Abstract
The Shape, Elongation, Division, and Sporulation (“SEDS”) proteins are a large family of ubiquitous and essential transmembrane enzymes with critical roles in bacterial cell wall biology. The exact function of SEDS proteins was long enigmatic, but recent work1–3 has revealed that the prototypical SEDS family member RodA is a peptidoglycan polymerase – a role previously attributed exclusively to members of the penicillin binding protein family4. This discovery has made RodA and other SEDS proteins promising targets for the development of next-generation antibiotics. However, little is known regarding the molecular basis for SEDS activity, and no structural data are available for RodA or any homolog thereof. Here, we report the crystal structure of Thermus thermophilus RodA at a resolution of 2.9 Å, determined using evolutionary covariance-based fold prediction to enable molecular replacement. The structure reveals a novel ten-pass transmembrane fold with large extracellular loops, one of which is partially disordered. The protein contains a highly conserved cavity in the transmembrane domain, reminiscent of ligand binding sites in transmembrane receptors. Mutagenesis experiments in Bacillus subtilis and Escherichia coli show that perturbation of this cavity abolishes RodA function both in vitro and in vivo, indicating it is catalytically essential. These results provide a framework for understanding bacterial cell wall synthesis and SEDS protein function.
Synthesis of a cell wall and maintenance of its integrity are essential processes in virtually all eubacteria, and targeted disruption of cell wall biogenesis is among the most effective therapeutic strategies in the treatment of bacterial infections. A central step in cell wall synthesis is the concatenation of a lipid II disaccharide pentapeptide headgroup onto a peptidoglycan chain through a glycosyl transfer reaction. This reaction has long been known to be catalyzed by the glycosyltransferase domains of class A penicillin binding proteins4 (aPBPs). However, deletion of all aPBPs is tolerated in Bacillus subtilis5, and bacteria lacking aPBPs are able to synthesize peptidoglycan6, implying the existence of non-aPBP glycoslytransferases. This mystery was resolved with the recent discovery that the highly conserved SEDS (shape, elongation, sporulation, and division) membrane proteins comprise a second class of peptidoglycan polymerases1–3 (Figure 1a). In fact, SEDS proteins are even more widely distributed than aPBPs1,7. Despite their importance and broad phylogenetic distribution however, SEDS protein function is not well understood and no SEDS protein has been characterized structurally.
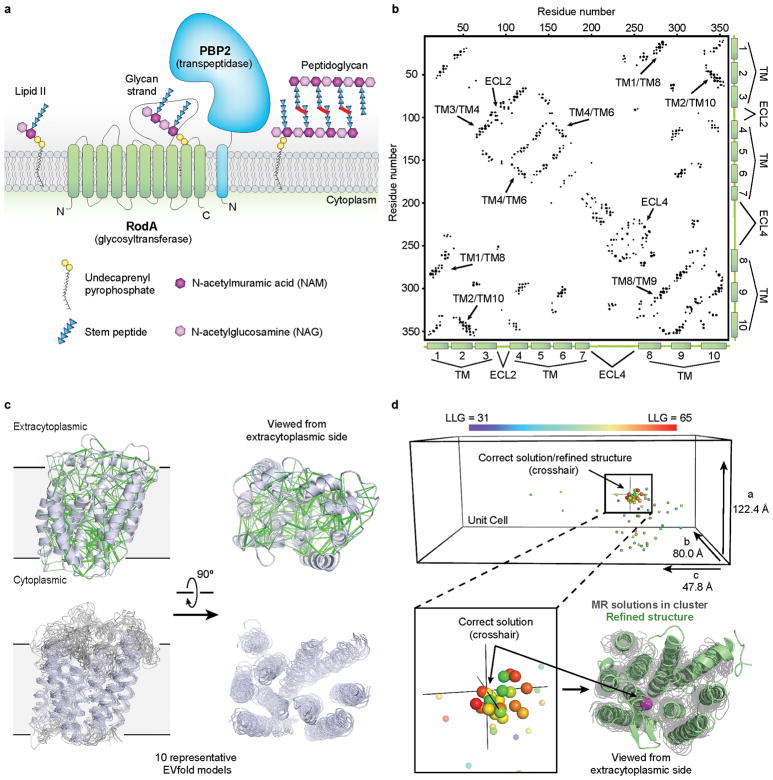
Figure 1. Biological role of RodA and evolutionary covariation fold prediction.
a, RodA is a peptidoglycan polymerase. b, Evolutionary covariation map showing 476 co-evolved residues in RodA. c, Evolutionary couplings (green lines) were used to generate models of RodA. Ten representative models are shown. d, Molecular replacement (MR) solutions obtained from 100 RodA models. As a reference point, the Cα atom of Glu108 is shown as a sphere colored according to LLG score for each solution. The corresponding position for the final refined RodA structure is shown as a crosshair. The refined structure (green) and the Cα atom of Glu108 (magenta sphere) is overlaid with the top MR solutions shown in gray.
To better understand SEDS protein function, we pursued structural studies of wild-type Thermus thermophilus RodA as well as a catalytically inactive RodA mutant (D255A). The proteins were expressed, purified and then crystallized by the lipidic cubic phase method. X-ray diffraction datasets were collected to a resolution of 2.9 Å and 3.2 Å for wild-type and D255A mutant proteins, respectively. Since RodA has no homologs of known structure, phase calculation by molecular replacement was impossible. A wide variety of heavy-atom phasing approaches were attempted without success. Having exhausted conventional methods, we sought to develop a new approach that requires neither experimental phase data nor a structure of a homologous protein.
Recent methodological advances in molecular replacement have expanded the range of suitable templates8, and evolutionary co-variation analysis now allows for fold prediction even in the absence of prior structural data9–11. This approach exploits the fact that residues that interact with one another structurally tend to co-evolve to maintain their interactions. Analysis of many sequences allows inference of spatial interactions between pairs of residues, providing restraints sufficient to define major features of protein structure. Analysis of RodA by this method showed extensive covariation throughout the protein (Figure 1b, c), and we reasoned that using evolutionary coupling restraints to build RodA models might provide suitable templates for molecular replacement phasing, an approach we call evolutionary coupling enabled molecular replacement (“EC-MR”).
In brief, our EC-MR approach consisted of parallel construction and sampling of many independent evolutionary coupling-derived models of RodA to identify suitable templates, followed by phase calculation and model building. First, 100 models of RodA were constructed on the basis of evolutionary restraints. These were each tested as single templates for molecular replacement in Phaser12. Of these, 22 models yielded a cluster of solutions that were high scoring and similar to one another (Figure 1d). An ensemble search model was constructed from a subset of these, producing maps suitable for manual rebuilding followed by ROSETTA refinement in Phenix13. The final refined structure showed normal crystallographic statistics (Extended Data Table 1; Extended Data Figure 1). While previous work has established that structural models constructed ab initio can in principle be suitable for molecular replacement phasing in select cases14,15, these analyses were conducted on very short proteins (<100 amino acids) with high resolution structural data (<2.1 Å). Other work has shown that at very high resolution, structures can be solved using even a single atom as a search model16. At lower resolution, symmetric α helical proteins can be phased from helical fragments, although this relies on symmetry conditions that are met only in a small minority of cases17. Determination of the structure of RodA by EC-MR establishes that evolutionary covariance-derived models can be suitable for phase determination of even a large (359 amino acid) asymmetric protein at modest resolution.
The structures of wild-type RodA and the D255A mutant are virtually identical (Extended Data Figure 2), and we focus here on the higher resolution wild-type RodA structure. The overall structure shows ten well-resolved transmembrane helices connected by loops, most of which are well ordered (Figure 2). Searches for proteins of similar fold with the DALI server18 yielded no hits, indicating that RodA possesses a unique overall fold. The transmembrane helices of RodA are largely straight and perpendicular to the membrane plane, with the exception of transmembrane helix 3 which runs diagonally through the membrane with a 45° kink at Pro71. While some studies have suggested that the SEDS protein FtsW could function as a lipid II flippase19, RodA lacks a transmembrane channel suitable for lipid II transport, and bears no notable structural similarity to transporters or flippases.
