Abstract
Nonhomologous DNA end-joining (NHEJ) is the predominant double-strand break (DSB) repair pathway throughout the cell cycle and accounts for nearly all DSB repair outside of the S and G2 phases. NHEJ relies on Ku to thread onto DNA termini and thereby improve the affinity of the NHEJ enzymatic components consisting of polymerases (Pol μ and Pol λ), a nuclease (the Artemis·DNA-PKcs complex), and a ligase (XLF·XRCC4·Lig4 complex). Each of the enzymatic components is distinctive for its versatility in acting on diverse incompatible DNA end configurations coupled with a flexibility in loading order, resulting in many possible junctional outcomes from one DSB. DNA ends can either be directly ligated or, if the ends are incompatible, processed until a ligatable configuration is achieved that is often stabilized by up to 4 bp of terminal microhomology. Processing of DNA ends results in nucleotide loss or addition, explaining why DSBs repaired by NHEJ are rarely restored to their original DNA sequence. Thus, NHEJ is a single pathway with multiple enzymes at its disposal to repair DSBs, resulting in a diversity of repair outcomes.
Keywords: DNA repair, nucleic acid enzymology, protein structure, DNA endonuclease, DNA-dependent serine/threonine protein kinase (DNA-PK), double-stranded DNA breaks, NHEJ
Introduction
Eukaryotic cells have evolved to repair multiple forms of DNA damage to maintain a high level of fidelity between cell divisions. Among types of damage, DNA double-strand breaks (DSBs)3 are particularly detrimental as they can result in insertions, deletions, or chromosomal translocations that are the primary transforming step in many human cancers. Pathological DSBs can arise from both exogenous (e.g. ionizing radiation or reactive oxygen species) or endogenous (e.g. DNA replication errors or incidental action by nuclear enzymes) sources. In some cases, DSBs are required as part of a physiological process, such as the breaks that occur during V(D)J recombination and immunoglobulin heavy chain (IgH) class switch recombination (1). Both pathological and physiological DSBs require efficient processes for repair that result in minimal to no change to the broken chromosome. Repair mechanisms can be largely divided between those that use extensive homology from a sister chromatid or homologous sequence elsewhere in the genome and those that use little to no homology. Both mechanisms require end processing by nucleases, utilization of DNA polymerases, and a final ligation step to complete repair of the broken DNA (Fig. 1). Nonhomologous DNA end-joining (NHEJ) was originally a phrase used to describe a type of illegitimate repair that utilizes little to no long homology (2) (we feel it unnecessary to include the word “canonical” or use the term “c-NHEJ” as we consider NHEJ a stand-alone pathway that does not need to be described in reference to separate alternative end-joining pathways that have their own distinct components). “Nonhomologous” could be misinterpreted as meaning completely homology-independent by a newcomer to the field, but up to 4 bp of microhomology during repair is common for NHEJ, and the term is simply meant to contrast with “homologous” recombination (HR), which can use several hundred base pairs of homology as a template for high-fidelity repair. In NHEJ, the DSB is first recognized by a heterodimer consisting of Ku70 and Ku80 (Ku). The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) has a high affinity for DNA ends, which is even tighter when Ku is bound to that end (3). The nuclease, Artemis, exists in tight complex with DNA-PKcs within the cell and is likely recruited along with DNA-PKcs (4). Nucleotide addition can occur by the Pol X family polymerases, Pol μ and Pol λ. Finally, the DNA ligase IV complex, including XRCC4, XLF, and perhaps PAXX, carries out the critical ligation step for either strand of the DSB.
