Abstract
DNA double-strand breaks (DSBs) arise regularly in cells and when left unrepaired cause senescence or cell death. Homologous recombination (HR) and nonhomologous end-joining (NHEJ) are the two major DNA-repair pathways. Whereas HR allows faithful DSB repair and healthy cell growth, NHEJ has higher potential to contribute to mutations and malignancy. Many regulatory mechanisms influence which of these two pathways is used in DSB repair. These mechanisms depend on the cell cycle, post-translational modifications, and chromatin effects. Here, we summarize current research into these mechanisms, with a focus on mammalian cells, and also discuss repair by “alternative end-joining” and single-strand annealing.
Keywords: DNA damage, DNA repair, BRCA1, ubiquitylation (ubiquitination), chromatin, homologous recombination, double-strand break, nonhomologous end-joining
Introduction
Many cancer cells show genomic abnormalities consistent with aberrant repair of DNA double-strand breaks (DSBs)2 (1). Mammalian DSBs can be repaired by homologous recombination (HR), “canonical” nonhomologous end-joining (C-NHEJ), “alternative” nonhomologous end-joining (A-EJ), or by single-strand annealing (SSA) (Fig. 1) (2, 3). Each of these pathways requires specific repair factors and produces different repair outcomes. HR is typically considered to be a “faithful” pathway, and deficiencies in HR contribute to mutations associated with malignancy and reduced cell health. Given these observations, a great deal of effort has been dedicated in recent years to understanding when and how DNA repair pathways are regulated.
Figure 1.

Overview of pathways for DNA DSB repair in mammals. Canonical nonhomologous end-joining (C-NHEJ) involves direct ligation at the break site, often with some number of base insertions or deletions that can cause mutation. A-EJ refers to NHEJ that does not use canonical end-joining factors. Homologous DNA sequences are indicated in red. A-EJ often involves some degree of resection, creating single-stranded regions that may pair at areas of micro-homology. Processing and excision of the intervening sequence is likely to cause mutation at the repair junction. SSA also involves resection and pairing of homologous regions but uses different repair machinery and is more likely to result in large deletion mutations. HR involves resection at the DSB and repair using a homologous DNA sequence as a template. It is usually error-free but may occasionally contribute to mutation. For more details see Refs. 2 and 3.
NHEJ often acts first to repair DSBs
Several lines of evidence indicate that C-NHEJ often acts first to attempt to repair DSBs (4–8). If NHEJ cannot be completed, then the DSB undergoes “resection,” in which one strand of the DNA duplex is degraded to produce a single-stranded DNA overhang suitable for alternative pathways of repair (2). Fluorescent reporter constructs integrated into the chromosomes of human cell lines revealed that NHEJ is much faster than HR, taking place within 30 min (versus several hours for HR), and accounts for ∼75% of repair events (6). Cell cycle-specific studies using knockout cell lines demonstrated that ∼80% of DSBs induced by ionizing radiation in G2 are repaired by NHEJ (4). According to these estimates, HR might be considered a pathway that acts in specific contexts, when NHEJ is not active or successful. In fact, a study in which DSBs were induced transiently using the I-SceI meganuclease revealed almost no HR-mediated repair (8). This low level of HR may be dependent on rapid ligation of cohesive ends formed by I-SceI cutting.
Structure of DNA breaks influences the pathway used for repair
Other evidence supports the idea that the structure of the DSB can influence DNA repair pathway choice. For example, DSBs induced using near-infrared microbeam irradiation, or high-linear energy transfer carbon ions, tend to have a more complex structure than those induced with I-SceI. In each case, the more complex DSBs generated using these approaches could not be repaired quickly by NHEJ and instead required additional processing or a greater use of HR for repair (9, 10). Use of different variants of Cas9 with site-specific gRNAs also allowed different DSB structures to be generated at break sites (11). Interestingly, DNA ends with a 47-nucleotide 5′ overhang were subject to HR-mediated gene conversion at a far higher frequency than “blunt” DSBs, which are repaired primarily by classical NHEJ.
Cell cycle is a major determinant of repair pathway choice
Although kinetics and end structure are undoubtedly important in determining what pathway is used for DSB repair, it is clear that different cell types use different repair pathways at different rates. For example, HR appears to be especially efficient in stem cells, whereas NHEJ is used more frequently in more differentiated lineages (12). DSB repair pathway choice must therefore be subject to regulation, which can take place at many different stages. A key determinant of repair pathway usage is the cell cycle phase (Fig. 2). Tracking live cells to measure what proportions of DSBs are repaired by HR indicated that HR reaches peak activity in mid-S phase, whereas NHEJ predominates in G1 and G2 (5). NHEJ is repressed during mitosis, however, by a mechanism involving phosphorylation of key DNA damage–response factors by the mitotic-specific kinase, CDK1 (13).
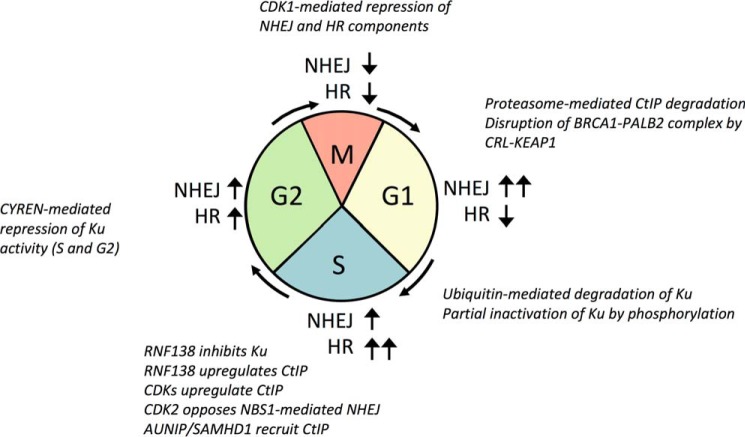
Figure 2.
Cell cycle-dependent regulation of DSB repair pathways in mammals. Pathways for repair of DSBs are active at different rates at different phases of the cell cycle. NHEJ dominates in G1, whereas HR is most active in S phase. NHEJ and HR appear to compete for repair of DSBs in G2, but both pathways are down-regulated during M phase. Changes in activity of different pathways depend on differential expression of regulatory factors and post-translational modification. Several regulatory mechanisms are shown. See text for details.
