Anaplastic lymphoma kinase (ALK) gene fusions drive approximately 5% of non-small cell lung cancers (NSCLC) (1). Fluorescence in situ hybridization (FISH) and immunohistochemistry are widely used to identify them based on ALK translocation and ALK overexpression, which are common in all cases and equally predict response to tyrosine kinase inhibitors (TKI) (2). On the other hand, the ALK fusion itself varies among patients. It can be typed by reverse-transcription polymerase chain reaction (RT-PCR) or next-generation sequencing (NGS) (3), but this is not currently required as part of the diagnostic workup (2,4). Even though different variants of the ALK fusion were first recognized over ten years ago (1) and have been extensively characterized in vitro (5,6), until very recently their clinical significance remained unclear. Major obstacles have been the complex management of ALK+ NSCLC patients, including highly variable sequences of TKI and local ablative treatments, as well as their long survival, currently exceeding 5 years in median after two ALK inhibitors (7), which have confounded and limited early studies. During the past months, however, several reports combining detailed clinical annotation with state-of-the-art molecular profiling in larger cohorts, have revealed a major impact of the specific ALK alteration on tumor biology and patient outcome. Echinoderm microtubule-associated protein-like 4 (EML4)-ALK fusion variant 3 (V3) in particular emerges as marker suitable for the selection of higher-risk cases under several therapeutic circumstances and calls for reconsideration of basic concepts and management strategies (8-11).
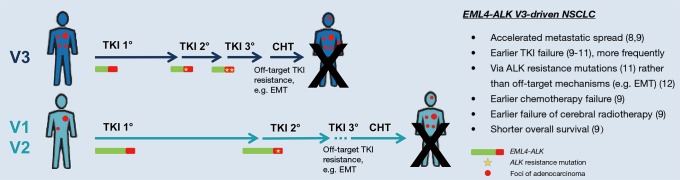
In the first line, EML4-ALK V3 appears to be associated with increased disease aggressiveness independent of treatment: Noh et al. have observed a higher frequency of metastatic disease among newly diagnosed ALK+ NSCLC cases harboring V3 vs. other variants (8), and own data have demonstrated an increased number of metastatic sites at initial diagnosis for stage IV ALK+ V3 NSCLC patients (9). In both studies, EML4-ALK V3 was associated with enhanced metastatic spread already before the start of treatment indicating higher clinical risk, which is present at baseline and not related to a specific therapy regimen (Figure 1). In addition, EML4-ALK V3 is associated with shorter progression-free survival (PFS) after non-TKI treatments, namely chemotherapy and cerebral radiotherapy (Figure 1) (9), while its polypeptide product shows a longer half-life and stronger oncogenic signaling in vitro (5,6,10). Collectively, these data suggest important and clinically relevant biological differences between tumors harboring V3 vs. other EML4-ALK variants regardless of TKI exposure.
Figure 1.
Clinical impact of EML4-ALK fusion variant V3. Differences in clinical course and patient outcome with various therapeutic modalities of lung adenocarcinoma driven by EML4-ALK V3 vs. V1/V2. The collective insight from several recent publications (8-12) is visualized, with arrow lengths roughly proportional to the corresponding progression-free and overall survival intervals as estimated by considering the different published series together. However, there is substantial heterogeneity, which probably reflects additional effects from further important biological factors that remain to be determined. Numbers in parentheses indicate references of the article; EML4, echinoderm microtubule-associated protein-like 4; ALK, anaplastic lymphoma kinase; TKI, tyrosine kinase inhibitor; EMT, epithelial-mesenchymal transition.
Moreover, there is accumulating evidence that EML4-ALK V3 is associated with a shorter PFS of patients receiving first- and second-generation ALK TKI in the first and second treatment lines (9,10), and even with a significantly worse overall survival (OS) compared to the other two common EML4-ALK variants V1 and V2 (Figure 1) (9). Interestingly, these observations are nicely explained by the increased propensity of V3- vs. V1-driven tumors to develop ALK resistance mutations as reported by Lin et al. (11), since sequential TKI administration is an important determinant of longer OS in ALK+ NSCLC (7).
Integrating these findings, it appears that V3-positive patients progress faster through TKI treatment lines (i.e., have a shorter PFS after first- and second-line TKI treatment, Figure 1), predominantly through development of TKI resistance mutations (11) and possibly facilitated by incomplete tumor cell suppression due to the higher IC50 of “wild-type” V3 (10). In contrast, V1 patients need longer to acquire resistance, but at the same time presumably progress rather due to more complex resistance mechanisms, which are not amenable to further TKI treatment (12). Thus, TKI-refractory V3 and V1 patients are expected to also differ in terms of several other important biological factors besides the frequency of ALK resistance mutations. These additional differences, which remain to be explored, will likely affect response to further ALK inhibitor treatment and probably account for the paradox of EML4-ALK V3 being unfavorable in all statistical analyses meticulously performed by several investigators (9-11)—except for patients receiving lorlatinib beyond the second treatment line in one study, where the V3 variant appears to confer longer PFS than V1 (11).