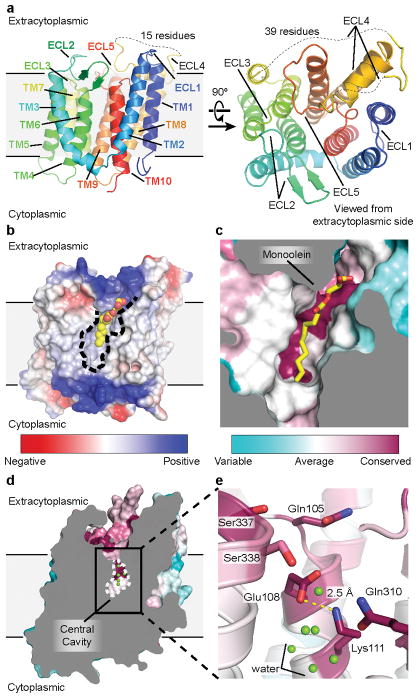
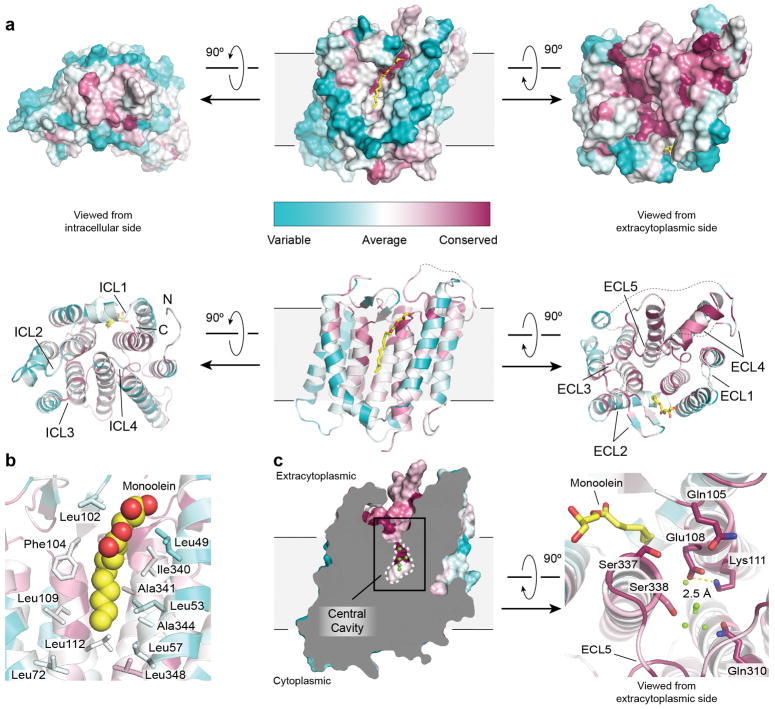
Figure 2. Structure of RodA.
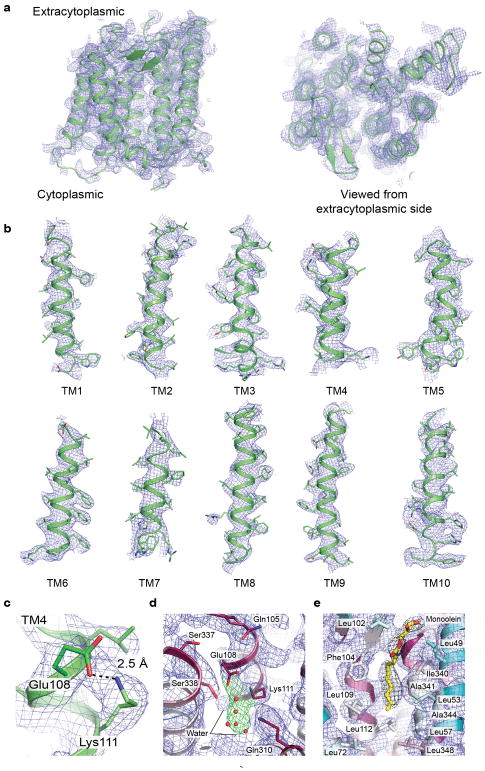
a, Structure of RodA viewed parallel to the membrane plane (left) and from the extracellular side (right). b, Surface view of RodA showing electrostatic potential and the position of a bound lipid between TM2 and TM3 (orientation is identical to that in panel a). c, Close up view of bound lipid (yellow). RodA surface is colored by sequence conservation. d, In the center of RodA is a water-filled cavity open to the extracytoplasmic surface. e, This cavity is flanked by highly conserved polar residues and a salt bridge between Glu108 and Lys111.
The intracellular loops (ICLs) of RodA are structured, but are very short and little polar surface area is exposed on the intracellular face of the enzyme. In contrast, extracellular loops (ECLs) 2, 4 and 5 are large and contain many functionally essential residues1, consistent with catalytic activity occurring at the extracytoplasmic face of the membrane. ECL2 includes a highly conserved β-hairpin, capped by Gly100 and Pro101. ECL4 is even larger at 80 amino acids in length, but is not resolved from residues 189 to 227 as well as from 237 to 251. These regions include essential residues as well as high-ranking evolutionary couplings between ECL4 and ECL1, ECL2, and ECL5, suggesting that they play functionally important roles despite not being resolved in the current structure (Extended Data Figures 3 and 4). It is possible that these regions become ordered only upon substrate binding or in complex with a peptidoglycan crosslinking enzyme. Unlike other ECLs, ECL5 is not exposed to the surface and instead is buried within the protein core.
Between TM2 and TM3 is a long hydrophobic groove containing electron density suggestive of a bound lipid molecule, which we tentatively modeled as monoolein due to its high concentration in the crystallization conditions (Figure 2b – c, Extended Data Figures 2 and 5). This groove is adjacent to a collection of highly conserved residues (Figure 2c), and may represent the binding site for the lipid-anchored substrates of RodA. Adjacent to this groove is a large water-filled cavity open to the extracellular face of the protein, flanked by Glu108, Met306, Leu307, Gln310 and Thr342 (Figure 2d). On the edge of this cavity, Glu108 and Lys111 form an absolutely conserved salt bridge (Figure 2e). Mapping the results of previous high-throughput mutagenesis in Bacillus subtilis RodA onto the structure shows that the salt bridge and other residues in the central cavity are intolerant of substitution1 (Figure 3a).