The activation of HR in S phase is dependent in part on activation of “resection” activities. Cyclin-dependent kinase (CDK)-mediated phosphorylation of CtIP, a key protein that stimulates resection, is one way that HR is activated during S phase (14). This enables HR-mediated repair of DSBs formed during S phase, which can use a sister chromatid as a template. Conversely, HR is disfavored during G1, when homologous templates for HR-mediated repair of DSBs are unlikely to be in close proximity. HR is suppressed during G1 by a variety of mechanisms. First, CtIP is substantially destabilized by proteasome-mediated degradation during G1, and it only becomes present at substantial amounts in the nucleus following activation of CDK activity (15). CtIP-mediated resection has nonetheless been reported during G1 (16). CtIP-mediated resection in G1 is dependent on phosphorylation of CtIP by Plk3 and leads to NHEJ-mediated repair with a high proportion of mutations. The potentially mutagenic impact of resection during G1 underscores the importance of additional mechanisms to regulate resection (discussed below). Recently, the RECQ family helicase, RECQL4, was identified as a regulator of repair pathway choice between the G1 and S phase of the cell cycle (17). RECQL4 is recruited to DSBs at all phases of the cell cycle, but it interacts with the NHEJ factors Ku70/80 during G1 and with MRE11 during S and G2. This switch in the binding partner of RECQL4 is mediated by phosphorylation by CDK1/2. A recent report shows that NBS1 is down-regulated by cell cycle-specific phosphorylation to determine the activity of NHEJ at telomeres (18). CDK2-mediated phosphorylation of NBS1–Ser-432 (Ser-433 in mice) promotes dissociation of NBS1 from TRF2-protected telomeres during S phase, preventing the formation of chromatid fusions by NHEJ.
Regulation of NHEJ
Several factors promote NHEJ during G1 and subsequently limit the use of NHEJ as the cell transitions into S phase. Recent reports indicate that a number of enzymes help remove the heterodimeric DNA-binding complex Ku70/80, which is a key component of the C-NHEJ pathway, from DSBs, thereby increasing the use of HR. Ku80 is a target for RNF138 E3 ubiquitin ligase activity, and in the absence of RNF138, Ku80 persists at the DNA end, inhibiting DSB resection and HR (19). The importance of RNF138 for determining repair pathway choice is further demonstrated by its ability to regulate the resection factor, CtIP (20). Ubiquitylation of CtIP by RNF138 increases its ability to promote resection, whereas ubiquitylation of Ku80 by RNF138 reduces its ability to promote NHEJ. The exact mechanism by which these ubiquitylations affect the activity of repair factors is not yet completely clear. The effect of RNF138 ubiquitylation on Ku80 abundance at break sites appears to be independent of proteasome-mediated degradation. However, RNF8, SCFFbxl12, VCP-p97, and RNF126 have all been implicated in targeting Ku to the proteasome, and thereby shifting the balance of repair from NHEJ to HR (21–24). The CRL4 E3 ubiquitin ligase, which contains either CUL4A or CUL4B, has additionally been implicated in removal of Ku via a ubiquitin-dependent mechanism that is also dependent on neddylation (25). At this point, it is not clear whether these ubiquitin-mediated processes act redundantly or in response to specific types of DNA damage.
In addition to ubiquitin-mediated mechanisms, the activity of Ku70/80 in NHEJ is regulated based on phosphorylation (26). Five serine/threonine residues in the central region of Ku70 are phosphorylated by the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which binds to Ku70/80 during G1. This phosphorylation alters the structure of the Ku heterodimer, reducing its affinity for DNA. Dissociation of Ku70/80 following phosphorylation allows resection to begin, increasing the rate of HR in S phase. DNA-PKcs also regulates HR by modulating the activity of the ATM kinase in response to DNA damage (27). ATM is a master regulator of DNA damage responses, with hundreds of known substrates. DNA-PKcs phosphorylates ATM at multiple sites, which reduces its kinase activity and therefore restricts its ability to induce DSB resection leading to HR.
In 2017, Karlsreder and co-workers (28) demonstrated that CYREN (cell cycle regulator of NHEJ) represses NHEJ during S and G2 phases of the cell cycle. CYREN is only expressed during these cell cycle phases, when it blocks NHEJ activity by binding to Ku. Knockout of CYREN produces a significant increase in NHEJ-mediated chromatid fusions at deprotected telomeres. This result underscores the importance of correct regulation of DSB repair pathways, to prevent mutations arising from “toxic NHEJ” (29).
Regulation of DSB resection
A major step committing a DSB to HR is 5′–3′ resection of the DNA end to form a 3′ single-stranded DNA overhang. The initial step in resection involves end processing by a complex of MRE11 and CtIP, followed by “long-range” resection either by BLM/DNA2 or EXO1 (2). MRE11 is present along with RAD50 and NBS1 as the “MRN” complex and has both endonuclease and exonuclease activity (30). MRE11 first cuts one DNA strand close to the break site, using its 5′–3′ endonuclease activity and then degrades the same strand using its processive 3′–5′ exonuclease activity. Ku70/80 is displaced from the break site by MRE11-mediated end processing, thereby preventing further NHEJ activity. Selective inhibition of the endonuclease activity of MRE11 can prevent subsequent resection, however, allowing further attempts to repair damage using NHEJ.
CtIP plays an important role in activation of MRE11 nuclease activities to initiate HR. As such, CtIP is itself subject to multiple layers of regulation. As mentioned previously, CDK-mediated phosphorylation of CtIP at Thr-847 facilitates resection by MRE11-CtIP upon transition to S phase (14). CtIP activity is also enhanced by constitutive sumoylation by CBX4 on Lys-896 and by SIRT6-mediated deacetylation (31, 32). Conversely, other post-translational modifications appear to limit CtIP-mediated resection. The E3 ubiquitin ligase, CRL3KLHL15, targets CtIP for proteasomal degradation (33). CtIP activity also appears to be limited by RNF111-mediated neddylation, although CtIP is not a direct substrate for RNF111 (34). In each of these cases, the exact mechanism by which KLHL15 levels or RNF111 activity are regulated in response to DNA damage to modulate CtIP activity has yet to be fully understood.
Formation of a complex of CtIP with BRCA1 was formerly considered to be an essential step leading to up-regulation of DSB resection activity during the transition to S phase. More recent work has challenged this model. Ser-327 in CtIP was proposed to be the critical interface for interaction with BRCA1, but mutation of this residue revealed that it is not necessary for normal HR (35, 36). Several other factors have been suggested to help recruit CtIP to DNA breaks. Recruitment of CtIP to S phase DNA damage sites appears to be increased by its interaction with AUNIP, a protein that has intrinsic DNA-binding capability, with particular affinity for DNA structures formed at stalled replication sites (37). The dNTP triphosphohydrolase, SAMHD1, also recruits CtIP to DNA break sites (38). SAMHD1 had previously been characterized as a factor that modifies the infectivity of HIV, by altering cellular pools of dNTPs required for reverse transcription. SAMHD1 also localizes to break sites, however, and cancer-associated mutations in SAMHD1 map to the CtIP-interaction region, instead of the dNTPase-active site. Formation of the complex of CtIP–MRE11 at break sites is also dependent on RECQL4, and levels of HR correlate with the expression level of RECQL4 (39).
Several new players have recently been suggested to regulate DSB resection in mammalian cells. PHF11 (plant homeodomain finger 11) was shown to interact with RPA to promote extensive resection by EXO1 (40). Conversely, the helicase HELB was shown to be an antagonist of resection, with an ability to inhibit long-range resection by EXO1 or BLM-DNA2 (41).
