Abstract
The three human RAS genes encode four proteins that play central roles in oncogenesis by acting as binary molecular switches that regulate signaling pathways for growth and differentiation. Each is subject to a set of posttranslational modifications (PTMs) that modify their activity or are required for membrane targeting. The enzymes that catalyze the various PTMs are potential targets for anti-RAS drug discovery. The PTMs of RAS proteins are the focus of this review.
Activating mutations of RAS genes have been implicated in human cancer for more than three decades. Modern cancer genome analysis has confirmed that the three human RAS genes, KRAS, NRAS, and HRAS, are mutated in cancer more frequently than any other oncogene (Forbes et al. 2011; Prior et al. 2012). Thus, the biochemical and cell biological properties of the products of RAS genes are a major focus for cancer biologists who hope to uncover vulnerabilities that can be exploited for drug discovery (Cox et al. 2014).
The three RAS genes give rise to four proteins because transcripts from the KRAS locus are alternately spliced to encode either KRAS4A or KRAS4B, which use alternate fourth exons. Although activating mutations of each RAS gene transforms rodent fibroblasts with similar efficiency (Maher et al. 1995), there is a strong and unexplained isoform difference in driving human cancer with the hierarchy KRAS (85%) >NRAS (12%) >HRAS (3%) (Hobbs et al. 2016). Each of the four RAS proteins is a member of a superfamily of small GTPases that includes the RAS, RHO, RAB, ARF, and RAN families. RAS, like the other members of this superfamily, is a guanine nucleotide-binding protein that functions as a binary molecular switch that is interconverted from an inactive to an active state by exchange of guanosine triphosphate (GTP) for guanosine diphosphate (GDP). GTP binding reorders two regions known as Switch I and Switch II on the surface of RAS, providing the structural basis for the activated state (Vetter and Wittinghofer 2001). The activation portion of the cycle is catalyzed by guanine nucleotide exchange factors (GEFs) that promote loss of GDP such that GTP, 10-fold more abundant in cells, can bind. Several proteins function as GEFs for RAS, including SOS, which transmits the signal from tyrosine kinase growth factor receptors to RAS (Bos et al. 2007). Signaling downstream from RAS is initiated by GTP-bound RAS binding to any of a dozen effectors that harbor RAS-binding domains (RBDs) that bind only GTP-bound RAS. The best-studied RAS effectors are RAF1 and PIK3CA that mediate mitogen-activated protein kinase (MAPK) and AKT/mechanistic target of rapamycin (mTOR) signaling, respectively (Marshall 1996; Cox and Der 2011). The inactivation cycle is mediated by the GTPase activity of RAS and other small GTPases, which hydrolyzes bound GTP to GDP and thereby returns the molecule to the off state. However, the intrinsic GTPase activity of RAS is quite low (Gibbs et al. 1984; Chung et al. 1993) such that an accessory protein, GTPase-activating protein (GAP), is required, which accelerates catalysis up to 105-fold (Ahmadian et al. 1997). Oncogenic mutations of RAS render it insensitive to GAP, allowing the GTPase to accumulate in the on state and sustain signaling (Trahey and McCormick 1987; Scheffzek et al. 1997).
The most straightforward approach to developing RAS inhibitors is to target the RAS protein directly. Efforts to inhibit RAS by interfering with GTP/GDP binding have proven fruitless because of the binding affinity of guanine nucleotides (John et al. 1990), although a thiol-reactive derivative of GTP has shown some in vitro efficacy in oncogenic RAS proteins with G12C mutations that afford a reactive cysteine in the guanine nucleotide-binding pocket (Lim et al. 2014). The crystal structure of RAS, first solved in 1989 (Santos and Nebreda 1989), revealed no pockets other than the guanine nucleotide-binding site that might be targeted by small molecules that could interrupt the switch function of RAS. However, recent, renewed efforts using this approach have yielded some fruit. Shokat and colleagues have exploited a shallow binding pocket under the Switch II region of RAS that is in proximity to the cysteine of G12C mutants (which account for 12% of RAS-driven tumors) and can be targeted with thiol-reactive compounds (Ostrem et al. 2013). These compounds have been shown to lock Switch II in a GDP-bound conformation thus abolishing signaling (Lito et al. 2016; Patricelli et al. 2016). Recently, Stockwell and colleagues reported on a small molecule that binds directly to all RAS proteins and interferes with signaling (Welsch et al. 2017), and Reddy and colleagues characterized rigosertib as a molecule that can bind to RBDs thereby interrupting RAS signaling (Athuluri-Divakar et al. 2016), although another study casts doubt on this mechanism of action (Ritt et al. 2016).
The difficulties in directly targeting RAS have led cancer biologists to look for alternate approaches. Among these have been synthetic lethal screens that have yet to bear fruit (Downward 2015; Wang et al. 2017) and targeting the kinases downstream from RAS, an approach that has been very successful, although has yet to produce durable clinical responses (Samatar and Poulikakos 2014). Another approach has been to target the many posttranslational modifications (PTMs) of RAS, among them those that remodel the carboxyl terminus of RAS proteins and thereby allow RAS to associate with membranes, a feature that is required for signaling. It is the PTMs of RAS proteins that are the subject of this review.
CONSTITUTIVE CARBOXY-TERMINAL MODIFICATIONS: MEMBRANE ASSOCIATION AND TRAFFICKING
CAAX Processing
RAS and RHO family small GTPases are members of a larger family of proteins that end with a CAAX sequence in which C is cysteine, A is usually aliphatic, and X is any amino acid. A CAAX motif signals for sequential modifications by three enzymes (Wright and Philips 2006). Primary translation products that terminate with a CAAX sequence immediately become substrates for one of two cytosolic prenyltransferases, farnesyltransferase (FTase) or geranylgeranyltransferase 1 (GGTase1) that add a 15- or 20-carbon, respectively, polyisoprene lipid via a stable thioether linkage to the sulphydryl group of the cysteine (Fu and Casey 1999). When a CAAX sequence ends in S or M, the protein is a primary substrate for FTase, whereas those CAAX motifs that end in L are modified by GGTase1 (Casey et al. 1989; Seabra et al. 1991). All four RAS proteins are modified by FTase (Fig. 1). Although CAAX prenylation has been considered immediately posttranslational and unregulated, recently, Williams and colleagues have found that RAS binding to splice variants of a promiscuous GEF, SmgGDS, can regulate entry into the CAAX processing pathway (Berg et al. 2010). More recently, regulation of KRAS prenylation via interactions with the noncoding, small nucleolar RNAs, SNORD50A and SNORD50B, was reported. However, regulation of Ras prenylation was shown indirectly using proximity ligation analysis and the identification of a presumptive prenylation-dependent electrophoretic mobility shift via western blotting (Siprashvili et al. 2016).
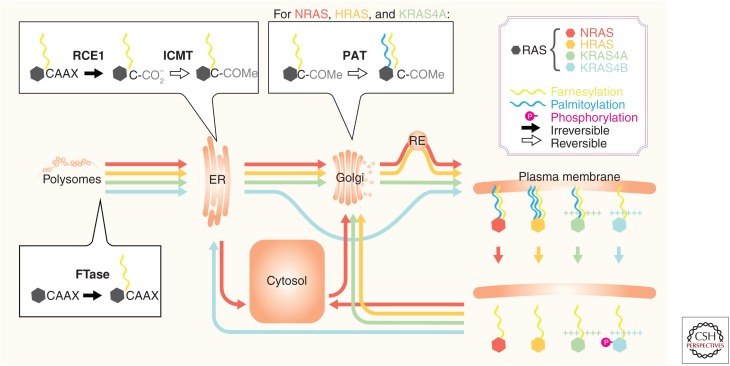
Figure 1.
Differential posttranslational processing regulates RAS isoform subcellular trafficking. The trafficking of RAS isoforms HRAS, NRAS, KRAS4A, and KRAS4B (color coded, see legend) are schematized for the nascent proteins following messenger RNA (mRNA) translation on polysomes in the cytosol. The enzymes that catalyze the posttranslational modifications (PTMs) that direct trafficking are highlighted in dialog boxes (FTase, farnesyltransferase; RCE1, Ras-converting enzyme 1 protease; ICMT, isoprenylcysteine carboxylmethyltransferase; PATs, DHHC family of palmitoyl acyl transferases) with irreversible modifications (farnesylation and proteolysis) indicated by solid arrows, and reversible modifications (carboxyl methylation, palmitoylation, and phosphorylation) indicated by outlined arrows (see legend). Lipid modifications (farnesylation and palmitoylation) are color coded (see legend). The subcellular locations at which these modifications occur (ER, endoplasmic reticulum; Golgi, Golgi apparatus; RE, recycling endosome; cytosol, plasma membrane) are also indicated. Between the indicated subcellular compartments, the color-coded arrows denote both the RAS isoform undergoing transit and the direction of trafficking. For KRAS4A and KRAS4B, (+) signs indicate polybasic residues that confer electrostatic affinity for phospholipid headgroups, which, in the case of KRAS4B, is interrupted by phosphorylation (via protein kinase C [PKC] and GMP-dependent protein kinase 2 [PKG2] as discussed in the text but not depicted here). For NRAS, arrows indicate transit to the cytosol, a compartment in which this isoform accumulates when unpalmitoylated, from both the ER and plasma membrane. Prenyl-dependent chaperone interactions for N/KRAS with PDE6δ or for NRAS with VPS35 are not depicted but are discussed in the text.