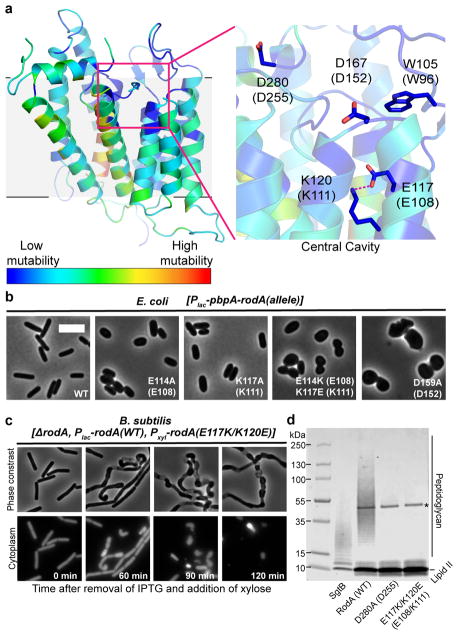
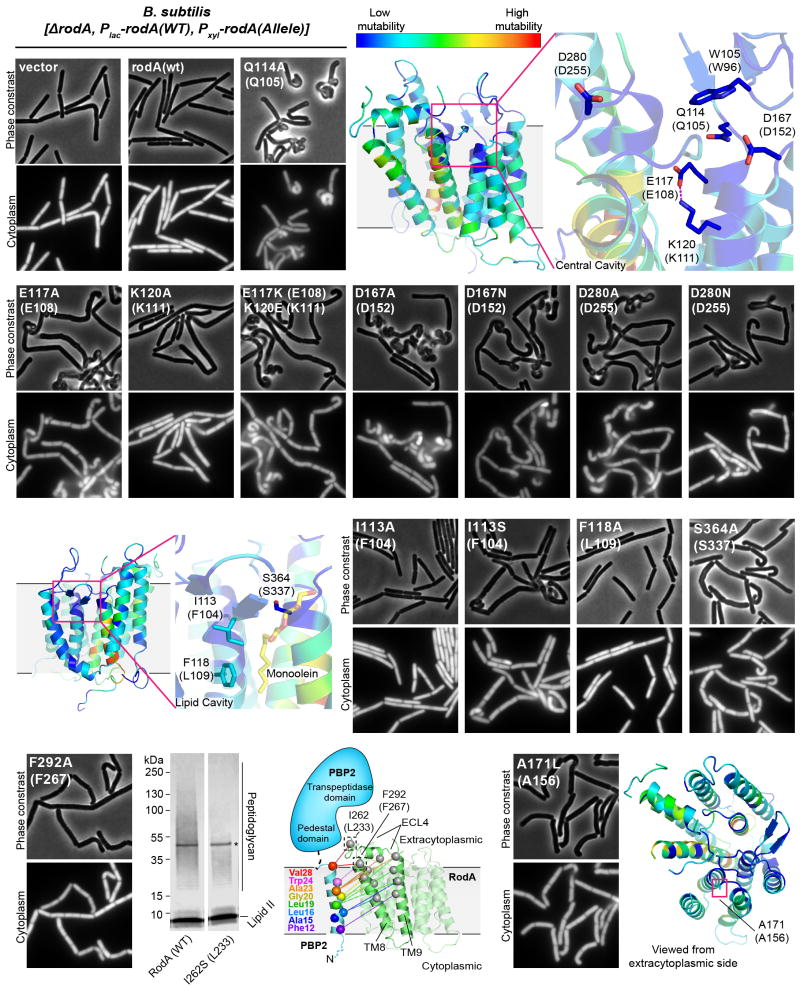
Figure 3. Central cavity is essential for RodA function.
a, Homology model of Bacillus subtilis RodA with each residue colored according to its tolerance for mutation (see Methods). Residues are numbered according to B. subtilis, with corresponding T. thermophilus numbering in parentheses. b, Wild-type E. coli cells harboring a plasmid with indicated RodA mutants. Scale bar 5 μm. c, Expression of RodA charge swap mutant is toxic in B. subtilis. Intracellular mCherry expression indicates cytosol. For both b and c, images are representative of 3 independent experiments. d, Catalytic competence of B. subtilis RodA mutants, representative of 2 independent experiments. Asterisk indicates the PBP4 labeling enzyme used for detection.
In order to explore the function this central cavity in more detail, we turned to site-directed mutagenesis followed by phenotypic characterization in the representative model Gram-negative and Gram-positive organisms E. coli and B. subtilis. First, we assessed the essentiality of the conserved salt bridge between Glu108 and Lys111 by monitoring the effect of the mutant enzyme when expressed in a wild-type background. Consistent with previous results, mutation of either of these residues to alanine resulted in a dominant-negative phenotype characteristic of Rod complex dysfunction, in which cells lose their elongated morphology and become enlarged and spherical prior to lysis (Figure 3b). This suggests that the mutant proteins are properly folded and able to interact with other members of the Rod complex, but are unable to promote cell elongation.
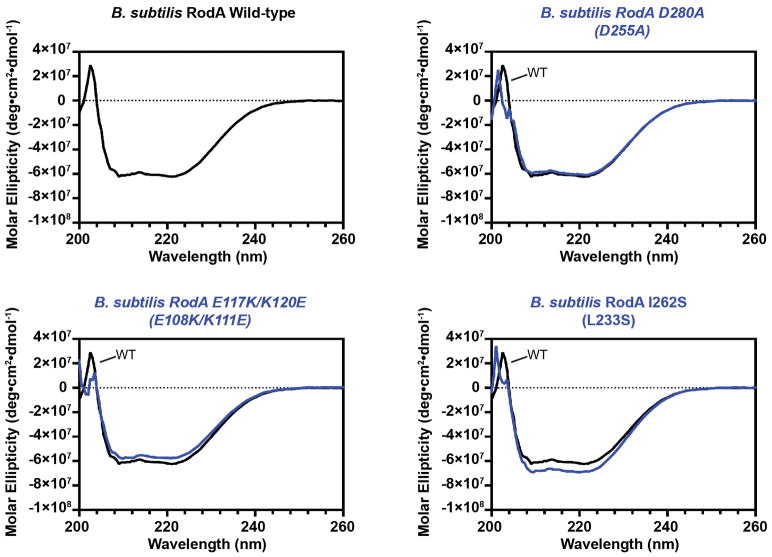
To ascertain whether the salt bridge is catalytic or merely important for proper folding, we constructed a salt bridge swap mutant (i.e., E108K and K111E double mutant). If the role of the salt bridge is purely structural, this swap should have minimal impact on RodA function since the salt bridge is maintained. However, if Glu108 plays a role in catalysis, then swapping the two amino acids would be expected to abrogate function. Indeed, the mutant protein folds properly as measured by circular dichroism spectroscopy (Extended Data Figure 6), but in cell assays shows a strong dominant-negative phenotype (Figure 3b, c, Extended Data Figure 7). Moreover, the mutant enzyme has no detectable peptidoglycan polymerization activity in vitro, confirming that its toxicity derives from a lack of catalytic activity (Figure 3d, Supplementary Figure 1). Other residues near the central cavity were similarly essential for RodA function, including Asp255 as reported previously1, as well as Asp152 (Figure 3b, Extended Data Figure 7). Analysis of evolutionary co-variation data with EVmutation20 likewise predicts these residues and the salt bridge to be immutable. Taken together, the high degree of sequence conservation, intolerance to mutation, and catalytic essentiality of residues surrounding the central cavity confirm that this portion of the protein plays critical role in peptidoglycan polymerization, making it a prime target for antibiotic discovery.