BRCA1 and 53BP1: A key regulatory partnership
The DNA damage–response factor 53BP1 was identified as a negative regulator of HR in 2010, and the outcomes of patients with BRCA1-mutant cancer were shown to be dependent on levels of 53BP1 expression (29, 42). 53BP1 limits resection of DSBs, steering repair toward NHEJ and away from HR. This effect is counteracted by BRCA1, which allows resection to continue in the presence of 53BP1. 53BP1 is recruited to chromatin around DSBs by binding to specific histone modifications (Fig. 3). The Tudor domain of 53BP1 binds to H4K20Me2, which is present throughout the genome, whereas the 53BP1 “ubiquitin-dependent recruitment” region binds to H2AK15ub, which is induced by RNF168 following damage signaling at DSBs (43, 44). Multiple factors influence the ability of 53BP1 to bind to these chromatin marks and therefore to affect repair pathway choices at break sites. For example, 53BP1 was shown to have reduced retention at DSBs in S phase, because H4K20Me2 becomes “diluted” as replication proceeds (45). This effect limits the ability of 53BP1 to bind around DSBs and restrict resection as S phase proceeds, favoring increased use of HR. Binding of 53BP1 to H4K20Me2 is reduced by the presence of JMJD2A and L3MBTL1, which compete for H4K20Me2-binding sites (46, 47). These proteins can be displaced by the E3 ubiquitin ligase, RNF8, but this activity is opposed by deubiquitinating enzymes (DUBs). Specifically, JMJD2A recruitment is enhanced by the DUB activity of POH1, whereas the DUB OTUB1 restricts ubiquitination-mediated removal of L3MBTL1 (48, 49). In each of these cases, the DUB activity favors HR, by making it harder for 53BP1 to bind to H4K20Me2 around DSBs. 53BP1 binding to H4K20Me2 is further repressed by the expression of TIRR, a protein that binds to the Tudor domain of 53BP1, preventing it from associating with chromatin (50). As it is often amplified in tumor cells, TIRR may promote malignancy by impacting regulation of DNA repair by 53BP1. The histone acetyltransferase complex, TIP60, also competes with 53BP1 for H4K20Me2 (51). NMR spectroscopy showed that acetylation of H4K16 by TIP60 to produce H4K16Ac disrupts 53BP1 binding to chromatin by interfering with the interaction between key residues in the 53BP1 Tudor domain and histone H4 in the core nucleosome (52). TIP60 therefore creates a chromatin environment that is more favorable for BRCA1 binding relative to 53BP1 binding, leading to increased use of HR instead of NHEJ.
Figure 3.
53BP1-BRCA1 network for regulation of repair pathway choice at DSBs. 53BP1 normally represses use of HR for repair of DSBs. This is achieved in part by 53BP1-mediated recruitment of the downstream regulators RIF1, REV7, and PTIP. These factors repress resection, including by recruitment of Artemis, which can remove potentially-recombinogenic tracts of single-stranded DNA at the break site. The effect of 53BP1 is antagonized by factors that compete for chromatin-binding sites (JMJD2A, L3MBTL, and RNF169), inhibit 53BP1 recruitment (TIP60 and TIRR), block recruitment of downstream modulators of 53BP1 (SCAI), or promote degradation of 53BP1 (UBCH7). BRCA1 recruited to break sites via RAP80 may not always be in a complex that supports HR, but interaction with ZMYM3 may contribute to activation of BRCA1 to promote HR. BRCA1 antagonizes 53BP1 in part through recruitment of UHRF, which ubiquitylates RIF1, preventing RIF1 from being stably retained at the break site. BRCA1 may also inhibit 53BP1 binding through E3 ubiquitin ligase activity at the break. See text for more details.
Just as JMJD2A and L3MBTL1 compete with 53BP1 for H4K20Me2 binding, the E3 ubiquitin ligases RNF169 and RAD18 also compete with 53BP1 for binding to H2AK15ub (53, 54). The ability of 53BP1 to bind to H2AK15ub is also limited by phosphorylation or acetylation of key residues within the ubiquitin-dependent recruitment region (13, 55, 56). In addition to its recruitment, the stability of 53BP1 is also regulated by proteasome-mediated degradation dependent on the E2 enzyme, UBCH7 (57). UBCH7 activity is up-regulated upon replication stress, leading to increased degradation of 53BP1 and therefore higher levels of HR.
In general, BRCA1 is considered a pro-HR factor, based on long-standing studies that demonstrated that it supports formation of foci of the key recombination mediator, RAD51, after DNA damage and that it enables HR in plasmid reporter assays (58). BRCA1 forms nuclear “foci” after DNA damage, and many of these foci are dependent on the presence of the adaptor protein, RAP80, and the activities of the E3 ubiquitin ligases, RNF8 and RNF168. Surprisingly, however, deletion of RAP80 increases the rate of HR, indicating that RAP80 may be sequestering BRCA1 in areas within the nucleus where it is not able to promote HR (59, 60). If RAP80-dependent foci of BRCA1 do not represent sites of active recombination, it follows that there must be some other way for BRCA1 to be recruited to DSBs that support its pro-HR function. One way that this may be achieved is by conversion of RAP80-containing BRCA1 complexes into a pro-recombinogenic form. The zinc finger protein, ZMYM3, may play this role, as it is found associated with BRCA1 and RAP80, and supports HR (61). Alternatively, some other factor may regulate recruitment of BRCA1 to support HR. Recently, the Cockayne Syndrome B protein was shown to mediate BRCA1 recruitment to damage sites, enabling higher rates of DSB resection and HR (62).
The mechanism by which BRCA1 prevents 53BP1-mediated inhibition of resection and HR is still not fully understood, but recent years have seen several studies that potentially shed light on this long-standing mystery. One mechanism by which BRCA1 relieves the block on DSB resection caused by RIF1 depends on the E3 ubiquitin ligase, UHRF1 (63). UHRF1 is phosphorylated by CDK2, which is active in S phase, allowing it to be recruited to DSBs by BRCA1. UHRF1 then ubiquitylates RIF1 bound to 53BP1 at the break site, causing RIF1 to be displaced and partially overcoming the block on resection caused by 53BP1. Another recent report suggested that BRCA1 displaces RIF1 by a different mechanism that involves PP4C-mediated dephosphorylation of 53BP1 (64). A third model for BRCA1-mediated reversal of the block on resection imposed by 53BP1 involves ubiquitination of H2AK127 by the BRCA1-BARD1 E3 ligase, which triggers SMARCAD1-mediated repositioning of nucleosomes and 53BP1 to allow processive DSB resection (65). This model is based on evidence from a point mutation in BARD1, which forms a heterodimeric E3 ubiquitin ligase with BRCA1. Longitudinal studies in genetically-targeted mice have however suggested that the E3 ligase activity of BRCA1 is dispensable for tumor suppression (66).