Prenylated CAAX sequences become substrates for the second enzyme—RAS converting enzyme 1 (RCE1)—an endoprotease that removes the AAX amino acids converting the prenylcysteine into the new carboxyl terminus (Boyartchuk et al. 1997; Freije et al. 1999; Otto et al. 1999). Carboxy-terminal prenylcysteines then become substrates for the final CAAX-processing enzyme—isoprenylcysteine carboxylmethyltransferase (ICMT)—that methylesterifies the α carboxyl group, thereby neutralizing the negative charge at the carboxyl terminus (Clarke et al. 1988; Gutierrez et al. 1989; Hancock et al. 1991b; Hrycyna et al. 1991; Pillinger et al. 1994). Whereas carboxylmethylation is reversible (although no specific esterase had been identified), prenylation is not and persists through the lifetime of the protein. The end result of CAAX processing is to remodel the hydrophilic carboxyl terminus of a globular hydrophilic protein and render it hydrophobic such that it has affinity for membranes (Fig. 1).
Proteins that are modified only with a prenyl lipid have affinity for the endoplasmic reticulum (ER) (Choy et al. 1999). The basis for this preference is not established but likely relates to the biophysical properties of a prenyl lipid and the ER membrane. Both RCE1 and ICMT are polytopic enzymes that are restricted to the ER with their active sites disposed toward the cytoplasm (Dai et al. 1998; Schmidt et al. 1998; Wright et al. 2009). Thus, the affinity of prenylated proteins for the ER is part and parcel of directing these proteins to the enzymes that will complete CAAX processing.
The elucidation of CAAX processing by 1990 and the demonstration that it is required for RAS transformation of fibroblasts (Willumsen et al. 1984; Bergo et al. 2002, 2004) pointed the way to the first rational design of anti-RAS drugs. FTase, the enzyme that catalyzes the first and rate-limiting step was targeted. Numerous FTase inhibitors (FTIs) were developed that proved relatively nontoxic, could be made orally available, and hit their target in vivo. Some of these went to phase III trials (Appels et al. 2005). Unfortunately, these trials, mostly involving tumors that are driven by mutant KRAS, failed. It was later observed that although under physiologic conditions all RAS proteins are modified by FTase, in the presence of an FTI, KRAS and NRAS but not HRAS were capable of modification by GGTase1, which allowed for full biological function (Whyte et al. 1997). Recently, an interest in FTIs for HRAS-driven tumors has been rekindled and clinical trials for head and neck tumors (ClinicalTrials.gov identifier: NCT02383927) and urothelial carcinoma (ClinicalTrials.gov identifier: NCT02535650) are underway. In principle, dual FTase/GGTase1 inhibitors would be expected to have effective anti-RAS activity but this combination, which should inhibit all RAS and RHO family proteins and some RAB proteins, is predicted to be too toxic. Some preclinical animal studies support this idea (Lobell et al. 2002; Liu et al. 2010) but the possibility of a therapeutic window has not been explored. It is important to keep in mind that FTIs did not fail because blocking RAS association with membranes is a poor strategy, but rather that FTIs did not block KRAS and NRAS association with membranes.
Although RCE1 and ICMT are required for the transformation of rodent fibroblasts (Bergo et al. 2002, 2004), studies of transgenic mice with conditional RCE1 and ICMT alleles have provided conflicting results. Whereas ICMT deficiency ameliorated KRAS-driven myeloproliferative disease (Wahlstrom et al. 2008), RCE1 deficiency exacerbated the disease (Wahlstrom et al. 2007), which is a result difficult to reconcile with the biology of CAAX processing. Surprisingly, when analyzed in the context of the LSL-KRASG12D;PDX1-CRE model of pancreatic adenocarcinoma, ICMT deficiency exacerbated the neoplastic disease (Court et al. 2013), and follow-up studies have implicated CAAX processing of RAB proteins that are required for NOTCH signaling (NOTCH is a tumor suppressor in this model) as the mechanism (Court et al. 2017). Thus, it is clear that the utility of targeting RCE1 or ICMT will be context dependent in cancer.
RCE1 and ICMT were recently reported to be synthetic lethal with oncogenic NRAS in Ba/F3 cells subjected to a genome-wide CRISPR/Cas9 screen (Wang et al. 2017), renewing interest in these postprenylation CAAX-processing enzymes. Like FTase, RCE1 and ICMT have been targeted for anti-RAS drug discovery. RCE1 inhibitors remain under development (Mohammed et al. 2016). Although they have been shown to mislocalize RAS proteins, preclinical evidence for inhibition of tumor growth has not been reported. The most potent ICMT inhibitor described to date also showed a strong effect on membrane association of RAS but relatively weak inhibition of tumor cell proliferation, although, in contrast to previous studies, these investigators found that KRAS-transformed cell lines were exquisitely sensitive to FTIs (Judd et al. 2011). Importantly, Casey and Wang have recently established a requirement for ICMT in human tumor cell lines and showed that an ICMT inhibitor inhibits tumor growth and induces cancer cell death (Lau et al. 2014, 2017; Sun et al. 2016; Manu et al. 2017). Even if therapeutics targeted to RCE1 or ICMT prove ineffective as single agents, the possibility that they might synergize with FTIs or other RAS therapeutics remains a scientifically sound possibility.
Palmitoylation/Depalmitoylation
CAAX processing is necessary but not sufficient for delivery of RAS proteins to the plasma membrane (PM) (Fig. 1). Also required is a so-called “second signal” immediately upstream of the CAAX motif (Hancock et al. 1991a). For KRAS4B, this consists of a polybasic region with eight lysines that allow for an electrostatic interaction with the negatively charged headgroups of phospholipids on the cytoplasmic face of the PM. The second signal for NRAS and HRAS consists of one or two, respectively, additional cysteines immediately upstream of the CAAX cysteine that become modified by palmitic acid (Hancock et al. 1990). KRAS4A is also palmitoylated but it has a hybrid second signal consisting of the palmitate that works in conjunction with a weaker polybasic region (Laude and Prior 2008; Tsai et al. 2015). Whereas the second signal of KRAS4B is constitutive and requires no further PTM, palmitoylation of NRAS, HRAS, and KRAS4A is regulated and reversible and therefore affords another opportunity for therapeutic intervention.
RAS proteins are among hundreds encoded in mammalian genomes that are modified by acylation, which includes both myristoylation and palmitoylation (Veit and Schmidt 2006). The study of protein acylation has exploded during the past 10 years as innovative, nonradioactive methods have been developed for detecting these modifications (Drisdel and Green 2004; Drisdel et al. 2006; Charron et al. 2009; Yang et al. 2010). The combination of acyl-exchange chemistry with alkyne and azido derivatives of fatty acids that permit covalent linkage to affinity tags through click chemistry has permitted exploration of the “palmitoylome,” which has proven to include hundreds and perhaps thousands of gene products (Sanders et al. 2015).
RAS palmitoylation occurs on cysteines at the carboxyl terminus via thioester linkages, referred to S-palmitoylation. For many other signaling molecules, including src family kinases, palmitoylation occurs at the amino terminus often in association with myristoylation of glycine. Many other proteins are palmitoylated at sites remote from the amino or carboxyl terminus, including transmembrane proteins that, by definition, do not require acylation for membrane association (Resh 1999). The protein palmitoylation machinery is comprised of a family of 25 protein acyltransferases (PATs) that each possess a DHHC sequence in their catalytic sites (Mitchell et al. 2006). Unlike other lipid modifications such as prenylation or myristoylation, no consensus signal has emerged that accounts for which surface-exposed cysteines can become substrates for PATs (Rocks et al. 2010). Genetic screens in yeast first identified Erf2 and Erf4 as the heterodimeric enzyme responsible for RAS palmitoylation in Saccharomyces cerevesiae (Lobo et al. 2002). In yeast, Erf2 is the DHHC-containing PAT subunit, whereas Erf4 regulates both the ubiquitin-mediated degradation of Erf2 and the stability of the palmitate-Erf2 catalytic intermediate (Mitchell et al. 2006). The mammalian orthologs of this complex are DDHC9 and GCP16 (Swarthout et al. 2005). Although HRAS is a substrate for DHHC9/GCP16, knockdown of DHHC9 does not block HRAS palmitoylation, suggesting that multiple PATs have activity toward the GTPase (Rocks et al. 2010). Indeed, the substrate promiscuity of PATs has made it difficult to assign enzyme–substrate pairs. Although no PAT has been characterized with regard to a full substrate repertoire, it is clear that each assumes a specific subcellular distribution, predominantly on the Golgi apparatus or PM. DHHC9 is restricted to the Golgi and may account for the localization of NRAS and HRAS on this compartment in addition to the PM (Swarthout et al. 2005). Interestingly, KRAS4A does not accumulate on Golgi, suggesting that this RAS protein is a substrate for another PAT (Tsai et al. 2014).
Upon palmitoylation, RAS acquires a 100-fold greater affinity for membranes than that of the prenylated-only protein (Shahinian and Silvius 1995; Schroeder et al. 1997). This increased affinity creates a kinetic trap that enriches NRAS and HRAS at the cytoplasmic face of Golgi membranes from where these RAS proteins can engage vesicular transport and gain access to other membranes, including the PM (Goodwin et al. 2005). Palmitoylation occurs on Cys 181 of NRAS, Cys 181 and 184 of HRAS, and on Cys 180 of KRAS4A. For both NRAS and HRAS, palmitoylation at Cys 181 directs sorting from the Golgi to recycling endosomes, and this is necessary for subsequent trafficking on the cytoplasmic face of exocytic vesicles destined for the PM (Choy et al. 1999; Apolloni et al. 2000). In contrast, palmitoylation of Cys 184 on HRAS is not absolutely required for delivery to the PM (Roy et al. 2005). Similarly, although palmitoylation of Cys 180 on KRAS4A dramatically increases the efficiency of PM targeting, it is not required because the polybasic regions can function without acylation (Tsai et al. 2015).