In daily clinical practice, this enhanced benefit from lorlatinib in later treatment lines will largely depend on whether an ALK mutation has emerged as the cause of TKI failure (i.e., on ALK sequencing results of a repeat biopsy at that time) rather than on the gene fusion variant as such, which does not change during therapy. However, detection of the unfavorable EML4-ALK variant V3 could be used to select patients for more aggressive surveillance and treatment strategies earlier in the course of their disease, which carries features of higher risk already at baseline (8,9). The recent approval of alectinib for first-line treatment of ALK+ NSCLC is probably not going to have a major impact in this regard, because in vitro data show a similar resistance of V3 expressing cells to alectinib, crizotinib and ceritinib with IC50 values >500 nM (10). Whether upfront administration of a third-generation ALK inhibitor with broader activity against ALK resistance mutations, such as lorlatinib (12), could to some extent negate the V3-associated risk, is unclear at present. Other strategies to consider currently are a closer monitoring of V3 ALK+ patients with radiologic studies and ctDNA assays as well as a more aggressive approach regarding local ablative treatments. We anticipate the development of novel management strategies and drugs against the higher-risk, V3-driven disease to become a main research objective in ALK+ NSCLC. While typing of the ALK fusion variant and ALK resistance mutation testing are not recommended by the current CAP/IASLC/AMP guidelines (2), data by several researchers illustrate the great potential of a more fine granular ALK analysis in clinical trials and eventually also in routine patient care (5,6,9-11).
Acknowledgements
None.
Footnotes
Conflicts of Interest: V Endris reports advisory board honoraria and lecture fees from AstraZeneca and ThermoFisher. A Stenzinger reports advisory board honoraria from Novartis, AstraZeneca, ThermoFisher, BMS, lecture fees from Illumina, AstraZeneca, Novartis, ThermoFisher and travel grants from Illumina, AstraZeneca, ThermoFisher. M Thomas reports advisory board honoraria from Novartis, Lilly, BMS, MSD, Roche, Celgene, Takeda, AbbVie, Boehringer, lecture fees from Lilly, MSD, Takeda, research funding from AstraZeneca, BMS, Celgene, Novartis, Roche and travel grants from BMS, MSD, Novartis, Boehringer. P Christopoulos and M Kirchner have no conflicts of interest to declare.
References
- 1.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- 2.Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn 2018;20:129-59. 10.1016/j.jmoldx.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 3.Pfarr N, Stenzinger A, Penzel R, et al. High-throughput diagnostic profiling of clinically actionable gene fusions in lung cancer. Genes Chromosomes Cancer 2016;55:30-44. 10.1002/gcc.22297 [DOI] [PubMed] [Google Scholar]
- 4.Kalemkerian GP, Narula N, Kennedy EB, et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J Clin Oncol 2018;36:911-9. 10.1200/JCO.2017.76.7293 [DOI] [PubMed] [Google Scholar]
- 5.Heuckmann JM, Balke-Want H, Malchers F, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res 2012;18:4682-90. 10.1158/1078-0432.CCR-11-3260 [DOI] [PubMed] [Google Scholar]
- 6.Richards MW, Law EW, Rennalls LP, et al. Crystal structure of EML1 reveals the basis for Hsp90 dependence of oncogenic EML4-ALK by disruption of an atypical beta-propeller domain. Proc Natl Acad Sci U S A 2014;111:5195-200. 10.1073/pnas.1322892111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duruisseaux M, Besse B, Cadranel J, et al. Overall survival with crizotinib and next-generation ALK inhibitors in ALK-positive non-small-cell lung cancer (IFCT-1302 CLINALK): a French nationwide cohort retrospective study. Oncotarget 2017;8:21903-17. 10.18632/oncotarget.15746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noh KW, Lee MS, Lee SE, et al. Molecular breakdown: a comprehensive view of anaplastic lymphoma kinase (ALK)-rearranged non-small cell lung cancer. J Pathol 2017;243:307-19. 10.1002/path.4950 [DOI] [PubMed] [Google Scholar]
- 9.Christopoulos P, Endris V, Bozorgmehr F, et al. EML4-ALK fusion variant V3 is a high-risk feature conferring accelerated metastatic spread, early treatment failure and worse overall survival in ALK+ NSCLC. Int J Cancer 2018;142:2589-98. 10.1002/ijc.31275 [DOI] [PubMed] [Google Scholar]
- 10.Woo CG, Seo S, Kim SW, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol 2017;28:791-7. [DOI] [PubMed] [Google Scholar]
- 11.Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J Clin Oncol 2018;36:1199-206. 10.1200/JCO.2017.76.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. 10.1158/2159-8290.CD-16-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]