Repair regulation downstream of 53BP1
53BP1 is phosphorylated after DNA damage, leading to association between 53BP1 and its downstream mediators, RIF1 and PTIP (67, 68). Another downstream effector of 53BP1, REV7, was identified in an shRNA screen for factors that control HR in the absence of BRCA1 (69, 70). Recent studies have provided additional insight into how these factors operate and are regulated. The interaction of 53BP1 with PTIP has the potential to be extremely significant, because PTIP can recruit the Artemis endonuclease to damage sites (71). Artemis is better known for processing DNA ends prior to NHEJ (3), but it appears that it also directs DNA repair pathway choice downstream of 53BP1 by cleaving single-stranded DNA overhangs, limiting the ability of RPA or RAD51 to bind and initiate HR. The interaction of 53BP1 with PTIP appears to be most relevant for producing aberrant NHEJ events in the absence of BRCA1, but its interaction with RIF1 is also important for a subset of NHEJ events, including class switch recombination at the immunoglobulin heavy chain locus in B lymphocytes (67). Interestingly, the SCAI protein (“suppressor of cancer cell invasion”) appears to compete with RIF1 for binding to phosphorylated 53BP1 (72). Whereas 53BP1-RIF1 promotes certain NHEJ events, 53BP1 bound to SCAI seems to be competent for HR, potentially representing a way in which the balance of repair pathways can be altered. Many HR factors are essential for embryonic development, whereas Scai−/− mice are viable but show a defect in 53BP1-dependent repair of heterochromatic DSBs (73). Clearly further work is required to fully understand the role of SCAI in DSB repair.
Regulation of RAD51 loading and unloading
After overcoming any block from 53BP1 and its downstream effectors, BRCA1 promotes efficient HR at the resected DSB through at least two processes. BRCA1 recruits first PALB2 (Partner and Localizer of BRCA2) and subsequently BRCA2 to the break site, where BRCA2 can load RAD51 (58). The interaction between BRCA1 and PALB2 is subject to regulation, representing another step where control of repair pathway choice can be exerted. Ubiquitylation of key lysine residues in PALB2 by the CRL3-KEAP1 E3 ubiquitin ligase complex prevents association of PALB2 and BRCA1 in G1, and therefore it pushes repair toward NHEJ (74). This effect of CRL3-KEAP1 is dependent on low levels of activity of the USP11 DUB, which becomes up-regulated during S phase, leading to deubiquitylation of PALB2 and formation of the pro-HR BRCA1–PALB2 complex. Phosphorylation of PALB2 also regulates its ability to form complexes with BRCA1, thereby regulating HR at specific phases of the cell cycle. Notably, ATR activation at resected DSBs leads to phosphorylation of Ser-59 in PALB2, which enhances its complex formation with BRCA1 (75).
A number of factors in mammalian cells have “anti-recombinase” activity, which means they antagonize the HR pathway (2, 76). These factors include FBH1, PARI, BLM, RECQL5, and DNA polymerase θ. They act either by destabilizing RAD51 binding to resected DNA ends or by disassembling nascent recombination intermediates. At this stage, it is not clear to what extent or how these “anti-recombinases” are regulated to give a particular repair outcome. The RPA-like protein, RADX, which was recently discovered through biochemical studies of factors present at stalled replication forks, acts to restrict the recombinogenic potential of RAD51 (77). In cells lacking BRCA2, this antagonistic effect of RADX on HR contributes to the cytotoxic effects of olaparib or camptothecin, which produce DSBs in S phase. RAD51 is further regulated by RFWD3, an E3 ubiquitin ligase that ubiquitylates RPA and RAD51, stimulating their removal by a VCP/p97-dependent pathway (78). Loss of RFWD3-mediated ubiquitylation results in a failure of HR and a form of Fanconi anemia.
Role of chromatin and nuclear position in regulation of repair pathway choice
Repair of DSBs in heterochromatic regions of the genome requires specialized signaling mechanisms (10), and accumulating evidence supports the idea that the choice of DSB repair pathway is also influenced by the nature of chromatin at the break site (Fig. 4). According to one estimate based on RPA immunofluorescence, 71% of DSBs committed to HR colocalized with the heterochromatin markers H3K9me3 or H4K20me3 (79). This supports the idea that heterochromatic DSBs are primarily repaired by HR. Three studies published in 2014 also demonstrated that HR is the primary pathway for repair of DSBs within transcriptionally-active regions (80–82). In this case, SETD2-mediated formation of H3K36Me3 at the sites of transcriptional elongation forms a binding site for the chromatin-binding factor, LEDGF, which binds after DNA breakage and helps recruit CtIP for DSB resection and HR (83). The histone demethylase, JMJD2A, can reduce the use of HR in these regions by reducing overall abundance of H3K36Me3. The presence of acetylated histones in transcriptionally-active regions, and demethylation by KDM5A, allows binding of the bromodomain protein, ZMYND8 (84). ZMYND8 favors HR, by recruitment of the chromatin-remodeling NuRD complex. Depletion of ZMYND8 reduces HR but has no effect on NHEJ. HR is also favored by the presence of the histone variant macroH2A (85). Interestingly, pericentromeric heterochromatin is repaired mainly by HR in the G2 phase of the cell cycle, whereas centromeric heterochromatins are repaired by both HR and NHEJ (86). This difference further supports the concept that the chromatin environment can influence DSB repair pathway choice. The nuclear microenvironment of a DSB can also influence choice of repair pathway. Site-specific DSBs induced at the nuclear periphery are repaired by end-joining pathways in preference to HR (87).
Figure 4.
Chromatin and nuclear positioning contribute to DSB repair pathway choice. H3K36Me3 and acetylated histones are present in transcriptionally-active regions. These marks promote use of HR for repair of DSBs by LEDGF-mediated recruitment of CtIP and ZMYND8-mediated recruitment of the NuRD complex, which remodels nucleosomes. The presence of macro-H2A also correlates with increased use of HR. Heterochromatic regions of the genome, particularly pericentromeric heterochromatin, are preferentially repaired by HR. Conversely, DSBs that occur near the nuclear periphery show increased repair by NHEJ. See text for details.
Regulation of A-EJ and SSA
Relative to the choice between HR and end-joining mechanisms in general, there has been less research on mechanisms regulating how A-EJ and SSA are regulated. Both of these pathways are dependent on some level of DSB resection, so factors promoting resection are likely to increase the rate of A-EJ and SSA relative to C-NHEJ. The absence of a sister chromatid for HR in the G1 phase of the cell cycle is likely to favor A-EJ and SSA for repair of resected DSBs in G1 (88). Using an RNAi screen, Howard et al. (89) identified 13 repair factors that promote A-EJ versus other end-joining events. These factors include CtIP and members of the Fanconi anemia family. Several of these factors are also involved in mediating HR and SSA. The RECQ helicase, WRN, was shown to regulate the choice between C-NHEJ and A-EJ by “shielding” DSBs from the resection activity of CtIP-MRE11 (90). This “shielding” effect of WRN appears to be independent of its helicase activity. The DNA polymerase Pol θ, which is frequently overexpressed in HR-deficient tumor cells, has emerged as a key regulator of A-EJ versus other pathways for the repair of DSBs (76, 91–94). The helicase domain of Pol θ appears to restrict HR and promote A-EJ by removing RPA from resected DNA ends (95). 53BP1 has been shown to act as an important suppressor of A-EJ in B cells (96). This effect of 53BP1 seems to stem from its ability to suppress resection of DSBs. Reduced resection of DSBs by 53BP1 also impacts the choice between A-EJ and SSA. When 53BP1 is absent or present in a limited amount, resection of DSBs is so great as to exhaust the available cellular stock of RAD51 (97). Under these conditions, RAD52-mediated SSA takes over from RAD51-mediated HR. 53BP1 therefore acts to “foster” the more “faithful” pathways of DNA repair, i.e. C-NHEJ and HR instead of A-EJ and SSA. A further determinant of the choice of HR versus SSA depends on the interaction of BRCA1 and PALB2. Mutations in either of these factors that prevent their normal association increase the use of SSA for repair of resected DSBs at the expense of HR (98).