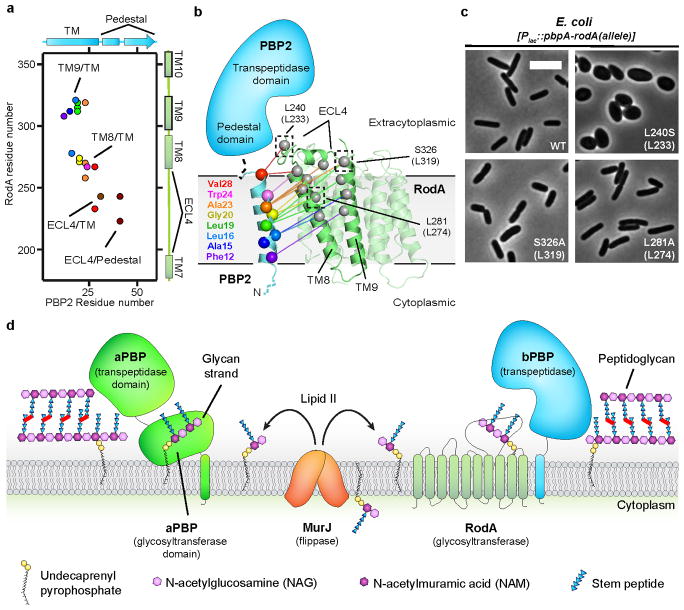
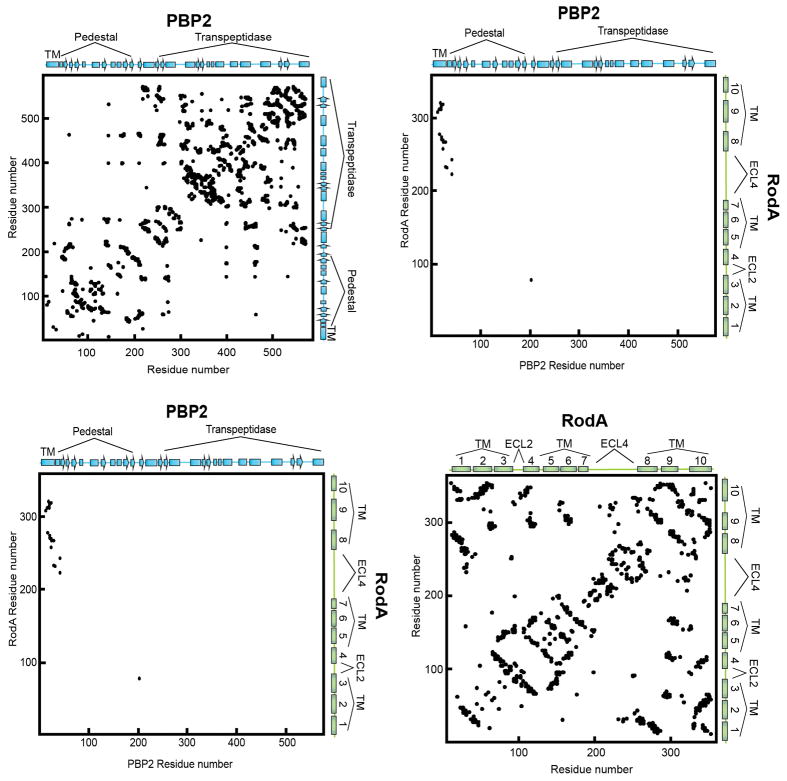
The glycan strand polymerization process catalyzed by RodA and aPBPs is essential but not sufficient to build a cell wall. A second key step is peptide crosslinking, catalyzed by the penicillin binding domains of both aPBPs and bPBPs. Cytological and protein-protein interaction studies indicate that SEDS proteins and bPBPs likely form a complex in cells1,2, and evolutionary coupling analysis shows strong co-variation between bPBP and SEDS protein sequences. This is sufficient to map the binding site between bPBPs and RodA to TM8 and TM9 (Figure 4a, b; Extended Data Figure 8), corresponding to the proposed interaction site between the divisome SEDS protein FtsW and its corresponding bPBP, FtsI10,21,22. RodA mutants in this interface exhibit a dominant negative effect in E. coli (Figure 4c). However, mutation of this site does not prevent RodA-mediated peptidoglycan polymerization in vitro (Extended Data Figure 7), confirming the functional importance of coordinated glycan strand elongation and peptide crosslinking in cells.
Figure 4. Interaction between RodA and its class B Penicillin Binding Protein (PBP2).
a, Evolutionary covariation map showing 19 evolutionary couplings between RodA and PBP2. b, Graphical representation of evolutionary covariation. c, Mutations in RodA/bPBP interface result in morphological abnormalities. Mutations were made in E. coli RodA, with T. thermophilus numbering in parentheses. Data representative of 2 independent experiments. Scale bar 5 μm. d, Model for peptidoglycan biogenesis. Lipid II is flipped across the membrane by MurJ, and polymerized and crosslinked by a complex of a SEDS protein (RodA) and bPBP, or by a bifunctional aPBP.
Complexes between SEDS proteins and bPBPs contain both glycan strand polymerization and peptide crosslinking active sites, recapitulating the dual catalytic activities found in aPBPs (Figure 4d). Understanding how SEDS proteins coordinate their activity with bPBPs to build a cell wall will be an important area for future investigation. The structure of RodA now provides a foundation for such work, serving as a framework for understanding the function of SEDS proteins.
Methods
Protein purification
RodA from Thermus thermophilus was cloned into pAM172 plasmid1 using EcoR1 and AvrII restriction enzymes (resulting in plasmids pMS211 and pMS224 for wild-type and D255A RodA, respectively). The expression plasmids contain an amino-terminal SUMO-fusion followed by a FLAG epitope tag and a 3C protease cleavage site (SUMO-FLAG-3C-RodA and SUMO-FLAG-3C-RodAD255A) were transformed into E. coli C43 derivative of BL21 (DE3) harboring an arabinose-inducible Ulp1 protease plasmid (pAM174) under the selection for both plasmids. Five fresh transformants from each were inoculated into 5 mL LB medium supplemented with 100 μg mL−1 ampicillin and 35 μg mL−1 chloramphenicol and allowed to grow overnight at 37°C in a rolling shaker. The 5 mL overnight culture was then diluted in 1 L of TB broth supplemented with 0.1% glucose, 2 mM MgCl2, 100 μg mL−1 ampicillin, and 35 μg mL−1 chloramphenicol. Cultures were grown at 37°C until an OD600 of 0.6 and shifted to 20°C. At an OD600 of 0.8, protein expression was induced by addition of IPTG (1 mM final) and arabinose (0.2% final) for RodA and ULP1, respectively. After a 16 h induction, cells were harvested and frozen at −80°C.
Cells were resuspended in lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 20 mM MgCl2, 100 μg mL−1 lysozyme, 1:100,000 (v:v) benzonase nuclease, and 2 mg mL−1 iodoacetamide), lysed by sonication and membranes were collected by ultracentrifugation at 100,000×g for 1 h at 4 °C. FLAG-3C-RodA was then extracted using a glass dounce tissue grinder in a solubilization buffer containing 20 mM HEPES pH 7.5, 500 mM NaCl, 20% (v/v) glycerol, 2 mg mL−1 iodoacetamide, and 1% (w/v) n-Dodecyl β-D-maltoside (DDM; Anatrace). Samples were stirred for 2 h at 4 °C, then centrifuged as before for 1 h. The supernatant containing solubilized RodA was supplemented with 2 mM CaCl2 and loaded by gravity flow onto 4 mL anti-Flag antibody affinity resin. The resin was washed extensively, first in 50 ml of buffer containing 20 mM HEPES pH 7.0, 500 mM NaCl, 2 mM CaCl2, 20% glycerol, 0.1% DDM, and then in 50 ml of the same buffer supplemented with 10 mM Adenosine 5′-triphosphate magnesium salt and 20 mM KCl to remove the bacterial chaperones, GroEL and DnaK. RodA was eluted in 20 mM HEPES pH 7.0, 500 mM NaCl, 20% glycerol, 0.1% DDM supplemented with 5 mM EDTA and 0.2 mg ml−1 Flag peptide. 3C protease was added (1:1000 w:w) and incubated with RodA at 4°C overnight. RodA was further purified by size exclusion chromatography (SEC) on a Sephadex S200 column (GE Healthcare) in buffer containing 20 mM HEPES pH 7.5, 500 mM NaCl, and 0.1% DDM. After preparative SEC, the protein was concentrated to 30–40 mg ml−1 and flash frozen with liquid nitrogen in aliquots of 8 μl. Samples were stored at −80 °C until use for crystallography. Purity and monodispersity of crystallographic samples was evaluated by SDS–PAGE and analytical SEC, respectively.