The requirement for palmitoylation in Ras-driven tumors has been tested in models of NRAS-driven leukemia, in which palmitoylation of NRASG12D was necessary for leukemogenesis (Cuiffo and Ren 2010; Liu et al. 2016). A similar approach to investigate palmitoylation of KRAS4AG12D in leukemogenesis revealed that even in the absence of palmitoylation, the polybasic region cooperates sufficiently with prenylation to promote disease, albeit with a notable delay in disease onset (Zhao et al. 2015).
In addition to prenylation and palmitoylation, the distribution of RAS molecules at the PM and the Golgi is also regulated by depalmitoylation. Although the thioester linkage of protein S-acylation is labile at physiologic pH, suggesting that at least some depalmitoylation could be noncatalytic, current evidence suggests that palmitoylation is reversed in a regulated manner by cellular thioesterases. The first nonlysosomal proteins found to have thioesterase activity toward RAS were acylprotein thioesterase 1 and 2 (APT1 and 2) (Duncan and Gilman 1998; Tomatis et al. 2010; Rusch et al. 2011). Studies of APT1/2 have been aided by the development of inhibitors of thioesterases, including palmostatin B and M (Dekker et al. 2010; Dekker and Hedberg 2011; Hedberg et al. 2011). However, these compounds have proven to possess broad inhibitory activity against serine hydrolases and cannot be used alone to implicate APT1. Some doubt exists as to whether RAS proteins are physiologic substrates for APT1/2. The yeast ortholog of APT1 has activity toward Giα1 but not RAS (Duncan and Gilman 2002). Knockdown or inhibition of APT1/2 blocks depalmitoylation of Huntingtin but not NRAS or PSD95 (Lin and Conibear 2015).
APT1 and 2 are serine hydrolases that are themselves palmitoylated (Kong et al. 2013) and this PTM affords some affinity for membranes, suggesting a mechanism whereby the esterase colocalizes with its substrates. However, it is the depalmitoylated, cytosolic pool of APT1 that manifests depalmitoylating activity (Vartak et al. 2014). Recently, activity profiling using palmostatin B identified the ABHD17 family of serine hydrolases as physiologically important thioesterases. Unlike APT1 or 2, ABHD17a shows robust NRAS depalmitoylating activity (Lin and Conibear 2015). Like APT1/2, ABHD17A is also palmitoylated (Kang et al. 2008; Martin and Cravatt 2009), but unlike APT1, this modification is required for both its membrane association and NRAS esterase activity. The full repertoire, specificity, and activity of cellular thioesterases that depalmitoylate RAS remains largely uncharacterized (Tabaczar et al. 2017). Nonetheless, disruption of the acylation cycle using inhibitors of depalmitoylation has been shown to limit cytokine-independent colony formation of NrasG12D but not Kras4BG12D, expressing fetal liver and bone marrow cells from mice (Xu et al. 2012).
To establish a physiologic role for the RAS palmitoylation/depalmitoylation cycle and to study the effect of it perturbation one must accurately measure the t1/2 of palmitate on RAS proteins in living cells and tissues. However, accurate measurement of the palmitate half-life on endogenous RAS has been limited by the poor sensitivity of detecting radiolabeled [3H] palmitoyl modification (Tsai et al. 2014). Iodinated derivatives of palmitate have been used to enhance sensitivity (Liang et al. 2001), but these reagents are not commercially available. Most reports have used a classical pulse-chase methodology with a relatively long (up to 1 h) pulse followed by a chase in the presence of cycloheximide to inhibit new protein synthesis (Lu and Hofmann 1995; Baker et al. 2000, 2003). This creates two problems. First, a long “pulse” leads to equilibration of cellular pools of palmitate, palmitoyl-CoA, and palmitoyl proteins. Because [3H]palmitoyl-CoA can be generated from both [3H]palmitate and [3H]palmitoyl proteins, it is not feasible to abruptly shut down [3H]palmitoylation by adding a great excess of unlabeled palmitate to the medium. Second, we have shown that cycloheximide markedly increases HRAS palmitoylation because it inhibits FKBP12, a prolyl isomerase that acts on a carboxy-terminal proline of HRAS and thereby promotes depalmitoylation (Ahearn et al. 2011). Baker et al. reported a t1/2 of 2.4 h of palmitate on HRAS in cells “pulsed” for 3 h in the presence of cycloheximide (Baker et al. 2000, 2003). Subsequently, Lu and Hofmann (1995) measured a t1/2 for HRAS of 90 min using a 30 min pulse and cycloheximide. Interestingly, singly palmitoylated HRAS mutants had a dramatically reduced t1/2 of 20 min. Although dual palmitoylation of HRAS could account for the discrepancy, it is more likely that a relatively long pulse and addition of cycloheximide increased the observed t1/2 because a subsequent study with a 5 min pulse without cycloheximide found the t1/2 of palmitate on HRAS to be less than 5 min (Ahearn et al. 2011). Using a pulse of 5 min without cycloheximide, Magee et al. reported that the t1/2 of palmitate on singly palmitoyated NRAS was 20 min (Magee et al. 1987). Fluorescence recovery after photobleaching studies of NRAS transport to and from the Golgi, a process regulated by the palmitotylation/depalmitlylation cycle, are consistent with a rapid turnover of seconds to a few minutes (Goodwin et al. 2005; Rocks et al. 2005). Thus, the kinetics of turnover of palmitate on RAS proteins is uncertain but is likely to be measured in a few minutes, whereas the t1/2 of the proteins is on the order of 24 h. It is unclear whether the newer techniques of measuring protein palmitoylation by click chemistry and acyl-exchange chemistry will improve the accuracy of measuring palmitate turnover, and there is reason to suspect that the additional steps and manipulations involved may complicate the analysis. Using the 17-ODYA palmitate analog and click chemistry, Lin and Conibear (2015) reported a t1/2 for the palmitate on NRAS of <0.5 h.
It is clear that the hypervariable regions (HVRs) of RAS proteins and the PTMs thereof encode information with regard to targeting to phospholipid bilayers and trafficking within the cell. The palmitate modification has long been known to promote association of both peripheral and intrinsic membrane proteins, such as HRAS and LAT, respectively, to liquid-ordered membrane domains, in the past referred to as lipid rafts (Webb et al. 2000; Liang et al. 2001; Lorent and Levental 2015). More recently, the “code” of the KRAS4B polybasic HVR has been read. Using gold particle labeling and electron microscopy spatial mapping, Zhou et al. have described how anionic lipid sorting of KRAS4B is encoded in combination by prenylation, the polybasic stretch of lysines, and by tertiary conformations revealed by molecular dynamic simulations (Zhou et al. 2017). In this model, these chemical and structural codes promote partitioning to PtdSer or PtdInositol bisphosphate-enriched domains (Maher et al. 1995; Cox et al. 2014). In turn, nanoclustering and differential effector pathway activation is established. The investigators also show how phosphorylation at serine 181 of KRAS4B (see below) operates to regulate this partitioning.
CONDITIONAL POSTTRANSLATIONAL MODIFICATIONS
Phosphorylation of KRAS4B and the Farnesyl Electrostatic Switch
Although KRAS4B requires no modification beyond CAAX processing to associate with membranes, the strength of the interaction is regulated by PTM. Phosphorylation of serine 181 within the polybasic region can diminish membrane affinity by partially neutralizing the positive charge (Fig. 1). Although phosphorylation of serine 181 does not completely inhibit association with the PM, the distribution of the GTPase between PM and the endomembrane is shifted in favor of the endomembrane (Bivona et al. 2006). In this way, the prenylated carboxyl terminus of KRAS4B mimics the myristoylated amino terminus of the MARKS protein that also requires a polybasic region in conjunction with the acyl lipid to effect membrane binding and harbors serines within the polybasic region that, when phosphorylated by protein kinase C (PKC), reverses membrane binding through a mechanism known as a myristoyl electrostatic switch (McLaughlin and Aderem 1995). By analogy, KRAS4B harbors a farnesyl electrostatic switch. Serine 181 of KRAS4B is a substrate for both PKCs (Bivona et al. 2006; Alvarez-Moya et al. 2010) and cyclic GMP-dependent protein kinase 2 (PKG2) (Cho et al. 2016). PKG2 is activated to phosphorylate KRAS4B downstream from AMP-activated protein kinase (AMPK) and requires endothelial nitric oxide synthase and soluble guanylyl cyclase. Phosphorylation of KRAS4B on serine 181 regulates association of the HVR with membrane microdomains (Barceló et al. 2013; Jang et al. 2015). Bryostatin, a potent agonist of PKC, was found to inhibit the growth of oncogenic KRAS4B-driven xenografts in an S181-dependent fashion, suggesting that phosphorylation is a negative regulatory event in tumor maintenance (Mohammad et al. 1998; Bivona et al. 2006; Kollar et al. 2014). Consistent with this observation, oncogenic KRAS4B with a phosphomimetic group at position 181, KRAS12V181E, was found to diminish cell survival (Bivona et al. 2006). One mechanism for decreased survival is the ability of KRAS12V181E to interfere with one of the prosurvival actions of Bcl-XL (Sung et al. 2013). A constant low level of calcium flux from ER stores to mitochondria is required for metabolic health and suppression of autophagy (White et al. 2005; Cárdenas et al. 2010). This calcium flux is delivered through inositol-tris-phosphate (IP3) receptors (IP3Rs) and their IP3-stimulated conductance is potentiated by Bcl-XL, thus accounting for one of the prosurvival actions of Bcl-XL. KRAS12V181E blocks this action of Bcl-XL (Sung et al. 2013). Wang et al. (2015) recently showed that phosphorylation of KRAS4B on serine 181 inhibits tumor initiation by blocking interaction with calmodulin and thereby abrogating suppression of noncanonical Wnt signaling. Thus, there are multiple independent lines of evidence suggesting that phosphorylation of KRAS4B is a negative regulatory event with regard to oncogenesis. One contrary report in the literature suggests that phosphorylation of KRAS4B on S181 is required for transformed rodent fibroblasts and colon cancer cell lines to grow subcutaneously in nude mice (Barceló et al. 2014). However, we have observed that LSL-KRAS12D181A double knockin mice that harbor an inducible KRAS4B oncogene that cannot be phosphorylated produce the same number of lung tumors on tracheal administration of adeno-Cre than do LSL-KRAS12D181S mice (unpubl.), suggesting that in vivo phosphorylation of S181 is not required for oncogenesis.