Perspective
Recent years have brought a great deal of new knowledge about cellular activities that regulate the choice of the DSB repair pathway in mammalian cells. In many cases, however, the exact mechanism by which these factors work is not clear. Future work will hopefully test the relative importance of different regulatory mechanisms and measure to what extent they are relevant in cells of different lineages and disease states. The hope is that with greater knowledge of how DNA repair is regulated, we will identify approaches to selectively disrupt certain repair pathways to reduce the frequency of mutations that lead to malignant cell growth or to kill cancer cells that become dependent on one particular repair pathway.
Acknowledgments
We are grateful to Drs. Bing Xia and Nina Peel for critical review of the manuscript.
This work was supported by National Institutes of Health Grant R01 CA190858 from NCI (to the S. F. B. laboratory) and an NJCCR postdoctoral fellowship (to J. H.). This is the first article in the Thematic Minireview series “DNA double-strand break repair and pathway choice.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DSB
- double-strand break
- HR
- homologous recombination
- NHEJ
- nonhomologous end-joining
- A-EJ
- alternative nonhomologous end-joining
- SSA
- single-strand annealing
- DUB
- deubiquitinating
- C-NHEJ
- canonical nonhomologous end-joining
- CDK
- cyclin-dependent kinase
- ATM
- ataxia telangiectasia-mutated
- RPA
- replication protein A.
References
- 1. Helleday T., Eshtad S., and Nik-Zainal S. (2014) Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15, 585–598 10.1038/nrg3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kowalczykowski S. C. (2015) An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7, a016410 10.1101/cshperspect.a016410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pannunzio N. R., Watanabe G., and Lieber M. R. (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 293, 10512–10523 10.1074/jbc.TM117.000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beucher A., Birraux J., Tchouandong L., Barton O., Shibata A., Conrad S., Goodarzi A. A., Krempler A., Jeggo P. A., and Lobrich M. (2009) ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 28, 3413–3427 10.1038/emboj.2009.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karanam K., Kafri R., Loewer A., and Lahav G. (2012) Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell 47, 320–329 10.1016/j.molcel.2012.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mao Z., Bozzella M., Seluanov A., and Gorbunova V. (2008) Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 7, 1765–1771 10.1016/j.dnarep.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rothkamm K., Krüger I., Thompson L. H., and Löbrich M. (2003) Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23, 5706–5715 10.1128/MCB.23.16.5706-5715.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shahar O. D., Raghu Ram E. V., Shimshoni E., Hareli S., Meshorer E., and Goldberg M. (2012) Live imaging of induced and controlled DNA double-strand break formation reveals extremely low repair by homologous recombination in human cells. Oncogene 31, 3495–3504 10.1038/onc.2011.516 [DOI] [PubMed] [Google Scholar]
- 9. Reynolds P., Anderson J. A., Harper J. V., Hill M. A., Botchway S. W., Parker A. W., and O'Neill P. (2012) The dynamics of Ku70/80 and DNA-PKcs at DSBs induced by ionizing radiation is dependent on the complexity of damage. Nucleic Acids Res. 40, 10821–10831 10.1093/nar/gks879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shibata A., Conrad S., Birraux J., Geuting V., Barton O., Ismail A., Kakarougkas A., Meek K., Taucher-Scholz G., Löbrich M., and Jeggo P. A. (2011) Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 30, 1079–1092 10.1038/emboj.2011.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bothmer A., Phadke T., Barrera L. A., Margulies C. M., Lee C. S., Buquicchio F., Moss S., Abdulkerim H. S., Selleck W., Jayaram H., Myer V. E., and Cotta-Ramusino C. (2017) Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat. Commun. 8, 13905 10.1038/ncomms13905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mujoo K., Pandita R. K., Tiwari A., Charaka V., Chakraborty S., Singh D. K., Hambarde S., Hittelman W. N., Horikoshi N., Hunt C. R., Khanna K. K., Kots A. Y., Butler E. B., Murad F., and Pandita T. K. (2017) Differentiation of human induced pluripotent or embryonic stem cells decreases the DNA damage repair by homologous recombination. Stem Cell Rep. 9, 1660–1674 10.1016/j.stemcr.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Orthwein A., Fradet-Turcotte A., Noordermeer S. M., Canny M. D., Brun C. M., Strecker J., Escribano-Diaz C., and Durocher D. (2014) Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science 344, 189–193 10.1126/science.1248024 [DOI] [PubMed] [Google Scholar]
- 14. Huertas P., and Jackson S. P. (2009) Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buis J., Stoneham T., Spehalski E., and Ferguson D. O. (2012) Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat. Struct. Mol. Biol. 19, 246–252 10.1038/nsmb.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Biehs R., Steinlage M., Barton O., Juhász S., Künzel J., Spies J., Shibata A., Jeggo P. A., and Löbrich M. (2017) DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol. Cell 65, 671–684 10.1016/j.molcel.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu H., Shamanna R. A., de Freitas J. K., Okur M., Khadka P., Kulikowicz T., Holland P. P., Tian J., Croteau D. L., Davis A. J., and Bohr V. A. (2017) Cell cycle-dependent phosphorylation regulates RECQL4 pathway choice and ubiquitination in DNA double-strand break repair. Nat. Commun. 8, 2039 10.1038/s41467-017-02146-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rai R., Hu C., Broton C., Chen Y., Lei M., and Chang S. (2017) NBS1 phosphorylation status dictates repair choice of dysfunctional telomeres. Mol. Cell 65, 801–817.e4 10.1016/j.molcel.2017.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ismail I. H., Gagné J. P., Genois M. M., Strickfaden H., McDonald D., Xu Z., Poirier G. G., Masson J. Y., and Hendzel M. J. (2015) The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat. Cell Biol. 17, 1446–1457 10.1038/ncb3259 [DOI] [PubMed] [Google Scholar]