Crystallography and Data Collection
Purified T. thermophilus wild-type and D255A RodA were reconstituted into lipidic cubic phase by mixing with a 10:1 (w:w) mix of monoolein (Hampton Research) with cholesterol (Sigma Aldrich) at a ratio of 1.0:1.5 protein:lipid by mass, using the coupled syringe reconstitution method23. All samples were mixed at least 100 times prior to dispensing. The resulting phase was dispensed in 15 – 40 nL drops onto a glass plate and overlaid with 600 nL of precipitant solution using a Gryphon LCP robot (Art Robbins Instruments). Crystals grew in precipitant solution containing 35 – 50% PEG 200, 100 mM NaCl, 100 mM MgCl2, and 100 mM Tris pH 7.6 – 8.2. Initial crystallization hits grew within 24 h, with diffraction-quality crystals reaching full size over the course of 2 – 4 weeks. Crystals were harvested using mesh loops and stored in liquid nitrogen until data collection. Data collection was carried out at Advanced Photon Source GM/CA beamline 23ID-B. An initial grid raster with 80 μm × 30 μm beam dimensions was performed to locate crystals within the loop. Additional fine-tuning rasters were performed using a 10 μm beam diameter to optimize the position of the crystal for data collection. Data were collected using a 10 μm beam and 0.2° oscillation width per frame at a wavelength of 1.033 Å and a 5-fold attenuation factor. For both wild-type and D255A RodA, a complete data set was obtained from a single crystal. Diffraction data were indexed and processed using XDS24. Both wild-type and D255A RodA crystallized in the C2 space group with one molecule in the asymmetric unit with a solvent content of approximately 60%.
Generation of Ab initio Evolutionary Coupling Derived Models of RodA
Multiple sequence alignments (MSA) of the full length T. thermophilus RodA (Uniprot ID Q5SIX3) were generated using the iterative hidden Markov model-based sequence search tool jackhmmer25 with 5 iterations. Alignments were built using the Uniref100 dataset26 released in April of 2017. An MSA was generated for 9 different bitscores, a sequence inclusion threshold normalized to length and expressed as the number of bits per residue, with values ranging from the most-inclusive 0.1 to the least-inclusive 0.9. The alignment depth was chosen to optimize the number of non-redundant sequences with the fewest gaps in the alignment as described elsewhere9,10, although that the alignment choice was robust with respect to consistency of predicted ECs over a wide range of alignment depths. Blindly optimizing the an alignment choice resulted in two alignments with the smaller one at 31,505 non-redundant sequences of length 346 residues (10 – 355) with an “effective number” of 8,729 sequences after down-weighting sequences with more than 80% identity and no more than 30% gaps in any columns used for the EC model computation.
Model folding criteria
For the chosen MSAs, evolutionary couplings (ECs) were determined using a pseudo-likelihood maximization (PLMC)27–29. A mixture model approach identified 99% percentile probability of being in contact. We used a combination of commonly used prediction methods to determine secondary structure predictions (PSIPRED30 and PolyPhobius31) together with secondary structure propensity computed directly from local ECs29 resulting in the identification 10 TM helices and 3 smaller helices in ECL4 and 2 beta-strands in ECL2. A total of 220 folded models for T. thermophilus RodA were generated for increasing numbers of EC restraints with using the folding protocol in EVfold9 which itself uses a distance geometry and simulated annealing protocol in CNS32,33. All models were ranked as described in previously11,34 and the 50 top-ranked models from each of the MSAs were used as as molecular replacement search models (vide infra). The full EVfold software package and ReadMe is available at https://github.com/debbiemarkslab/EVcouplings. To generate additional models used for further ensemble-based phasing runs, 5 additional models were generated using the top-ranked fold prediction from bitscore 0.8 with an additional round of dedicated simulated annealing in CNS. Cartesian dynamics were used for heating, torsion for cooling, and 600 final minimization steps with 50 cycles were used with other parameters kept as defaults. In addition to evolutionary couplings scores, folded models EVfold generates an EC enrichment score for each residues that reflects how constrained each residue is, and can be thought of as conservation of “coupling”.
Phasing and refinement
100 evolutionary coupling derived models of wild-type RodA were primed for molecular replacement using Sculptor35, which included limited side-chain pruning and B-factor assignment based on accessible surface area36. Each model was tested as a single search template for molecular replacement in Phaser12, resulting in 98 out of the 100 initial models providing candidate solutions. The solutions were sorted by translation function Z-score (TF-Z) and the top 32 solutions manually inspected in PyMOL37. A total of 22 solutions were highly similar and distinct from all other solutions. Sorting the solutions by log-likelihood gain (LLG) provided a similar result where all 22 solutions were among the top 40% of solutions. However, most of these were not well separated from other potential solutions in their respective searches, typically with two to four other candidate solutions giving LLG values at least 75% that of the top solution. The models giving the top 2 solutions (as judged by both TF-Z and LLG metrics) were used as a single ensemble for phase calculation followed by manual building using COOT38 and reciprocal space refinement using phenix.refine39. Manual refinement was complemented by the use of ROSETTA refinement in Phenix13. Subsequently, additional cycles of manual building and reciprocal space refinement led to the final refined structure.
Verification of sequence register was straightforward and unambiguous due to the relatively high resolution and frequency of bulky amino acid side chains (RodA is roughly 10% tryptophan, tyrosine, and phenylalanine). Representative composite omit map density is shown in Extended Data Figure 1. The structure of RodAD255A was solved using wild-type RodA as the molecular replacement search model and the resulting refined structure is nearly identical to that of wild-type RodA (0.1 Å RSMD between all Cα atoms). After refinement, the quality of both structures was assessed using MolProbity to calculate Ramachandran statistics and other parameters40, and figures were prepared in PyMOL. All crystallographic data processing, refinement, and analysis software was compiled and supported by the SBGrid Consortium41.
Predicting residue contacts between RodA and PBP2
EVComplex10 was used to predict inter-protein contacts between T. thermophilus full-length RodA and full-length PBP2 (Uniprot ID Q5SJ23). We constructed alignments for RodA (33670 sequences) and PBP2 (40764 sequences) as described above for RodA alone, using the April 2017 Uniprot release42 for clarity on species identifiers. We concatenated RodA and PBP2A sequences from each species when they were within 10,000 nucleotides of each other based on European Nucleotide Archive (ENA) data downloaded in February 201743, and evolutionary couplings were computed on the complex alignment as previously described44. A mixture model approach was used to identify top-scoring contacts, both within individual monomers and between RodA and PBP2, and to assign a probability to each contact of being within the tail of the distribution of coupling scores as described previously29. This scoring method is accurate for predicting whether or not proteins interact as well as which residues are in contact10. The distribution of ECs is approximated by a Gaussian-lognormal mixture model, and we defined the tail of the distribution as those scores that have >95% probability of belonging to the lognormal component. ECs in this tail defined as high probability resulted in 26 residue pairs predicted as contacts between RodA and PBP2.
Sequence and structure conservation analysis
Sequence conservation analysis in Figure 3 and Extended Data Figure 5 was computed using the ConSurf server45. In brief, a multiple sequence alignment of T. thermophilus RodA to its closest 150 homologues was generated using the HHMER algorithm provided by ConSurf, with conservation scores plotted in PyMOL. Additionally, a multiple sequence alignment of RodA to 506 homologues from representative bacterial taxa was generated using a protein sequence BLAST search on the NCBI public database using T. thermophilus RodA protein sequence as query and plotted in Extended Data Figure 3.
Homology Modeling and Mutability Index
A homology model of Bacillus subtilis RodA based on the structure of Thermus thermophilus RodA was constructed using MODELLER46. In brief, due to the low sequence identity (~26%) between the two enzymes, a multiple sequence alignment of 10 RodA homologs from diverse bacterial taxa was performed (Thermus thermophilus, Bacillus subtilis, Escherichia coli, Staphylococcus aureus, Streptococcus pneumoniae, Shigella flexneri, Haemophilus influenza, Deinococcus marmois, Bacillus safensis, and Lysinibacillus odysseyi). The resulting alignment was used as the search template in MODELLER in which disordered residues 204–256 and 266–276 were omitted from the model generation. A total of 10 models were generated and the top scoring model was chosen for further analysis.