Ubiquitination and Sumoylation
Despite intensive study of RAS proteins for three decades, little is known about their catabolism. HRAS has been shown to be polyubiquitinated when co-overexpressed with FLAG-Ub (Jeong et al. 2012). The E3 ligase was identified as β-TrCP (Kim et al. 2009). Zeng et al. reported interactions of H,N and KRAS4B with the E3 ligase Nedd4-1 and showed polyubiquitination of KRAS4B on Lys5 using HA-Ub. Signaling via growth factor receptors and oncogenic RAS both impaired Nedd4-1-mediated RAS degradation and in turn enhanced Nedd4-1-mediated PTEN degradation to drive cancer growth (Zeng et al. 2014). Mono- and diubiquitination of RAS proteins is also established. HRAS, NRAS, and KRAS4B have all been shown to be either mono- or diubiquitinated (Jura et al. 2006; Sasaki et al. 2010). These modifications were first reported on NRAS and HRAS and modification of the latter promoted association with endosomes. The E3 ligase for HRAS was identified as RABEX5 (Xu et al. 2010), a protein that also has exchange activity for RAB5. Interestingly, the RAS effector RIN1 was required for the ubiquitination reaction. Later mass spectrometry analysis identified Lys147 on both HRAS and KRAS as a site of mono-/diubiquitination (Sasaki et al. 2010). The same study identified Lys117 and Lys170 as additional sites on HRAS and Campbell’s group added Lys104 of KRAS as an additional site (Baker et al. 2013). Importantly, Sasaki et al. (2010) reported that mono- and diubiquitination of KRAS on Lys 147 enhanced GTP loading and interaction with effectors, and Baker et al. (2013) reported that mono- or diubiquitination of KRAS on Lys117 accelerated nucleotide exchange through a mechanism distinct from that of modified Lys147. More recently, we found that HRAS, NRAS, and KRAS4B are modified on Lys42 by the ubiquitin-like protein SUMO3 (BH Choi, C Chen, M Philips, and W Dai, unpubl.). We have identified the E3 ligase as PIASγ and found that sumoylation promotes MAPK signaling and tumor progression. The weakness of all of these studies is that the stoichiometry of modification has not been determined and the physiologic significance is uncertain.
Nitrosylation
The concept of redox regulation of cellular signaling prompted investigation into direct modification of RAS proteins by reactive oxygen species. The most surface-exposed cysteine, Cy118, conserved in all RAS isoforms, was shown to be nitrosylated (Lander et al. 1995a). Two mechanisms have been described for this modification, one involving direct modification with •NO2, a product of NO and O2, and one involving a glutathionyl radical intermediate (Lander et al. 1996; Heo and Campbell 2004). S-nitrosylation of RAS on cys118 has been shown to increase GDP/GTP exchange (Lander et al. 1995b; Williams et al. 2003) and has been implicated in breast cancer in which nitric oxide synthase 2 has been shown to be up-regulated (Switzer et al. 2012). Most compelling are studies by Counter and colleagues who showed that activation of eNOS by AKT is required for xenograft initiation and maintenance, a pathway mediated by S-nitrosylation of wild-type RAS in human tumor cells harboring an oncogenic RAS allele (Lim et al. 2008). Interestingly, eNOS is palmitoylated, highlighting how nistrosylation of RAS may be partially regulated by compartmentalization (Garcia-Cardena et al. 1996).
Acetylation
KRAS can be acetylated on Lys104, which acts as a negative regulatory modification by interfering with GEF-catalyzed nucleotide exchange and thereby reducing transforming capacity (Yang et al. 2012). Acetylation of Lys104 of KRAS is reversed by HDAC6 or SIRT2 (Yang et al. 2013). Thus, among the antineoplastic activities of inhibitors of these deacetylating enzymes may be removal of the acetyl group from KRAS.
Bacterial Toxins and Exoenzymes
Pathogenic bacteria have evolved a plethora of virulence factors, many of which are enzymes that modify signaling molecules, including members of the RAS superfamily of small GTPases (Aktories 2011). Although the majority of these target RHO family proteins, some are active against RAS. Exoenzyme S (ExoS) is an ADP-ribosyl transferase produced by Pseudomonas aeruginosa and injected directly into eukaryotic cells by type III secretion. ExoS ADP ribosylates Arg41 and Arg 128 on RAS, which attenuates GTP loading and inhibits interaction with effectors (Ganesan et al. 1999). Lethal toxin in Clostridium sordeli is a monoglucosyltranferase closely related to toxin B in Clostridium difficile, which uses UDP-glucose to modify Thr35 and thereby inhibits MAPK signaling (Just et al. 1996; Herrmann et al. 1998).
CONSEQUENCES OF RAS PTMs
PTMs of RAS proteins can be divided into those that modify amino acids in the guanine nucleotide binding (G) domain and those that modify the carboxy-terminal HVR (Fig. 2). Not surprisingly, modifications of the G domain (mono- or diubiquitination, sumoylation, acetylation, glucosylation, and nitrosylation) are conditional and modulate GDP/GTP exchange and engagement of effectors. One exception to this rule is mono- and diubiquitination of HRAS that promotes trafficking to endosomes (Jura et al. 2006). Ubiquitination and sumoylation should markedly alter the electrophoretic mobility of RAS proteins. The fact that immunodetectable endogenous cellular RAS proteins run at 21 kDa suggests that the stoichiometry of these modifications is relatively low. The stoichiometries of RAS acetylation and nitrosylation are unknown but mass spectrometry analysis suggests that they too are relatively low (Lander et al. 1997; Yang et al. 2012). Accordingly, although small pools of modified RAS might have significant biological consequences, it is not clear what are the physiologic roles of these modifications in the biology of RAS. Clarification of this issue is of great importance in that the enzymes that catalyze the PTMs become rational targets for anti-RAS drug discovery once a physiologic role is established.
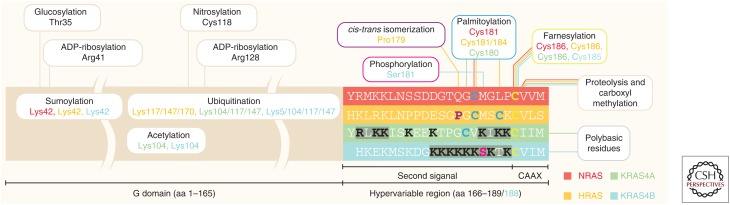
Figure 2.
Posttranslational modifications (PTMs) of RAS isoforms. The full length of RAS is depicted schematically and includes partitioning of the G domain (aa1–165) and the hypervariable region (HVR) (aa166–188/189). Color coding of the HVR distinguishes each RAS isoform (HRAS, NRAS, KRAS4A, and KRAS4B). PTMs occurring within the G domain are indicated (glucosylation, sumolyation, ADP-ribosylation, acetylation, ubiquitination, and nitrosylation). For the HVR, unique color coding highlights residues modified by the various PTMs (phosphorylation, cis-trans [prolyl] isomerization, palmitoylation, farnesylation, proteolysis, and carboxyl methylation). Polybasic residues in the HVRs of KRAS4A and KRAS4B are indicated by black text. HVR modifications are correlated to the RAS isoforms on which they occur via isoform color-coded linkers, and also via the isoform color-coded text for the modified amino acids listed within the modifications dialog boxes.
In contrast to modifications of the G domain, the PTMs of the CaaX sequence (prenylation, proteolysis, and carboxyl methylation) are constitutive and immediately posttranslational such that the stoichiometry is high and the vast majority of endogenous RAS proteins are fully processed. Whereas CaaX processing is irreversible, palmitoylation of NRAS, HRAS, and KRAS4A is dynamic and reversible (Bijlmakers and Marsh 2003). Although it appears that initial palmitoylation is constitutive in that it occurs immediately posttranslationally and stoichiometrically, sending a first wave of nascent RAS to the PM, the lifetimes of palmitoylated RAS proteins are characterized by many cycles of palmitoylation/depalmitoylation. Likewise, phosphorylation of KRAS4B on ser181 is reversible and the stoichiometry is unknown.
It is clear that the carboxy-terminal PTMs control affinity for membranes in general and trafficking of the proteins between cellular compartments. For example, whereas palmitoylated HRAS is efficiently targeted to the PM, a depalmitoylated pool accumulates on the ER and Golgi (Choy et al. 1999). In addition, the addition and subtraction of palmitate influences the microdomains into which RAS proteins partition (Plowman and Hancock 2005). This realization has given rise to the idea of compartmentalized signaling of RAS, whereby one set of pathways might be engaged most efficiently form one intracellular compartment and another set from another location. Compartmentalized signaling increases the complexity of RAS signaling and greatly increases the repertoire of outcomes, a feature consistent with the requirement for RAS signaling in a very large number of cellular processes that must be independently regulated (Fehrenbacher et al. 2009). Differential signaling outcomes of stringent, artificial targeting of activated RAS proteins to different cellular compartments supports this idea (Chiu et al. 2002) as does evidence for compartmentalized RAS signaling in Schizosaccharomyces pombe (Onken et al. 2006). Perhaps the most compelling example of compartmentalized RAS signaling is in lymphocyte development in which RAS signaling is required for both positive and negative selection of nascent lymphocytes in the thymus, two diametrically opposed cellular outcomes (proliferation vs. apoptosis). Palmer and colleagues showed that whereas negative selection was associated with RAS/MAPK signaling from the PM, positive selection was associated with signaling from internal membranes (Daniels et al. 2006).