- 20. Schmidt C. K., Galanty Y., Sczaniecka-Clift M., Coates J., Jhujh S., Demir M., Cornwell M., Beli P., and Jackson S. P. (2015) Systematic E2 screening reveals a UBE2D-RNF138-CtIP axis promoting DNA repair. Nat. Cell Biol. 17, 1458–1470 10.1038/ncb3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng L., and Chen J. (2012) The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat. Struct. Mol. Biol. 19, 201–206 10.1038/nsmb.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishida N., Nakagawa T., Iemura S. I., Yasui A., Shima H., Katoh Y., Nagasawa Y., Natsume T., Igarashi K., and Nakayama K. (2017) Ubiquitylation of Ku80 by RNF126 promotes completion of nonhomologous end joining-mediated DNA repair. Mol. Cell. Biol. 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Postow L., and Funabiki H. (2013) An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle 12, 587–595 10.4161/cc.23408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van den Boom J., Wolf M., Weimann L., Schulze N., Li F., Kaschani F., Riemer A., Zierhut C., Kaiser M., Iliakis G., Funabiki H., and Meyer H. (2016) VCP/p97 extracts sterically trapped Ku70/80 rings from DNA in double-strand break repair. Mol. Cell 64, 189–198 10.1016/j.molcel.2016.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown J. S., Lukashchuk N., Sczaniecka-Clift M., Britton S., le Sage C., Calsou P., Beli P., Galanty Y., and Jackson S. P. (2015) Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites. Cell Rep. 11, 704–714 10.1016/j.celrep.2015.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee K. J., Saha J., Sun J., Fattah K. R., Wang S. C., Jakob B., Chi L., Wang S. Y., Taucher-Scholz G., Davis A. J., and Chen D. J. (2016) Phosphorylation of Ku dictates DNA double-strand break (DSB) repair pathway choice in S phase. Nucleic Acids Res. 44, 1732–1745 10.1093/nar/gkv1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Y., Lee J. H., Jiang W., Crowe J. L., Zha S., and Paull T. T. (2017) Regulation of the DNA damage response by DNA-PKcs inhibitory phosphorylation of ATM. Mol. Cell 65, 91–104 10.1016/j.molcel.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arnoult N., Correia A., Ma J., Merlo A., Garcia-Gomez S., Maric M., Tognetti M., Benner C. W., Boulton S. J., Saghatelian A., and Karlseder J. (2017) Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552 10.1038/nature24023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bunting S. F., Callén E., Wong N., Chen H. T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., Xu X., Deng C. X., Finkel T., Nussenzweig M., Stark J. M., and Nussenzweig A. (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shibata A., Moiani D., Arvai A. S., Perry J., Harding S. M., Genois M. M., Maity R., van Rossum-Fikkert S., Kertokalio A., Romoli F., Ismail A., Ismalaj E., Petricci E., Neale M. J., Bristow R. G., et al. (2014) DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18 10.1016/j.molcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaidi A., Weinert B. T., Choudhary C., and Jackson S. P. (2010) Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329, 1348–1353 10.1126/science.1192049 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Soria-Bretones I., Cepeda-García C., Checa-Rodriguez C., Heyer V., Reina-San-Martin B., Soutoglou E., and Huertas P. (2017) DNA end resection requires constitutive sumoylation of CtIP by CBX4. Nat. Commun. 8, 113 10.1038/s41467-017-00183-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferretti L. P., Himmels S. F., Trenner A., Walker C., von Aesch C., Eggenschwiler A., Murina O., Enchev R. I., Peter M., Freire R., Porro A., and Sartori A. A. (2016) Cullin3-KLHL15 ubiquitin ligase mediates CtIP protein turnover to fine-tune DNA-end resection. Nat. Commun. 7, 12628 10.1038/ncomms12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jimeno S., Fernández-Ávila M. J., Cruz-García A., Cepeda-García C., Gómez-Cabello D., and Huertas P. (2015) Neddylation inhibits CtIP-mediated resection and regulates DNA double strand break repair pathway choice. Nucleic Acids Res. 43, 987–999 10.1093/nar/gku1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Polato F., Callen E., Wong N., Faryabi R., Bunting S., Chen H. T., Kozak M., Kruhlak M. J., Reczek C. R., Lee W. H., Ludwig T., Baer R., Feigenbaum L., Jackson S., and Nussenzweig A. (2014) CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med. 211, 1027–1036 10.1084/jem.20131939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reczek C. R., Szabolcs M., Stark J. M., Ludwig T., and Baer R. (2013) The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 201, 693–707 10.1083/jcb.201302145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lou J., Chen H., Han J., He H., Huen M. S. Y., Feng X. H., Liu T., and Huang J. (2017) AUNIP/C1orf135 directs DNA double-strand breaks towards the homologous recombination repair pathway. Nat. Commun. 8, 985 10.1038/s41467-017-01151-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Daddacha W., Koyen A. E., Bastien A. J., Head P. E., Dhere V. R., Nabeta G. N., Connolly E. C., Werner E., Madden M. Z., Daly M. B., Minten E. V., Whelan D. R., Schlafstein A. J., Zhang H., Anand R., et al. (2017) SAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep. 20, 1921–1935 10.1016/j.celrep.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu H., Shamanna R. A., Keijzers G., Anand R., Rasmussen L. J., Cejka P., Croteau D. L., and Bohr V. A. (2016) RECQL4 promotes DNA end resection in repair of DNA double-strand breaks. Cell Rep. 16, 161–173 10.1016/j.celrep.2016.05.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gong Y., Handa N., Kowalczykowski S. C., and de Lange T. (2017) PHF11 promotes DSB resection, ATR signaling, and HR. Genes Dev. 31, 46–58 10.1101/gad.291807.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tkáč J., Xu G., Adhikary H., Young J. T. F., Gallo D., Escribano-Díaz C., Krietsch J., Orthwein A., Munro M., Sol W., Al-Hakim A., Lin Z. Y., Jonkers J., Borst P., Brown G. W., et al. (2016) HELB is a feedback inhibitor of DNA end resection. Mol. Cell 61, 405–418 10.1016/j.molcel.2015.12.013 [DOI] [PubMed] [Google Scholar]
- 42. Bouwman P., Aly A., Escandell J. M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., Haffty B. G., Tommiska J., Blomqvist C., Drapkin R., Adams D. J., Nevanlinna H., et al. (2010) 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., and Mer G. (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fradet-Turcotte A., Canny M. D., Escribano-Díaz C., Orthwein A., Leung C. C., Huang H., Landry M. C., Kitevski-LeBlanc J., Noordermeer S. M., Sicheri F., and Durocher D. (2013) 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pellegrino S., Michelena J., Teloni F., Imhof R., and Altmeyer M. (2017) Replication-coupled dilution of H4K20me2 guides 53BP1 to pre-replicative chromatin. Cell Rep. 19, 1819–1831 10.1016/j.celrep.2017.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Acs K., Luijsterburg M. S., Ackermann L., Salomons F. A., Hoppe T., and Dantuma N. P. (2011) The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 18, 1345–1350 10.1038/nsmb.2188 [DOI] [PubMed] [Google Scholar]
- 47. Mallette F. A., Mattiroli F., Cui G., Young L. C., Hendzel M. J., Mer G., Sixma T. K., and Richard S. (2012) RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31, 1865–1878 10.1038/emboj.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Butler L. R., Densham R. M., Jia J., Garvin A. J., Stone H. R., Shah V., Weekes D., Festy F., Beesley J., and Morris J. R. (2012) The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 31, 3918–3934 10.1038/emboj.2012.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kato K., Nakajima K., Ui A., Muto-Terao Y., Ogiwara H., and Nakada S. (2014) Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol. Cell 53, 617–630 10.1016/j.molcel.2014.01.030 [DOI] [PubMed] [Google Scholar]
