes at CD3. Immunity. 2007;26(1):43–54. doi: 10.1016/j.immuni.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 30.Tolar P, Sohn HW, Pierce SK. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat. Immunol. 2005;6(11):1168–1176. doi: 10.1038/ni1262. [DOI] [PubMed] [Google Scholar]
- 31.Shah NH, Wang Q, Yan QR, Karandur D, Kadlecek TA, et al. An electrostatic selection mechanism controls sequential kinase signaling downstream of the T cell receptor. eLife. 2016;5 doi: 10.7554/eLife.20105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su X, Ditlev JA, Rosen MK, Vale RD. Reconstitution of TCR Signaling Using Supported Lipid Bilayers. Methods Mol. Biol. 2017;1584:65–76. doi: 10.1007/978-1-4939-6881-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stone MB, Shelby SA, Nunez MF, Wisser K, Veatch SL. Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. eLife. 2017;6 doi: 10.7554/eLife.19891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, et al. The Immunological Synapse: A Molecular Machine Controlling T Cell Activation. Science. 1999;285(5425):221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 35.Lohmuller T, Triffo S, O'Donoghue GP, Xu Q, Coyle MP, et al. Supported membranes embedded with fixed arrays of gold nanoparticles. Nano Lett. 2011;11(11):4912–4918. doi: 10.1021/nl202847t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manz BN, Jackson BL, Petit RS, Dustin ML, Groves J. T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc. Natl. Acad. Sci. U S A. 2011;108(22):9089–9094. doi: 10.1073/pnas.1018771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor MJ, Husain K, Gartner ZJ, Mayor S, Vale RD. A DNA-Based T Cell Receptor Reveals a Role for Receptor Clustering in Ligand Discrimination. Cell. 2017;169(1):108–119. e120. doi: 10.1016/j.cell.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen MC, Riddell SR. Designing chimeric antigen receptors to effectively and safely target tumors. Current Opinion in Immunology. 2015;33:9–15. doi: 10.1016/j.coi.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guest RD, Hawkins RE, Kirillova N, Cheadle EJ, Arnold J, et al. The Role of Extracellular Spacer Regions in the Optimal Design of Chimeric Immune Receptors: Evaluation of Four Different scFvs and Antigens. Journal of Immunotherapy. 2005;28(3):203–211. doi: 10.1097/01.cji.0000161397.96582.59. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Stagg NJ, Johnston J, Harris MJ, Menzies SA, et al. Membrane-Proximal Epitope Facilitates Efficient T Cell Synapse Formation by Anti-FcRH5/CD3 and Is a Requirement for Myeloma Cell Killing. Cancer Cell. 2017;31(3):383–395. doi: 10.1016/j.ccell.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gradisar H, Bozic S, Doles T, Vengust D, Hafner-Bratkovic I, et al. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nat. Chem. Biol. 2013;9(6):362–366. doi: 10.1038/nchembio.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodman RP, Schaap IA, Tardin CF, Erben CM, Berry RM, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310(5754):1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 43.Dustin ML, Starr T, Varma R, Thomas VK. Current Protocols in Immunology. JohnWiley & Sons; New York: 2007. Supported Planar Bilayers for Study of the Immunological Synapse; pp. 18.13.11–18.13.35. [DOI] [PubMed] [Google Scholar]
- 44.Cai H, Depoil D, Muller J, Sheetz MP, Dustin ML, et al. Spatial Control of Biological Ligands on Surfaces Applied to T Cell Activation. Methods Mol. Biol. 2017;1584:307–331. doi: 10.1007/978-1-4939-6881-7_18. [DOI] [PubMed] [Google Scholar]
- 45.Vasiliver-Shamis G, Cho MW, Hioe CE, Dustin ML. Human immunodeficiency virus type 1 envelope gp120-induced partial T-cell receptor signaling creates an F-actin-depleted zone in the virological synapse. J. Virol. 2009;83(21):11341–11355. doi: 10.1128/JVI.01440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.