The carboxy terminal PTMs of RAS control affinity not only for membranes and membrane microdomains but also for cytosolic prenyl-binding proteins. RHO family small GTPases undergo the same PTMs at their carboxyl termini as do RAS proteins. Because they are modified by a 20-carbon geranylgeranyl polyisoprene lipid rather than the 15-carbon farnesyl lipid of RAS proteins, they have even greater intrinsic affinity for membranes. Nevertheless, the bulk of endogenous RAC, RHO, and CDC42 are present in the aqueous environment of the cytosol (Michaelson et al. 2001). This phenomenon is a consequence of a prenyl-binding protein knows as RhoGDI. RhoGDI sequesters the geranylgeranyl moiety of RHO proteins and allows delivery of the GTPase to the relevant membrane comportment on which signaling takes place (Hoffman et al. 2000). The subcellular trafficking of RAB GTPases is accomplished in the same manner through the action of RabGDI (Ullrich et al. 1993). Importantly, carboxylmethylation of RAB proteins has been shown to inhibit binding to RabGDI (Leung et al. 2007), demonstrating how PTMs can regulate the interactions with prenyl-binding proteins.
This ability of GDP dissociation inhibitors (GDIs) to regulate RHO and RAB proteins has caused RAS biologists to explore the possibility that cytosolic farnesyl-binding proteins are required for trafficking of RAS. Several such proteins have been reported. We reported that the vast majority of endogenous, farnesylated NRAS resides in the cytosol in a high molecular weight pool and that at least a portion of that pool includes NRAS bound to VPS35, a component of the coat of the retromer (Zhou et al. 2016). Other proteins, including PRA1 (Figueroa et al. 2001) and SmgGDS (Berg et al. 2010), have been described as prenyl-binding proteins capable of binding RAS. However, the prenyl-binding protein that has attracted the most attention is the δ subunit of phosphodiesterase type 6 (PDE6δ). PDE6δ was first described in photoreceptors as a component of the PDE6 complex that bound the prenylated α and β subunits (Cook et al. 2000; Zhang et al. 2004). The crystal structure of PDE6δ revealed structural homology with RhoGDI with a prenyl-binding grove between β sheets (Hanzal-Bayer et al. 2002). PDE6δ is a promiscuous carrier protein accepting a wide variety of prenylated proteins as cargo. In an elegant series of studies, Wittinghoffer and colleagues showed that active ARL2/3, a nonprenylated small GTPase, acts as a releasing factor for farnesylated RHEB bound to PDE6δ (Ismail et al. 2011). Overexpression of PDE6δ can extract green fluorescent protein (GFP)-tagged NRAS and KRAS4B, but not HRAS or KRAS4A, from the PM (Nancy et al. 2002; Tsai et al. 2015). Bastiaens and colleagues have proposed that PDE6δ is absolutely required for proper trafficking of KRAS4B (Chandra et al. 2012; Schmick et al. 2014), a conclusion at odds with the fact that, whereas KRAS is essential for mouse development (Johnson et al. 1997), PDE6δ null mice are small but otherwise normal (Zhang et al. 2007). Nevertheless, the importance of PDE6δ in KRAS biology was confirmed by Waldmann’s group that developed a series of inhibitors of RAS binding to PDE6δ (e.g., deltarasin), and showed that these compounds inhibit the growth of KRAS-dependent pancreatic adenocarcinoma cells (Zimmermann et al. 2013, 2014). A recent crystal structure of PDE6δ in complex with KRAS4B reveals that binding requires, in addition to the protein–lipid interaction that can accommodate either a farnesyl or geranylgeranyl lipid, some protein–protein interaction with the HVR of KRAS4B (Dharmaiah et al. 2016). Moreover, this study revealed that carboxylmethylation of KRAS4B was required for efficient binding to PDE6δ. The latter observation makes sense because the prenylcysteine enters the hydrophobic pocket of PDE6δ but is surprisingly contrary to the results with RAB proteins and RabGDI in which carboxylmethylation inhibits the interaction. Either way, these are examples of PTMs regulating interactions of small GTPases not only with membranes but also with cytosolic chaperones.
CONCLUSION
Because RAS mutations are so frequently drivers in human cancer, there is a great desire to develop direct inhibitors of the small GTPases that are the products of the RAS oncogenes. Because RAS proteins bind guanine nucleotides with picomolar affinity inhibitors of GTP binding are not practical, other approaches have been sought. RAS proteins are substrates for a wide variety of PTMs discussed above. Because RAS PTMs are catalyzed by specific enzymes, each one represents a potential target for anticancer drug discovery. The G domain of RAS proteins is modified by mono- and diubiquitination, sumoylation, acetylation, and nitrosylation, each of which is reversible and, in principle, susceptible to pharmacological manipulation. However, it is not clear what is the physiologic role of these modifications in RAS signaling, either downstream from growth factor receptors or as a consequence of oncogenic mutation. In contrast, the constitutive PTMs of the carboxy-terminal HVRs of RAS proteins that regulate association with membrane and prenyl binding proteins are clearly essential for RAS protein function and remain rational targets for drug discovery.
Footnotes
Editors: Linda VanAelst, Julian Downward, and Frank McCormick
Additional Perspectives on Ras and Cancer in the 21st Century available at www.perspectivesinmedicine.org
REFERENCES
- Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M, Fehrenbacher N, Linder ME, Philips MR. 2011. FKBP12 binds to acylated H-Ras and promotes depalmitoylation. Mol Cell 41: 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. 1997. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686–689. [DOI] [PubMed] [Google Scholar]
- Aktories K. 2011. Bacterial protein toxins that modify host regulatory GTPases. Nat Rev Microbiol 9: 487–498. [DOI] [PubMed] [Google Scholar]
- Alvarez-Moya B, Lopez-Alcala C, Drosten M, Bachs O, Agell N. 2010. K-Ras4B phosphorylation at Ser181 is inhibited by calmodulin and modulates K-Ras activity and function. Oncogene 29: 5911–5922. [DOI] [PubMed] [Google Scholar]
- Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. 2000. H-Ras but not K-Ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol 20: 2475–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appels NM, Beijnen JH, Schellens JH. 2005. Development of farnesyl transferase inhibitors: A review. Oncologist 10: 565–578. [DOI] [PubMed] [Google Scholar]
- Athuluri-Divakar SK, Vasquez-Del Carpio R, Dutta K, Baker SJ, Cosenza SC, Basu I, Gupta YK, Reddy MV, Ueno L, Hart JR, et al. 2016. A small molecule RAS-mimetic disrupts RAS association with effector proteins to block signaling. Cell 165: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TL, Booden MA, Buss JE. 2000. S-Nitrosocysteine increases palmitate turnover on Ha-Ras in NIH 3T3 cells. J Biol Chem 275: 22037–22047. [DOI] [PubMed] [Google Scholar]
- Baker TL, Zheng H, Walker J, Coloff JL, Buss JE. 2003. Distinct rates of palmitate turnover on membrane-bound cellular and oncogenic H-Ras. J Biol Chem 278: 19292–19300. [DOI] [PubMed] [Google Scholar]
- Baker R, Wilkerson EM, Sumita K, Isom DG, Sasaki AT, Dohlman HG, Campbell SL. 2013. Differences in the regulation of K-Ras and H-Ras isoforms by monoubiquitination. J Biol Chem 288: 36856–36862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceló C, Paco N, Beckett AJ, Alvarez-Moya B, Garrido E, Gelabert M, Tebar F, Jaumot M, Prior I, Agell N. 2013. Oncogenic K-Ras segregates at spatially distinct plasma membrane signaling platforms according to its phosphorylation status. J Cell Sci 126: 4553–4559. [DOI] [PubMed] [Google Scholar]
- Barceló C, Paco N, Morell M, Alvarez-Moya B, Bota-Rabassedas N, Jaumot M, Vilardell F, Capella G, Agell N. 2014. Phosphorylation at Ser-181 of oncogenic KRAS is required for tumor growth. Cancer Res 74: 1190–1199. [DOI] [PubMed] [Google Scholar]
- Berg TJ, Gastonguay AJ, Lorimer EL, Kuhnmuench JR, Li R, Fields AP, Williams CL. 2010. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J Biol Chem 285: 35255–35266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Ambroziak P, Gregory C, George A, Otto JC, Kim E, Nagasae H, Casey PJ, Balmain A, Young SG. 2002. Absence of the CAAX endoprotease Rce1: Effects on cell growth and transformation. Mol Cell Biol 22: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Gavino BJ, Hong C, Beigneux AP, McMahon M, Casey PJ, Young SG. 2004. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J Clin Invest 113: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlmakers MJ, Marsh M. 2003. The on–off story of protein palmitoylation. Trends Cell Biol 13: 32–42. [DOI] [PubMed] [Google Scholar]
- Bivona TG, Quatela SE, Bodemann BO, Ahearn IO, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, et al. 2006. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell 21: 481–493. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. 2007. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 129: 865–877. [DOI] [PubMed] [Google Scholar]
- Boyartchuk VL, Ashby MN, Rine J. 1997. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275: 1796–1800. [DOI] [PubMed] [Google Scholar]
- Cárdenas C, Miller RA, Smith I, Bui T, Molgó J, Müller M, Vais H, Cheung KH, Yang J, Parker I, et al. 2010. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 142: 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey PJ, Solski PA, Der CJ, Buss JE. 1989. p21ras is modified by a farnesyl isoprenoid. Proc Natl Acad Sci 86: 8323–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, et al. 2012. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol 14: 148–158. [DOI] [PubMed] [Google Scholar]
- Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. 2009. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc 131: 4967–4975. [DOI] [PubMed] [Google Scholar]
- Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL II, Cox AD, Philips MR. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol 4: 343–350. [DOI] [PubMed] [Google Scholar]
- Cho KJ, Casteel DE, Prakash P, Tan L, van der Hoeven D, Salim AA, Kim C, Capon RJ, Lacey E, Cunha SR, et al. 2016. AMPK and endothelial nitric oxide synthase signaling regulates K-Ras plasma membrane interactions via cyclic GMP-dependent protein kinase 2. Mol Cell Biol 36: 3086–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. 1999. Endomembrane trafficking of Ras: The CAAX motif targets proteins to the ER and Golgi. Cell 98: 69–80. [DOI] [PubMed] [Google Scholar]
- Chung HH, Benson DR, Cornish VW, Schultz PG. 1993. Probing the role of loop 2 in Ras function with unnatural amino acids. Proc Natl Acad Sci 90: 10145–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S, Vogel JP, Deschenes RJ, Stock J. 1988. Posttranslational modification of the Ha-Ras oncogene protein: Evidence for a third class of protein carboxyl methyltransferases. Proc Natl Acad Sci 85: 4643–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook TA, Ghomashchi F, Gelb MH, Florio SK, Beavo JA. 2000. Binding of the δ subunit to rod phosphodiesterase catalytic subunits requires methylated, prenylated C-termini of the catalytic subunits. Biochemistry 39: 13516–13523. [DOI] [PubMed] [Google Scholar]
- Court H, Amoyel M, Hackman M, Lee KE, Xu R, Miller G, Bar-Sagi D, Bach EA, Bergö MO, Philips MR. 2013. Isoprenylcysteine carboxylmethyltransferase deficiency exacerbates KRAS-driven pancreatic neoplasia via Notch suppression. J Clin Invest 123: 4681–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court H, Ahearn IM, Amoyel M, Bach E, Philips MR. 2017. Regulation of NOTCH signaling by RAB7 and RAB8 require carboxyl methylation by ICMT. J Cell Biol DOI: 10.1083/jcb.201701053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ. 2011. Ras history: The saga continues. Small GTPases 1: 2–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. 2014. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 13: 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuiffo B, Ren R. 2010. Palmitoylation of oncogenic NRAS is essential for leukemogenesis. Blood 115: 3598–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR. 1998. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem 273: 15030–15034. [DOI] [PubMed] [Google Scholar]
- Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Holländer GA, Gascoigne NR, Palmer E. 2006. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444: 724–729. [DOI] [PubMed] [Google Scholar]
- Dekker FJ, Hedberg C. 2011. Small molecule inhibition of protein depalmitoylation as a new approach towards downregulation of oncogenic Ras signalling. Bioorg Med Chem 19: 1376–1380. [DOI] [PubMed] [Google Scholar]
- Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Schölermann B, et al. 2010. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol 6: 449–456. [DOI] [PubMed] [Google Scholar]
- Dharmaiah S, Bindu L, Tran TH, Gillette WK, Frank PH, Ghirlando R, Nissley DV, Esposito D, McCormick F, Stephen AG, et al. 2016. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc Natl Acad Sci 113: E6766–E6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. 2015. RAS synthetic lethal screens revisited: Still seeking the elusive prize? Clin Cancer Res 21: 1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisdel RC, Green WN. 2004. Labeling and quantifying sites of protein palmitoylation. BioTechniques 36: 276–285. [DOI] [PubMed] [Google Scholar]
- Drisdel RC, Alexander JK, Sayeed A, Green WN. 2006. Assays of protein palmitoylation. Methods 40: 127–134. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Gilman AG. 1998. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein α subunits and p21RAS. J Biol Chem 273: 15830–15837. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Gilman AG. 2002. Characterization of Saccharomyces cerevisiae acyl-protein thioesterase 1, the enzyme responsible for G protein α subunit deacylation in vivo. J Biol Chem 277: 31740–31752. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher N, Bar-Sagi D, Philips M. 2009. Ras/MAPK signaling from endomembranes. Mol Oncol 3: 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa C, Taylor J, Vojtek AB. 2001. Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J Biol Chem 276: 28219–28225. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. 2011. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39(Database issue): D945–D950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije JM, Blay P, Pendás AM, Cadiñanos J, Crespo P, López-Otín C. 1999. Identification and chromosomal location of two human genes encoding enzymes potentially involved in proteolytic maturation of farnesylated proteins. Genomics 58: 270–280. [DOI] [PubMed] [Google Scholar]
- Fu HW, Casey PJ. 1999. Enzymology and biology of CaaX protein prenylation. Recent Prog Horm Res 54: 315–342; discussion 42–43. [PubMed] [Google Scholar]
- Ganesan AK, Vincent TS, Olson JC, Barbieri JT. 1999. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange. J Biol Chem 274: 21823–21829. [DOI] [PubMed] [Google Scholar]
- Garcia-Cardena G, Oh P, Liu J, Schnitzer JE, Sessa WC. 1996. Targeting of nitric oxide synthase to endothelial cell caveolae via palmitoylation: Implications for nitric oxide signaling. Proc Natl Acad Sci 93: 6448–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JB, Sigal IS, Poe M, Scolnick EM. 1984. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci 81: 5704–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK. 2005. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol 170: 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Magee AI, Marshall CJ, Hancock JF. 1989. Post-translational processing of p21ras is two-step and involves carboxyl-methylation and carboxyl terminal proteolysis. EMBO J 8: 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, Marshall CJ. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to loacalize p21ras to the plasma membrane. Cell 63: 133–139. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Cadwallader K, Paterson H, Marshall CJ. 1991a. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J 10: 4033–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Cadwallader K, Marshall CJ. 1991b. Methylation and proteolysis are essential for efficient membrane binding of prenylated p21K-ras(B). EMBO J 10: 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. 2002. The complex of Arl2-GTP and PDEδ: From structure to function. EMBO J 21: 2095–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg C, Dekker FJ, Rusch M, Renner S, Wetzel S, Vartak N, Gerding-Reimers C, Bon RS, Bastiaens PI, Waldmann H. 2011. Development of highly potent inhibitors of the Ras-targeting human acyl protein thioesterases based on substrate similarity design. Angew Chem Int Ed Engl 50: 9832–9837. [DOI] [PubMed] [Google Scholar]
- Heo J, Campbell SL. 2004. Mechanism of p21Ras S-nitrosylation and kinetics of nitric oxide-mediated guanine nucleotide exchange. Biochemistry 43: 2314–2322. [DOI] [PubMed] [Google Scholar]
- Herrmann C, Ahmadian MR, Hofmann F, Just I. 1998. Functional consequences of monoglucosylation of Ha-Ras at effector domain amino acid threonine 35. J Biol Chem 273: 16134–16139. [DOI] [PubMed] [Google Scholar]
- Hobbs GA, Der CJ, Rossman KL. 2016. RAS isoforms and mutations in cancer at a glance. J Cell Sci 129: 1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GR, Nassar N, Cerione RA. 2000. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100: 345–356. [DOI] [PubMed] [Google Scholar]
- Hrycyna CA, Sapperstein SK, Clarke S, Michaelis S. 1991. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J 10: 1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, Wittinghofer A. 2011. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol 7: 942–949. [DOI] [PubMed] [Google Scholar]
- Jang H, Abraham SJ, Chavan TS, Hitchinson B, Khavrutskii L, Tarasova NI, Nussinov R, Gaponenko V. 2015. Mechanisms of membrane binding of small GTPase K-Ras4B farnesylated hypervariable region. J Biol Chem 290: 9465–9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WJ, Yoon J, Park JC, Lee SH, Lee SH, Kaduwal S, Kim H, Yoon JB, Choi KY. 2012. Ras stabilization through aberrant activation of Wnt/β-catenin signaling promotes intestinal tumorigenesis. Sci Signal 5: ra30. [DOI] [PubMed] [Google Scholar]
- John J, Sohmen R, Feuerstein J, Linke R, Wittinghofer A, Goody RS. 1990. Kinetics of interaction of nucleotides with nucleotide-free H-Ras p21. Biochemistry 29: 6058–6065. [DOI] [PubMed] [Google Scholar]
- Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bonson RT, Umanoff H, Edelmann W, Kucherlapati R, et al. 1997. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev 11: 2468–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd WR, Slattum PM, Hoang KC, Bhoite L, Valppu L, Alberts G, Brown B, Roth B, Ostanin K, Huang L, et al. 2011. Discovery and SAR of methylated tetrahydropyranyl derivatives as inhibitors of isoprenylcysteine carboxyl methyltransferase (ICMT). J Med Chem 54: 5031–5047. [DOI] [PubMed] [Google Scholar]
- Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. 2006. Differential modification of Ras proteins by ubiquitination. Mol Cell 21: 679–687. [DOI] [PubMed] [Google Scholar]
- Just I, Selzer J, Hofmann F, Green GA, Aktories K. 1996. Inactivation of Ras by Clostridium sordellii lethal toxin-catalyzed glucosylation. J Biol Chem 271: 10149–10153. [DOI] [PubMed] [Google Scholar]
- Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al. 2008. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456: 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SE, Yoon JY, Jeong WJ, Jeon SH, Park Y, Yoon JB, Park YN, Kim H, Choi KY. 2009. H-Ras is degraded by Wnt/β-catenin signaling via β-TrCP-mediated polyubiquitylation. J Cell Sci 122: 842–848. [DOI] [PubMed] [Google Scholar]
- Kollar P, Rajchard J, Balounova Z, Pazourek J. 2014. Marine natural products: Bryostatins in preclinical and clinical studies. Pharm Biol 52: 237–242. [DOI] [PubMed] [Google Scholar]
- Kong E, Peng S, Chandra G, Sarkar C, Zhang Z, Bagh MB, Mukherjee AB. 2013. Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-Ras product and growth-associated protein-43. J Biol Chem 288: 9112–9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander HM, Ogiste JS, Teng KK, Novogrodsky A. 1995a. p21ras as a common signaling target of reactive free radicals and cellular redox stress. J Biol Chem 270: 21195–21198. [DOI] [PubMed] [Google Scholar]
- Lander HM, Ogiste JS, Pearce SF, Levi R, Novogrodsky A. 1995b. Nitric oxide-stimulated guanine nucleotide exchange on p21ras. J Biol Chem 270: 7017–7020. [DOI] [PubMed] [Google Scholar]
- Lander HM, Milbank AJ, Tauras JM, Hajjar DP, Hempstead BL, Schwartz GD, Kraemer RT, Mirza UA, Chait BT, Burk SC, et al. 1996. Redox regulation of cell signalling. Nature 381: 380–381. [DOI] [PubMed] [Google Scholar]
- Lander HM, Hajjar DP, Hempstead BL, Mirza UA, Chait BT, Campbell S, Quilliam LA. 1997. A molecular redox switch on p21ras. Structural basis for the nitric oxide-p21ras interaction. J Biol Chem 272: 4323–4326. [DOI] [PubMed] [Google Scholar]
- Lau HY, Ramanujulu PM, Guo D, Yang T, Wirawan M, Casey PJ, Go ML, Wang M. 2014. An improved isoprenylcysteine carboxylmethyltransferase inhibitor induces cancer cell death and attenuates tumor growth in vivo. Cancer Biol Ther 15: 1280–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau HY, Tang J, Casey PJ, Wang M. 2017. Isoprenylcysteine carboxylmethyltransferase is critical for malignant transformation by all RAS isoforms. Oncogene 36: 3934–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude AJ, Prior IA. 2008. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J Cell Sci 121: 421–427. [DOI] [PubMed] [Google Scholar]
- Leung KF, Baron R, Ali BR, Magee AI, Seabra MC. 2007. Rab GTPases containing a CAAX motif are processed post-geranylgeranylation by proteolysis and methylation. J Biol Chem 282: 1487–1497. [DOI] [PubMed] [Google Scholar]
- Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. 2001. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem 276: 30987–30994. [DOI] [PubMed] [Google Scholar]
- Lim KH, Ancrile BB, Kashatus DF, Counter CM. 2008. Tumour maintenance is mediated by eNOS. Nature 452: 646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Westover K, Ficarro S, Harrison R, Choi H, Pacold M, Carrasco M, Hunter J, Kim ND, Xie T, et al. 2014. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew Chem Int Ed Engl 53: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DT, Conibear E. 2015. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife 4: e11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Solomon M, Li LS, Hansen R, Rosen N. 2016. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351: 604–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Sjogren AK, Karlsson C, Ibrahim MX, Andersson KM, Olofsson FJ, Wahlstrom AM, Dalin M, Yu H, Chen Z, et al. 2010. Targeting the protein prenyltransferases efficiently reduces tumor development in mice with K-RAS-induced lung cancer. Proc Natl Acad Sci 107: 6471–6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jiao B, Zhang R, Zhao H, Zhang C, Wu M, Li D, Zhao X, Qiu Q, Li J, et al. 2016. Palmitoylacyltransferase Zdhhc9 inactivation mitigates leukemogenic potential of oncogenic Nras. Leukemia 30: 1225–1228. [DOI] [PubMed] [Google Scholar]
- Lobell RB, Liu D, Buser CA, Davide JP, DePuy E, Hamilton K, et al. 2002. Preclinical and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol Cancer Ther 1: 747–758. [PubMed] [Google Scholar]
- Lobo S, Greentree WK, Linder ME, Deschenes RJ. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem 21: 21. [DOI] [PubMed] [Google Scholar]
- Lorent JH, Levental I. 2015. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem Phys Lipids 192: 23–32. [DOI] [PubMed] [Google Scholar]
- Lu JY, Hofmann SL. 1995. Depalmitoylation of CAAX motif proteins. Protein structural determinants of palmitate turnover rate. J Biol Chem 270: 7251–7256. [DOI] [PubMed] [Google Scholar]
- Magee AI, Gutierrez L, McKay IA, Marshall CJ, Hall A. 1987. Dynamic fatty acylation of p21N-ras. EMBO J 6: 3353–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher J, Baker DA, Manning M, Dibb NJ, Roberts IA. 1995. Evidence for cell-specific differences in transformation by N-, H- and K-Ras. Oncogene 11: 1639–1647. [PubMed] [Google Scholar]
- Manu KA, Chai TF, Teh JT, Zhu WL, Casey PJ, Wang M. 2017. Inhibition of isoprenylcysteine carboxylmethyltransferase induces cell-cycle arrest and apoptosis through p21 and p21-regulated BNIP3 induction in pancreatic cancer. Mol Cancer Ther 16: 914–923. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. 1996. Ras effectors. Curr Opin Cell Biol 8: 197–204. [DOI] [PubMed] [Google Scholar]
- Martin BR, Cravatt BF. 2009. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods 6: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Aderem A. 1995. The myristoyl-electrostatic switch: A modulator of reversible protein–membrane interactions. Trends Biochem Sci 20: 272–276. [DOI] [PubMed] [Google Scholar]
- Michaelson D, Silletti J, Murphy G, D’Eustachio P, Rush M, Philips MR. 2001. Differential localization of Rho GTPases in live cells. Regulation by hypervariable regions and rhogdi binding. J Cell Biol 152: 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. 2006. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res 47: 1118–1127. [DOI] [PubMed] [Google Scholar]
- Mohammad RM, Dugan MC, Mohamed AN, Almatchy VP, Flake TM, Dergham ST, Sheilds AF, Al-Katib AA, Vaitkevicius VK, Sarkar FH. 1998. Establishment of a human pancreatic tumor xenograft model: Potential application for preclinical evaluation of novel therapeutic agents. Pancreas 16: 19–25. [DOI] [PubMed] [Google Scholar]
- Mohammed I, Hampton SE, Ashall L, Hildebrandt ER, Kutlik RA, Manandhar SP, Floyd BJ, Smith HE, Dozier JK, Distefano MD, et al. 2016. 8-Hydroxyquinoline-based inhibitors of the Rce1 protease disrupt Ras membrane localization in human cells. Bioorg Med Chem 24: 160–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nancy V, Callebaut I, El Marjou A, de Gunzburg J. 2002. The δ subunit of retinal rod cGMP phosphodiesterase regulates the membrane association of Ras and Rap GTPases. J Biol Chem 277: 15076–15084. [DOI] [PubMed] [Google Scholar]
- Onken B, Wiener H, Philips M, Chang EC. 2006. Compartmentalized signaling of Ras in fission yeast. Proc Natl Acad Sci 103: 9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. 2013. K-RasG12C inhibitors allosterically control GTP affinity and effector interactions. Nature 503: 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto JC, Kim E, Young SG, Casey PJ. 1999. Cloning and characterization of a mammalian prenyl protein-specific protease. J Biol Chem 274: 8379–8382. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, et al. 2016. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov 6: 316–329. [DOI] [PubMed] [Google Scholar]
- Pillinger MH, Volker C, Stock JB, Weissmann G, Philips MR. 1994. Characterization of a plasma membrane-associated prenylcysteine-directed α carboxyl methyltransferase in human neutrophils. J Biol Chem 269: 1486–1492. [PubMed] [Google Scholar]
- Plowman SJ, Hancock JF. 2005. Ras signaling from plasma membrane and endomembrane microdomains. Biochim Biophys Acta 1746: 274–283. [DOI] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. 2012. A comprehensive survey of Ras mutations in cancer. Cancer Res 72: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. 1999. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta 1451: 1–16. [DOI] [PubMed] [Google Scholar]
- Ritt DA, Abreu-Blanco MT, Bindu L, Durrant DE, Zhou M, Specht SI, Stephen AG, Holderfield M, Morrison DK. 2016. Inhibition of Ras/Raf/MEK/ERK pathway signaling by a stress-induced phospho-regulatory circuit. Mol Cell 64: 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307: 1746–1752. [DOI] [PubMed] [Google Scholar]
- Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, et al. 2010. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141: 458–471. [DOI] [PubMed] [Google Scholar]
- Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, Parton RG, Henis YI, Kloog Y, Hancock JF. 2005. Individual palmitoyl residues serve distinct roles in H-Ras trafficking, microlocalization, and signaling. Mol Cell Biol 25: 6722–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch M, Zimmermann TJ, Bürger M, Dekker FJ, Görmer K, Triola G, Brockmeyer A, Janning P, Böttcher T, Sieber SA, et al. 2011. Identification of acyl protein thioesterases 1 and 2 as the cellular targets of the Ras-signaling modulators palmostatin B and M. Angew Chem Int Ed Engl 50: 9838–9842. [DOI] [PubMed] [Google Scholar]
- Samatar AA, Poulikakos PI. 2014. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat Rev Drug Discov 13: 928–942. [DOI] [PubMed] [Google Scholar]
- Sanders SS, Martin DD, Butland SL, Lavallée-Adam M, Calzolari D, Kay C, Yates JR III, Hayden MR. 2015. Curation of the mammalian palmitoylome indicates a pivotal role for palmitoylation in diseases and disorders of the nervous system and cancers. PLoS Comput Biol 11: e1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos E, Nebreda AR. 1989. Structural and functional properties of ras proteins. FASEB J 3: 2151–2163. [DOI] [PubMed] [Google Scholar]
- Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER, Haviv S, Asara JM, Pandolfi PP, Cantley LC. 2010. Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci Signal 4: ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A. 1997. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic mutants. Science 277: 5324. [DOI] [PubMed] [Google Scholar]
- Schmick M, Vartak N, Papke B, Kovacevic M, Truxius DC, Rossmannek L, Bastiaens PIH. 2014. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 157: 459–471. [DOI] [PubMed] [Google Scholar]
- Schmidt WK, Tam A, Fujimura-Kamada K, Michaelis S. 1998. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc Natl Acad Sci 95: 11175–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H, Leventis R, Rex S, Schelhaas M, Nägele E, Waldmann H, Silvius JR. 1997. S-Acylation and plasma membrane targeting of the farnesylated carboxyl-terminal peptide of N-ras in mammalian fibroblasts. Biochemistry 36: 13102–13109. [DOI] [PubMed] [Google Scholar]
- Seabra MC, Reiss Y, Casey PJ, Brown MS, Goldstein JL. 1991. Protein farnesyltransferase and geranylgeranyltransferase share a common α subunit. Cell 65: 429–434. [DOI] [PubMed] [Google Scholar]
- Shahinian S, Silvius JR. 1995. Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry 34: 3813–3822. [DOI] [PubMed] [Google Scholar]
- Siprashvili Z, Webster DE, Johnston D, Shenoy RM, Ungewickell AJ, Bhaduri A, Flockhart R, Zarnegar BJ, Che Y, Meschi F, et al. 2016. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet 48: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun WT, Xiang W, Ng BL, Asari K, Bunte RM, Casey PJ, Wang M, Chuah C. 2016. Inhibition of isoprenylcysteine carboxylmethyltransferase augments BCR-ABL1 tyrosine kinase inhibition-induced apoptosis in chronic myeloid leukemia. Exp Hematol 44: 189–193 e2. [DOI] [PubMed] [Google Scholar]
- Sung PJ, Tsai FD, Vais H, Court H, Yang J, Fehrenbacher N, Foskett JK, Philips MR. 2013. Phosphorylated K-Ras limits cell survival by blocking Bcl-xL sensitization of inositol trisphosphate receptors. Proc Natl Acad Sci 110: 20593–20598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. 2005. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem 280: 31141–31148. [DOI] [PubMed] [Google Scholar]
- Switzer CH, Cheng RY, Ridnour LA, Glynn SA, Ambs S, Wink DA. 2012. Ets-1 is a transcriptional mediator of oncogenic nitric oxide signaling in estrogen receptor-negative breast cancer. Breast Cancer Res 14: R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabaczar S, Czogalla A, Podkalicka J, Biernatowska A, Sikorski AF. 2017. Protein palmitoylation: Palmitoyltransferases and their specificity. Exp Biol Med (Maywood) 242: 1150–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatis VM, Trenchi A, Gomez GA, Daniotti JL. 2010. Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS ONE 5: e15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M, McCormick F. 1987. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238: 542–545. [DOI] [PubMed] [Google Scholar]
- Tsai FD, Wynne JP, Ahearn IM, Philips MR. 2014. Metabolic labeling of Ras with tritiated palmitate to monitor palmitoylation and depalmitoylation. Methods Mol Biol 1120: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FD, Lopes MS, Zhou M, Court H, Ponce O, Fiordalisi JJ, Gierut JJ, Cox AD, Haigis KM, Philips MR. 2015. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci 112: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Stenmark H, Alexandrov K, Huber LA, Kaibuchi K, Sasaki T, Takai Y, Zerial M. 1993. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem 268: 18143–18150. [PubMed] [Google Scholar]
- Vartak N, Papke B, Grecco HE, Rossmannek L, Waldmann H, Hedberg C, Bastiaens PI. 2014. The autodepalmitoylating activity of APT maintains the spatial organization of palmitoylated membrane proteins. Biophysical J 106: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M, Schmidt MFG. 2006. Encyclopedic reference of genomics and proteomics in molecular medicine. Springer, Berlin. [Google Scholar]
- Vetter IR, Wittinghofer A. 2001. The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304. [DOI] [PubMed] [Google Scholar]
- Wahlstrom AM, Cutts BA, Karlsson C, Andersson KM, Liu M, Sjogren AK, Swolin B, Young SG, Bergo MO. 2007. Rce1 deficiency accelerates the development of K-RAS-induced myeloproliferative disease. Blood 109: 763–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstrom AM, Cutts BA, Liu M, Lindskog A, Karlsson C, Sjogren AK, Andersson KM, Young SG, Bergo MO. 2008. Inactivating Icmt ameliorates K-RAS-induced myeloproliferative disease. Blood 112: 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MT, Holderfield M, Galeas J, Delrosario R, To MD, Balmain A, McCormick F. 2015. K-Ras promotes tumorigenicity through suppression of non-canonical Wnt signaling. Cell 163: 1237–1251. [DOI] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, Sabatini DM. 2017. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168: 890–903.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb Y, Hermida-Matsumoto L, Resh MD. 2000. Inhibition of protein palmitoylation, raft localization, and T-cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem 275: 261–270. [DOI] [PubMed] [Google Scholar]
- Welsch ME, Kaplan A, Chambers JM, Stokes ME, Bos PH, Zask A, Zhang Y, Sanchez-Martin M, Badgley MA, Huang CS, et al. 2017. Multivalent small-molecule pan-RAS inhibitors. Cell 168: 878–89 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C, Li C, Yang J, Petrenko NB, Madesh M, Thompson CB, Foskett JK. 2005. The endoplasmic reticulum gateway to apoptosis by Bcl-XL modulation of the InsP3R. Nat Cell Biol 7: 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. 1997. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem 272: 14459–14464. [DOI] [PubMed] [Google Scholar]
- Williams JG, Pappu K, Campbell SL. 2003. Structural and biochemical studies of p21Ras S-nitrosylation and nitric oxide-mediated guanine nucleotide exchange. Proc Natl Acad Sci 100: 6376–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willumsen BM, Norris K, Papageorge AG, Hubbert NL, Lowy DR. 1984. Harvey murine sarcoma virus p21 ras protein: Biological and biochemical significance of the cysteine nearest the carboxy terminus. EMBO J 3: 2581–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright LP, Philips MR. 2006. Thematic review series: Lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J Lipid Res 47: 883–891. [DOI] [PubMed] [Google Scholar]
- Wright LP, Court H, Mor A, Ahearn IM, Casey PJ, Philips MR. 2009. Topology of mammalian isoprenylcysteine carboxyl methyltransferase determined in live cells with a fluorescent probe. Mol Cell Biol 29: 1826–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Lubkov V, Taylor LJ, Bar-Sagi D. 2010. Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination. Curr Biol 20: 1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hedberg C, Dekker FJ, Li Q, Haigis KM, Hwang E, Waldmann H, Shannon K. 2012. Inhibiting the palmitoylation/depalmitoylation cycle selectively reduces the growth of hematopoietic cells expressing oncogenic Nras. Blood 119: 1032–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Ascano JM, Hang HC. 2010. Bioorthogonal chemical reporters for monitoring protein acetylation. J Am Chem Soc 132: 3640–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Nickerson S, Kim ET, Liot C, Laurent G, Spang R, Philips MR, Shan Y, Shaw DE, Bar-Sagi D, et al. 2012. Regulation of RAS oncogenicity by acetylation. Proc Natl Acad Sci 109: 10843–10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Laurent G, Bause AS, Spang R, German N, Haigis MC, Haigis KM. 2013. HDAC6 and SIRT2 regulate the acetylation state and oncogenic activity of mutant K-RAS. Mol Cancer Res 11: 1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T, Wang Q, Fu J, Lin Q, Bi J, Ding W, Qiao Y, Zhang S, Zhao W, Lin H, et al. 2014. Impeded Nedd4-1-mediated Ras degradation underlies Ras-driven tumorigenesis. Cell Rep 7: 871–882. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu XH, Zhang K, Chen CK, Frederick JM, Prestwich GD, Baehr W. 2004. Photoreceptor cGMP phosphodiesterase δ subunit (PDEδ) functions as a prenyl-binding protein. J Biol Chem 279: 407–413. [DOI] [PubMed] [Google Scholar]
- Zhang H, Li S, Doan T, Rieke F, Detwiler PB, Frederick JM, Baehr W. 2007. Deletion of PrBP/δ impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc Natl Acad Sci 104: 8857–8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Liu P, Zhang R, Wu M, Li D, Zhao X, Zhang C, Jiao B, Chen B, Chen Z, et al. 2015. Roles of palmitoylation and the KIKK membrane-targeting motif in leukemogenesis by oncogenic KRAS4A. J Hematol Oncol 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wiener H, Su W, Zhou Y, Liot C, Ahearn I, Hancock JF, Philips MR. 2016. VPS35 binds farnesylated N-Ras in the cytosol to regulate N-Ras trafficking. J Cell Biol 214: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Prakash P, Liang H, Cho KJ, Gorfe AA, Hancock JF. 2017. Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell 168: 239–251 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, et al. 2013. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 497: 638–642. [DOI] [PubMed] [Google Scholar]
- Zimmermann G, Schultz-Fademrecht C, Küchler P, Murarka S, Ismail S, Triola G, Nussbaumer P, Wittinghofer A, Waldmann H. 2014. Structure guided design and kinetic analysis of highly potent benzimidazole inhibitors targeting the PDEδ prenyl binding site. J Med Chem 57: 5435–5448. [DOI] [PubMed] [Google Scholar]