The mutability index of each residue in the homology model of B. subtilis RodA was calculated based on the MutSeq data reported previously1. First, for each residue the nonsynonymous mutation rate was defined as the number of mutations observed after excluding synonymous and nonsense mutations. To calculate a raw mutability index, the nonsynonymous mutation rate was divided by the summed rate of silent mutations observed for a given nucleotide position. Finally, the raw mutability index for each residue was normalized with respect to the expected silent mutation rate for each general class of nucleotide change, resulting in a percent mutability index for each residue. A low mutability index (i.e., low percentage) represents a residue that is relatively intolerable to mutation at that position and a high mutability index (high percentage) represents a residue that can relatively tolerate changes at that position.
RodA mutant plasmid and strain construction
Mutants of E. coli RodA were introduced using QuikChange mutagenesis into a plasmid containing the allele Plac-pbpA-rodA(WT) (pHC857)1,2. Wild-type E.coli cells (strain TB28) were then transformed with the mutant plasmids to generate strains each harboring mutations in the central cavity (E114A, K117A, E114K/K117E, and D159A) and at the RodA-PBP2 interface (L240S, S326A, and L281A).
Mutants of Bacillus Subtilis RodA was introduced using QuikChange mutagenesis into the pER174a plasmid (amyE::Pxyl-rodA(WT)-Hisx10)1. The resulting mutagenic plasmids (amyE::Pxyl-rodA-(Allele)-Hisx10) was directly transformed into the B. subtilis strain (rodA::kan, Pspank-rodA(WT)-Hisx10, and PpensacA::mCherry) to generate strains each harboring mutations in the central cavity (Q114A, E117A, K120A, E117K/K120E, D280N, and S364A), the proposed lipid-binding cavities (I113A, I113S, and F118A, and A171L), and the RodA-PBP2 interface (F292A). The B. subtilis strains harboring amyE::Pxyl-rodA-(D167A), amyE::Pxyl-rodA-(D167N), and amyE::Pxyl-rodA-(D280A) were previously described by Meeske et al1.
Mutational Analysis in vivo
All E. coli cultures were grown at 37°C in M9 minimal medium supplemented with 0.2% (w/v) maltose, 0.2% (w/v) casamino acids, and 25 μg mL−1 chloramphenicol. Overnight cultures were diluted to an OD600 of 0.05 and grown to OD600 = 0.25 in the absence of inducer. These cultures were then further diluted to an OD600 of 0.005 in medium containing 1 mM IPTG to induce expression of the pbpA-rodA construct. Cells were grown for five generations in the presence of inducer and fixed when the OD600 reached between 0.15 and 0.2. Fixative solution contained 0.04% glutaraldehyde, 4% formaldehyde, 32 mM sodium phosphate, pH 7.5. Widefield phase contrast microscopy was performed on a Nikon TE2000 microscope equipped with a 100x Plan Apo 1.4 NA objective and a CoolSNAP HQ2 monochrome camera.
B. subtilis strains were derived from the PY79 prototrophic strain47. Cells were grown in LB medium at 37°C in the presence of 10 μM IPTG to express wild-type RodA. When cultures reached mid-log, cells were washed three times with plain LB and resuspended in 20 mL of LB to an OD600 of 0.02, then supplemented with 10 mM xylose to induce expression of RodA wild-type and variants. Cells were analyzed by phase contrast microscopy 90 minutes later. Microscopy was performed on a Nikon Ti microscope equipped with Plan Apo 100x/1.4NA phase contrast oil objective and a CoolSnapHQ2 camera. Cells were immobilized using 2% agarose pads containing growth medium. Images were cropped and adjusted using MetaMorph software (Molecular Devices).
Mutant analysis in vitro
Mutants of B. subtilis RodA were transformed into E. coli strain CAM333, a derivative of strain C43 with deletions in ponB, pbpC, and mgtA, and were expressed and purified by immunoaffinity chromatography as described previously1.
Prior to circular dichroism spectroscopy experiments, each RodA mutant was dialyzed into a buffer consisting of 10 mM sodium phosphate pH 7.5, 500 mM potassium fluoride, 0.5% (w/v) CHAPS, and 0.05% (w/v) DDM. Spectra were acquired on a Jasco J-815 spectropolarimeter. CD spectra were recorded between 200–260 nm using a quartz cuvette with a path length of 1 mm, a 50 nm/min scanning speed, and a bandwidth of 1 nm. Five spectra were measured at 25°C, averaged, and corrected for buffer contribution. Secondary structure assessment was not performed due to high absorbance contributions at wavelengths less than 205 nm from the detergent mixture necessary for protein stability and function (0.5% CHAPS, 0.05% DDM).
For assessment of enzymatic activity, lipid II substrate was purified from E. coli as described48. Peptidoglycan polymerization reactions were adapted from previously described methods49. Briefly, purified B. subtilis wild-type and mutant RodA proteins were incubated with lipid II in reaction buffer containing 50 mM HEPES pH 7.0, 20 mM CaCl2, 20 mM MgCl2, and 20% DMSO. The working concentration of RodA and lipid II were 1 μM and 20 μM, respectively. RodA was purified in 0.5% CHAPS and 0.05% DDM, therefore the working concentration for CHAPS and DDM was 0.05% and 0.005%, respectively. As a positive control lipid II was also polymerized with SgtBY181D (1 μM) in reaction buffer containing 12.5 mM HEPES pH 7.5, 2 mM MnCl2, 0.25 mM Tween-80, and 20% DMSO. All reactions were incubated at 25°C for 1 h and quenched by incubation at 95°C for 2 min. Peptidoglycan biotinylation of each reaction mixture was performed by addition of biotinylated D-lysine (BDL) (1.5 mM, final concentration) and PBP4 (3.8 μM, final concentration) followed by incubation a 25°C for 1 h. The biotinylation reaction was then quenched by addition of 13 μL 2x SDS loading dye. The samples were then loaded into a 4–20% gradient polyacrylamide gel and run at 180 V. The products were transferred onto a PVDF membrane (BioRad) and fixed in 0.4% paraformaldehyde diluted in PBS for 30 min at room temperature. The membrane was blocked with SuperBlock TBS blocking buffer (ThermoFisher) for 1 h at room temperature and the biotinylated products were detected by incubation with fluorescently-tagged streptavidin (IRDye 800-CW streptavidin (Li-Cor Biosciences, 1:5,000 in SuperBlock) for an additional 30 min. Membranes were washed 4 × 10 min with TBST (0.01% Tween 20) and a final 10 min wash in PBS. Blots were then visualized using an Oydessy CLx imaging system (Li-Cor Biosciences).
Data availability
Structure factors and refined atomic coordinates for RodA wild-type and D255A mutant are deposited in the Protein Data Bank under accession codes 6BAR and 6BAS, respectively.
Code Availability
The full EVfold software package is available at https://github.com/debbiemarkslab/EVcouplings.
Extended Data
Extended Data Figure 1. Representative electron density.
a–c, Simulated annealing composite omit 2Fo−Fc electron density map of Thermus thermophilus RodA contoured at 1.0 σ within a 2.0 Å radius of atoms shown. d, The same map contoured at 1.0 σ and colored blue and green for RodA and water molecules, respectively. The modeled waters are shown in red spheres. e, The same map contoured at 1.0 σ within a 3.0 Å radius RodA (shown in blue) and the same map contoured at 1.0 σ for monoolein (shown in black).
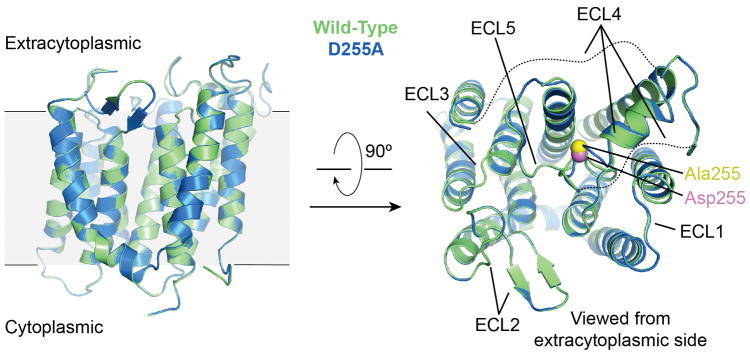
Extended Data Figure 2. Comparison of RodA wild-type and D255A structures.