- 50. Drané P., Brault M. E., Cui G., Meghani K., Chaubey S., Detappe A., Parnandi N., He Y., Zheng X. F., Botuyan M. V., Kalousi A., Yewdell W. T., Münch C., Harper J. W., Chaudhuri J., et al. (2017) TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature 543, 211–216 10.1038/nature21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jacquet K., Fradet-Turcotte A., Avvakumov N., Lambert J. P., Roques C., Pandita R. K., Paquet E., Herst P., Gingras A. C., Pandita T. K., Legube G., Doyon Y., Durocher D., and Côté J. (2016) The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol. Cell 62, 409–421 10.1016/j.molcel.2016.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tang J., Cho N. W., Cui G., Manion E. M., Shanbhag N. M., Botuyan M. V., Mer G., and Greenberg R. A. (2013) Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20, 317–325 10.1038/nsmb.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu Q., Botuyan M. V., Cui G., Zhao D., and Mer G. (2017) Mechanisms of ubiquitin-nucleosome recognition and regulation of 53BP1 chromatin recruitment by RNF168/169 and RAD18. Mol. Cell 66, 473–487.e9 10.1016/j.molcel.2017.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kitevski-LeBlanc J., Fradet-Turcotte A., Kukic P., Wilson M. D., Portella G., Yuwen T., Panier S., Duan S., Canny M. D., van Ingen H., Arrowsmith C. H., Rubinstein J. L., Vendruscolo M., Durocher D., and Kay L. E. (2017) The RNF168 paralog RNF169 defines a new class of ubiquitylated histone reader involved in the response to DNA damage. eLife 6, e23872 10.7554/eLife.23872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo X., Bai Y., Zhao M., Zhou M., Shen Q., Yun C. H., Zhang H., Zhu W. G., and Wang J. (2018) Acetylation of 53BP1 dictates the DNA double strand break repair pathway. Nucleic Acids Res. 46, 689–703 10.1093/nar/gkx1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee D. H., Acharya S. S., Kwon M., Drane P., Guan Y., Adelmant G., Kalev P., Shah J., Pellman D., Marto J. A., and Chowdhury D. (2014) Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol. Cell 54, 512–525 10.1016/j.molcel.2014.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mayca Pozo F., Tang J., Bonk K. W., Keri R. A., Yao X., and Zhang Y. (2017) Regulatory cross-talk determines the cellular levels of 53BP1 protein, a critical factor in DNA repair. J. Biol. Chem. 292, 5992–6003 10.1074/jbc.M116.760645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Prakash R., Zhang Y., Feng W., and Jasin M. (2015) Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7, a016600 10.1101/cshperspect.a016600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Coleman K. A., and Greenberg R. A. (2011) The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J. Biol. Chem. 286, 13669–13680 10.1074/jbc.M110.213728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hu Y., Scully R., Sobhian B., Xie A., Shestakova E., and Livingston D. M. (2011) RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 25, 685–700 10.1101/gad.2011011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Leung J. W., Makharashvili N., Agarwal P., Chiu L. Y., Pourpre R., Cammarata M. B., Cannon J. R., Sherker A., Durocher D., Brodbelt J. S., Paull T. T., and Miller K. M. (2017) ZMYM3 regulates BRCA1 localization at damaged chromatin to promote DNA repair. Genes Dev. 31, 260–274 10.1101/gad.292516.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Batenburg N. L., Thompson E. L., Hendrickson E. A., and Zhu X. D. (2015) Cockayne syndrome group B protein regulates DNA double-strand break repair and checkpoint activation. EMBO J. 34, 1399–1416 10.15252/embj.201490041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang H., Liu H., Chen Y., Yang X., Wang P., Liu T., Deng M., Qin B., Correia C., Lee S., Kim J., Sparks M., Nair A. A., Evans D. L., Kalari K. R., et al. (2016) A cell cycle-dependent BRCA1-UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nat. Commun. 7, 10201 10.1038/ncomms10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Isono M., Niimi A., Oike T., Hagiwara Y., Sato H., Sekine R., Yoshida Y., Isobe S. Y., Obuse C., Nishi R., Petricci E., Nakada S., Nakano T., and Shibata A. (2017) BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep. 18, 520–532 10.1016/j.celrep.2016.12.042 [DOI] [PubMed] [Google Scholar]
- 65. Densham R. M., Garvin A. J., Stone H. R., Strachan J., Baldock R. A., Daza-Martin M., Fletcher A., Blair-Reid S., Beesley J., Johal B., Pearl L. H., Neely R., Keep N. H., Watts F. Z., and Morris J. R. (2016) Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 23, 647–655 10.1038/nsmb.3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shakya R., Reid L. J., Reczek C. R., Cole F., Egli D., Lin C. S., deRooij D. G., Hirsch S., Ravi K., Hicks J. B., Szabolcs M., Jasin M., Baer R., and Ludwig T. (2011) BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334, 525–528 10.1126/science.1209909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Callen E., Di Virgilio M., Kruhlak M. J., Nieto-Soler M., Wong N., Chen H. T., Faryabi R. B., Polato F., Santos M., Starnes L. M., Wesemann D. R., Lee J. E., Tubbs A., Sleckman B. P., Daniel J. A., et al. (2013) 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153, 1266–1280 10.1016/j.cell.2013.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A. D., Feldhahn N., Resch W., Oliveira T. Y., Chait B. T., Nussenzweig A., Casellas R., Robbiani D. F., and Nussenzweig M. C. (2013) Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339, 711–715 10.1126/science.1230624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Boersma V., Moatti N., Segura-Bayona S., Peuscher M. H., van der Torre J., Wevers B. A., Orthwein A., Durocher D., and Jacobs J. J. (2015) MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature 521, 537–540 10.1038/nature14216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xu G., Chapman J. R., Brandsma I., Yuan J., Mistrik M., Bouwman P., Bartkova J., Gogola E., Warmerdam D., Barazas M., Jaspers J. E., Watanabe K., Pieterse M., Kersbergen A., Sol W., et al. (2015) REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544 10.1038/nature14328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang J., Aroumougame A., Lobrich M., Li Y., Chen D., Chen J., and Gong Z. (2014) PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev. 28, 2693–2698 10.1101/gad.252478.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Isobe S. Y., Nagao K., Nozaki N., Kimura H., and Obuse C. (2017) Inhibition of RIF1 by SCAI allows BRCA1-mediated repair. Cell Rep. 20, 297–307 10.1016/j.celrep.2017.06.056 [DOI] [PubMed] [Google Scholar]