Structures of wild-type (green) and D255A (blue) RodA are shown viewed parallel to the membrane and from the extracytoplasmic side in the left and right panels, respectively. The Cα atom of residue 255 for each structure is shown in spheres and colored pink and yellow for wild-type and D255A, respectively. The dashed lines represent the disordered residues 189 – 227 and 237 – 251 in both structures. The two structures are essentially identical with a Cα RMSD of 0.1 Å.
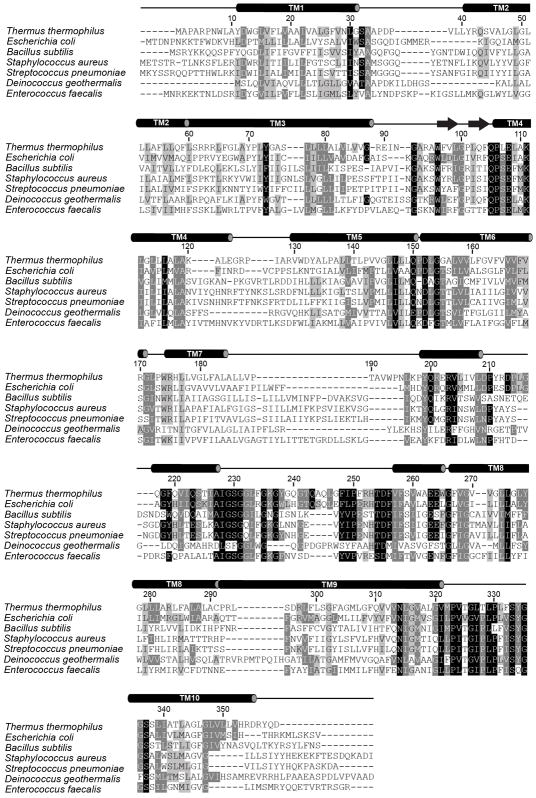
Extended Data Figure 3. RodA sequence conservation.
The results of an alignment of 506 RodA sequences from diverse bacterial taxa with representative examples displayed. Residues with 98%, 80%, and 60% similarity across all 506 sequences are shown in black, grey, and light grey respectively. Secondary structure elements are shown above the alignment on the basis of T. thermophilus RodA crystal structure and JPRED analysis of the portions of ECL4 that were not modeled in the structure.
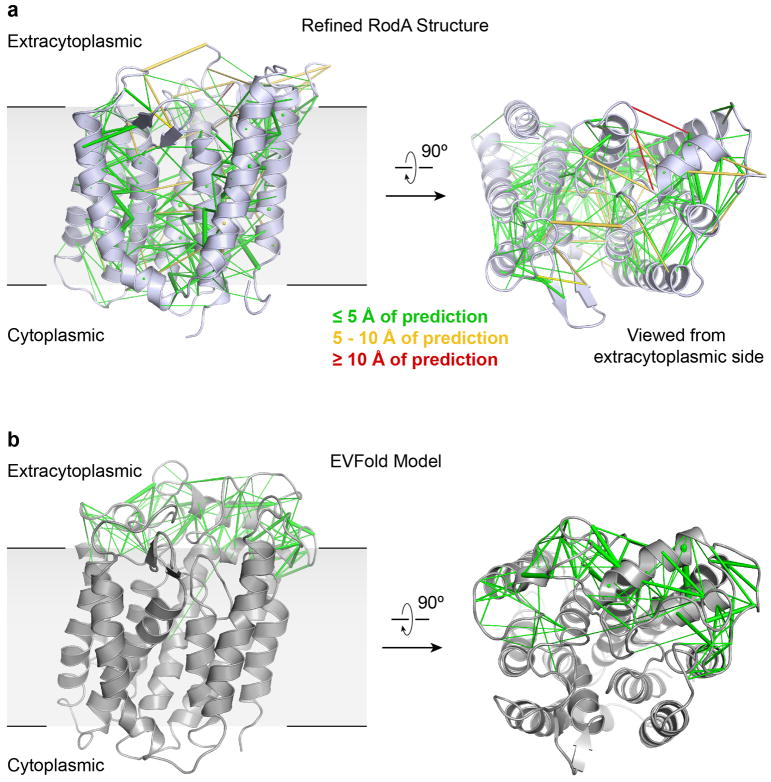
Extended Data Figure 4. RodA evolutionary couplings.
a, The refined crystal structure of T. thermophilus RodA (shown in light blue) is in close agreement with its evolutionary couplings (ECs). Green, yellow, and red lines between residues represent regions of the structure that are less than 5 Å, between 5 and 10 Å, and greater than 10 Å of the predicted ECs, respectively. b, The partially disordered extracellular loop 4 (ECL4) in the refined structure is strongly coupled evolutionarily to the TM domain of RodA. A graphical representation of predicted intra- and inter-domain ECs for ECL4 is mapped onto a EVFold model (shown in grey).
Extended Data Figure 5. Sequence conservation of T. thermophilus RodA.
a, Surface and ribbon representation of RodA (top and bottom panels, respectively). Analysis was performed using Consurf, colored in a scale from teal (poorly conserved) to magenta (highly conserved). A bound lipid modelled as monolien is shown in yellow sticks. The dashed lines represent disordered residues 189 – 227 and 237 – 251 in extracellular loop 4. b, The bound lipid (shown as spheres) is surrounded by many aliphatic amino acids (shown as sticks). c, Extracytoplasmic view of the water-filled central cavity and its proximity to the bound lipid molecule.
Extended Data Figure 6. Circular dichroism spectroscopy analysis of purified B. subtilis RodA variants.
Circular dichroism measurements of wild-type RodA as well as D280A, E117K/K120E, and I262S indicate that the overall folds of all four forms are comparable and display characteristic alpha-helical peaks at 208 and 222 nm. For each panel, the CD spectra of wild-type and the indicated mutant RodA are shown in black and blue, respectively. The corresponding T. thermophilus numbering for each mutant is shown in parentheses. Experiments were repeated independently 2 times with similar results.
Extended Data Figure 7. Mutagenic Analysis of B. subtilis RodA function in vivo.
Micrographs of B. subtilis strains harboring an IPTG-inducible allele of rodA(WT) and PxylA-rodA(WT) or PxylA-rodA(mut). Expression was induced by growing cells in the presence of 10 μM IPTG and 10 mM xylose. The bacterial cytosol is indicated by intracellular mCherry expression (Ppen-mCherry). Mutants in the central cavity show particularly deleterious phenotypes in this dominant negative assay. Experiments were repeated independently 2–3 times with similar results. Mutation made in the predicted RodA/bPBP interface does not prevent peptidoglycan polymerization in vitro (lower left panel), representative of 2 independent experiments.
Extended Data Figure 8. Evolutionary coupling analysis for the RodA-PBP2 complex.
Evolutionary covariation maps highlighting 693 couplings for PBP2 (top left panel), and 362 couplings for RodA (bottom right panel) using a 95% confidence threshold. These maps display good correlation to the crystal structure of RodA and homology models of PBP2. The top right and lower bottom panels depict the 19 predicted inter-protein contacts between RodA and PBP2 using the same 95% confidence threshold.
Extended Data Table 1.
Data collection and refinement statistics.
|
T. thermophilus RodA Wild-type |
T. thermophilus RodA D255A |
|
|---|---|---|
| Data collection | ||
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 122.4, 80.0, 47.8 | 121.2, 79.2, 47.4 |
| α, β, γ(°) | 90.0, 91.1, 90.0 | 90.0, 91.1, 90.0 |
| Resolution (Å) | 40.0 - 2.9 (3.1 - 2.9)* | 40.0 - 3.2 (3.4 - 3.2)* |
| Rsym (%) | 10.6 (147.9) | 17.1 (204.9) |
| <I /σ (I)> | 6.69 (0.69) | 4.08 (0.44) |
| CC1/2 (%) | 99.7 (45.8) | 99.6 (41.7) |
| Completeness (%) | 98.4 (98.8) | 99.6 (99.5) |
| Multiplicity | 3.02 (3.15) | 3.43 (3.35) |
| Refinement | ||
| Resolution (Å) | 40.0 - 2.91 (2.99 - 2.91) | 40.0 - 3.19 (3.31 - 3.19) |
| No. reflections | 18684 (1851 in test set) | 13756 (1360 in test set) |
| Rwork/Rfree (%) | 22.9/27.4 | 27.7/30.2 |
| No. atoms | ||
| Protein | 2229 | 2201 |
| Solvent ions/lipids | 44 | 1 |
| Water | 52 | 8 |
| Average B-factors (Å2) | ||
| Protein | 104.2 | 111.0 |
| Lipids | 109.6 | - |
| Solvent ions | 105.4 | 125.9 |
| Waters | 87.6 | 89.9 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.005 | 0.002 |
| Bond angles (°) | 0.825 | 0.751 |
| Ramachandran Statistics | ||
| Favored (%) | 94.5 | 93.5 |
| Allowed (%) | 5.5 | 6.5 |
| Outliers (%) | 0.0 | 0.0 |
Each datasets was collected from a single crystal.
Values in parentheses are for highest resolution shell.
Supplementary Material
Acknowledgments
Financial support for the work was provided by NIH grant U19AI109764 (A.C.K., D.Z.R., T.G.B., S.W., and D. K.), NIH grant R01GM106303 (D.S.M.) and a CIHR doctoral research award to P.D.A.R. We thank Advanced Photon Source GM/CA beamline staff for excellent technical support during X-ray data collection, and Dr. Kelly Arnett (Harvard Center for Macromolecular Interactions) for excellent support of circular dichroism experiments.
Footnotes
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
Author Contributions M.S. and A.J.M. performed expression screening experiments, and M.S. performed large scale purification and crystallization of RodA as well as enzyme assays and circular dichroism spectroscopy. Additional input regarding enzyme assays was provided by P.D.A.R, V.S., D.K., and S.W. The structure was solved and refined by M.S. and A.C.K. using evolutionary coupling-derived models developed by K.B., A.G.G., T.A.H, and D.S.M. Assessment of RodA mutant phenotypes was conducted by G.D. and P.D.A.R. with supervision from T.G.B. and D.Z.R. Overall project supervision was performed by A.C.K. with input from T.G.B. and D.Z.R. The manuscript was written by M.S. and A.C.K. with input from other authors.
Supplementary Information is available in the online version of the paper.
References
- 1.Meeske AJ, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016 doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho H, et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nature microbiology. 2016:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emami K, et al. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nature microbiology. 2017;2:16253. doi: 10.1038/nmicrobiol.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiology and molecular biology reviews : MMBR. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McPherson DC, Popham DL. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. Journal of bacteriology. 2003;185:1423–1431. doi: 10.1128/JB.185.4.1423-1431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Packiam M, Weinrick B, Jacobs WR, Jr, Maurelli AT. Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly”. Proc Natl Acad Sci U S A. 2015;112:11660–11665. doi: 10.1073/pnas.1514026112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otten C, Brilli M, Vollmer W, Viollier PH, Salje J. Peptidoglycan in Obligate Intracellular Bacteria. Molecular microbiology. 2017 doi: 10.1111/mmi.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiMaio F, et al. Improved molecular replacement by density- and energy-guided protein structure optimization. Nature. 2011;473:540–543. doi: 10.1038/nature09964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopf TA, et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopf TA, et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife. 2014;3 doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marks DS, et al. Protein 3D Structure Computed from Evolutionary Sequence Variation. PLOS ONE. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/s0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiMaio F, et al. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat Methods. 2013;10:1102–1104. doi: 10.1038/nmeth.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian B, et al. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rigden DJ, Keegan RM, Winn MD. Molecular replacement using ab initio polyalanine models generated with ROSETTA. Acta Crystallogr D Biol Crystallogr. 2008;64:1288–1291. doi: 10.1107/s0907444908033192. [DOI] [PubMed] [Google Scholar]
- 16.McCoy AJ, et al. Ab initio solution of macromolecular crystal structures without direct methods. Proc Natl Acad Sci U S A. 2017;114:3637–3641. doi: 10.1073/pnas.1701640114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strop P, Brzustowicz MR, Brunger AT. Ab initio molecular-replacement phasing for symmetric helical membrane proteins. Acta Crystallogr D Biol Crystallogr. 2007;63:188–196. doi: 10.1107/s0907444906045793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids research. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohammadi T, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. The EMBO journal. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopf TA, et al. Mutation effects predicted from sequence co-variation. Nat Biotechnol. 2017;35:128–135. doi: 10.1038/nbt.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovchinnikov S, et al. Large-scale determination of previously unsolved protein structures using evolutionary information. Elife. 2015;4:e09248. doi: 10.7554/eLife.09248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leclercq S, et al. Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Scientific reports. 2017;7:43306. doi: 10.1038/srep43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protocols. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/s0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekeberg M, Lövkvist C, Lan Y, Weigt M, Aurell E. Improved contact prediction in proteins: Using pseudolikelihoods to infer Potts models. Phys Rev E. 2013;87:012707. doi: 10.1103/PhysRevE.87.012707. [DOI] [PubMed] [Google Scholar]
- 28.Balakrishnan S, Kamisetty H, Carbonell JG, Lee S-I, Langmead CJ. Learning generative models for protein fold families. Proteins: Structure, Function, and Bioinformatics. 2011;79:1061–1078. doi: 10.1002/prot.22934. [DOI] [PubMed] [Google Scholar]
- 29.Toth-Petroczy A, et al. Structured States of Disordered Proteins from Genomic Sequences. Cell. 2016;167:158–170.e112. doi: 10.1016/j.cell.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 31.Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucl Acids Res. 2007;35:W429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 33.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Marks DS, Hopf TA, Sander C. Protein structure prediction from sequence variation. Nat Biotech. 2012;30:1072–1080. doi: 10.1038/nbt.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunkoczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr. 2011;67:303–312. doi: 10.1107/s0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarzenbacher R, Godzik A, Grzechnik SK, Jaroszewski L. The importance of alignment accuracy for molecular replacement. Acta Crystallogr D Biol Crystallogr. 2004;60:1229–1236. doi: 10.1107/s0907444904010145. [DOI] [PubMed] [Google Scholar]
- 37.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/s0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/s0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/s0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morin A, et al. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.UniProt: the universal protein knowledgebase. Nucl Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pakseresht N, et al. Assembly information services in the European Nucleotide Archive. Nucl Acids Res. 2014;42:D38–D43. doi: 10.1093/nar/gkt1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hopf TA, et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife. 2014;3 doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic acids research. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Current protocols in protein science. 2016;86:2.9.1–2.9.37. doi: 10.1002/cpps.20. [DOI] [PubMed] [Google Scholar]
- 47.Youngman PJ, Perkins JB, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiao Y, et al. Lipid II overproduction allows direct assay of transpeptidase inhibition by beta-lactams. Nat Chem Biol. 2017;13:793–798. doi: 10.1038/nchembio.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiao Y, et al. Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J Am Chem Soc. 2014;136:14678–14681. doi: 10.1021/ja508147s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure factors and refined atomic coordinates for RodA wild-type and D255A mutant are deposited in the Protein Data Bank under accession codes 6BAR and 6BAS, respectively.