- 73. Hansen R. K., Mund A., Poulsen S. L., Sandoval M., Klement K., Tsouroula K., Tollenaere M. A., Räschle M., Soria R., Offermanns S., Worzfeld T., Grosse R., Brandt D. T., Rozell B., Mann M., et al. (2016) SCAI promotes DNA double-strand break repair in distinct chromosomal contexts. Nat. Cell Biol. 18, 1357–1366 10.1038/ncb3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Orthwein A., Noordermeer S. M., Wilson M. D., Landry S., Enchev R. I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G., Xia B., Peter M., and Durocher D. (2015) A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 10.1038/nature16142 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75. Buisson R., Niraj J., Rodrigue A., Ho C. K., Kreuzer J., Foo T. K., Hardy E. J., Dellaire G., Haas W., Xia B., Masson J. Y., and Zou L. (2017) Coupling of homologous recombination and the checkpoint by ATR. Mol. Cell 65, 336–346 10.1016/j.molcel.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ceccaldi R., Liu J. C., Amunugama R., Hajdu I., Primack B., Petalcorin M. I., O'Connor K. W., Konstantinopoulos P. A., Elledge S. J., Boulton S. J., Yusufzai T., and D'Andrea A. D. (2015) Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 10.1038/nature14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dungrawala H., Bhat K. P., Le Meur R., Chazin W. J., Ding X., Sharan S. K., Wessel S. R., Sathe A. A., Zhao R., and Cortez D. (2017) RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol. Cell 67, 374–386.e5 10.1016/j.molcel.2017.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Inano S., Sato K., Katsuki Y., Kobayashi W., Tanaka H., Nakajima K., Nakada S., Miyoshi H., Knies K., Takaori-Kondo A., Schindler D., Ishiai M., Kurumizaka H., and Takata M. (2017) RFWD3-mediated ubiquitination promotes timely removal of both RPA and RAD51 from DNA damage sites to facilitate homologous recombination. Mol. Cell 66, 622–634 10.1016/j.molcel.2017.04.022 [DOI] [PubMed] [Google Scholar]
- 79. Kakarougkas A., Ismail A., Klement K., Goodarzi A. A., Conrad S., Freire R., Shibata A., Lobrich M., and Jeggo P. A. (2013) Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 41, 9719–9731 10.1093/nar/gkt729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Aymard F., Bugler B., Schmidt C. K., Guillou E., Caron P., Briois S., Iacovoni J. S., Daburon V., Miller K. M., Jackson S. P., and Legube G. (2014) Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 10.1038/nsmb.2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Carvalho S., Vítor A. C., Sridhara S. C., Martins F. B., Raposo A. C., Desterro J. M., Ferreira J., and de Almeida S. F. (2014) SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife 3, e02482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pfister S. X., Ahrabi S., Zalmas L. P., Sarkar S., Aymard F., Bachrati C. Z., Helleday T., Legube G., La Thangue N. B., Porter A. C., and Humphrey T. C. (2014) SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7, 2006–2018 10.1016/j.celrep.2014.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Daugaard M., Baude A., Fugger K., Povlsen L. K., Beck H., Sørensen C. S., Petersen N. H., Sorensen P. H., Lukas C., Bartek J., Lukas J., Rohde M., and Jäättelä M. (2012) LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 19, 803–810 10.1038/nsmb.2314 [DOI] [PubMed] [Google Scholar]
- 84. Gong F., Clouaire T., Aguirrebengoa M., Legube G., and Miller K. M. (2017) Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 216, 1959–1974 10.1083/jcb.201611135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Khurana S., Kruhlak M. J., Kim J., Tran A. D., Liu J., Nyswaner K., Shi L., Jailwala P., Sung M. H., Hakim O., and Oberdoerffer P. (2014) A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 8, 1049–1062 10.1016/j.celrep.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tsouroula K., Furst A., Rogier M., Heyer V., Maglott-Roth A., Ferrand A., Reina-San-Martin B., and Soutoglou E. (2016) Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol. Cell 63, 293–305 10.1016/j.molcel.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 87. Lemaître C., Grabarz A., Tsouroula K., Andronov L., Furst A., Pankotai T., Heyer V., Rogier M., Attwood K. M., Kessler P., Dellaire G., Klaholz B., Reina-San-Martin B., and Soutoglou E. (2014) Nuclear position dictates DNA repair pathway choice. Genes Dev. 28, 2450–2463 10.1101/gad.248369.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bhargava R., Onyango D. O., and Stark J. M. (2016) Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32, 566–575 10.1016/j.tig.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Howard S. M., Yanez D. A., and Stark J. M. (2015) DNA damage–response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 11, e1004943 10.1371/journal.pgen.1004943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shamanna R. A., Lu H., de Freitas J. K., Tian J., Croteau D. L., and Bohr V. A. (2016) WRN regulates pathway choice between classical and alternative non-homologous end joining. Nat. Commun. 7, 13785 10.1038/ncomms13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kent T., Chandramouly G., McDevitt S. M., Ozdemir A. Y., and Pomerantz R. T. (2015) Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 22, 230–237 10.1038/nsmb.2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mateos-Gomez P. A., Gong F., Nair N., Miller K. M., Lazzerini-Denchi E., and Sfeir A. (2015) Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 10.1038/nature14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wyatt D. W., Feng W., Conlin M. P., Yousefzadeh M. J., Roberts S. A., Mieczkowski P., Wood R. D., Gupta G. P., and Ramsden D. A. (2016) Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell 63, 662–673 10.1016/j.molcel.2016.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yousefzadeh M. J., Wyatt D. W., Takata K., Mu Y., Hensley S. C., Tomida J., Bylund G. O., Doublié S., Johansson E., Ramsden D. A., McBride K. M., and Wood R. D. (2014) Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10, e1004654 10.1371/journal.pgen.1004654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mateos-Gomez P. A., Kent T., Deng S. K., McDevitt S., Kashkina E., Hoang T. M., Pomerantz R. T., and Sfeir A. (2017) The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol. 24, 1116–1123 10.1038/nsmb.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Panchakshari R. A., Zhang X., Kumar V., Du Z., Wei P. C., Kao J., Dong J., and Alt F. W. (2018) DNA double-strand break response factors influence end-joining features of IgH class switch and general translocation junctions. Proc. Natl. Acad. Sci. U.S.A. 115, 762–767 10.1073/pnas.1719988115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ochs F., Somyajit K., Altmeyer M., Rask M. B., Lukas J., and Lukas C. (2016) 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol. 23, 714–721 10.1038/nsmb.3251 [DOI] [PubMed] [Google Scholar]
- 98. Anantha R. W., Simhadri S., Foo T. K., Miao S., Liu J., Shen Z., Ganesan S., and Xia B. (2017) Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. eLife 6, e21350 10.7554/eLife.21350 [DOI] [PMC free article] [PubMed] [Google Scholar]