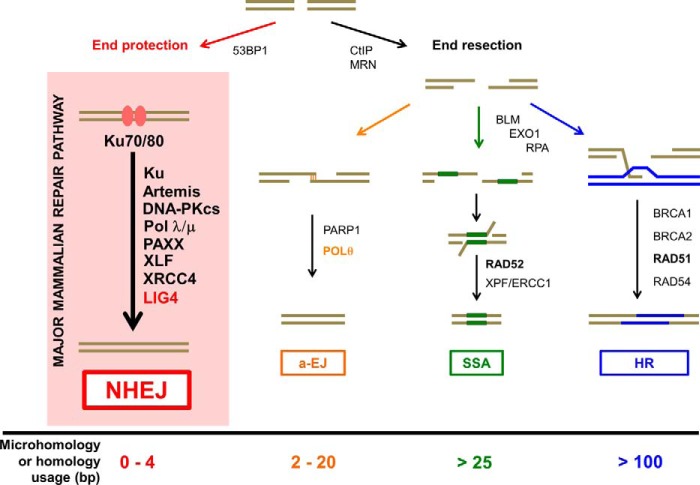
Figure 1.
NHEJ in the context of other double-strand break repair pathways. DNA double-strand breaks (DSBs) can be repaired by NHEJ, alternative end-joining (a-EJ), single-strand annealing (SSA), or homologous recombination (HR). Pathway choice and pathways other than NHEJ are discussed in other Minireviews in this thematic series. The name NHEJ originally arose to distinguish it from repair that requires extensive DNA homology (i.e. HR and SSA). Lengths of terminal microhomology (MH) between 1 and 4 bp are common in NHEJ. a-EJ is also called microhomology-mediated end joining (MMEJ) or Pol θ-mediated end joining (TMEJ). The major difference in the pathways is the requirement for significant DNA end resection. The p53-binding protein 1 (53BP1) is a chromatin remodeler and a positive regulator for NHEJ. Although Artemis·DNA-PKcs can carry out some nucleolytic resection (typically <20 nt), the NHEJ pathway does not require extensive end resection, and the ends are protected from deeper resection by the binding of the Ku heterodimer (Ku70–80) to the DNA ends. By contrast, the C-terminal binding protein-interacting protein (CtIP) and the MRN (MRE11 (meiotic recombination 11)·RAD50·NBS1 (Nijmegen breakage syndrome protein 1)) complexes are involved in extensive 5′ to 3′ resection of regions of the duplex, and this generates stretches of ssDNA at DNA ends for a-EJ, SSA, and HR. SSA typically requires >25 bp of microhomology, whereas the requirement for a-EJ is typically <20 bp. Poly(ADP-ribose) polymerase 1 (PARP1) and Pol θ are important for a-EJ. Bloom syndrome RecQ-like helicase (BLM) and exonuclease 1 (EXO1) account for additional resection, and replication protein A (RPA) binds to ssDNA to promote the SSA and HR pathways. RAD52-mediated annealing of homologous sequence is key for the SSA pathway. XPF-ERCC1 cuts the remaining 3′ nonhomologous ssDNA prior to ligation by DNA ligase 1. By contrast, RAD51-mediated strand exchange with its association with BRCA1, BRCA2, and RAD54 is essential for facilitating the HR pathway.
Importantly, NHEJ is an iterative process, where each of the DNA ends involved in the break can be acted upon by these components multiple times and in a different order (Fig. S1). Other important factors that dictate repair are the differential requirements for the various NHEJ proteins depending on the configuration of the DNA ends, which can include blunt ends, 5′ or 3′ overhangs, or ends containing adducts refractory to processing or ligation. Recent work has begun to systematically examine how various DNA end configurations are processed differently (5, 6). We briefly mention how NHEJ relates to the other pathways of double-strand break repair, but our major focus is the NHEJ process. Therefore, readers are directed to the other works in this Thematic Minireview series for a detailed explanation of other DSB repair mechanisms.
Overview of NHEJ in humans and its relationship with other pathways of double-strand break repair
In human cells, NHEJ appears to repair nearly all DSBs outside of S and G2 cell cycle phases and even about 80% of DSBs within S and G2 that are not proximal to a replication fork (Fig. 1) (7). In late S and G2, HR is another major pathway for DSB repair, relying on more extensive homology tracts as a template for repair (8). When NHEJ is compromised due to the absence of one or more key protein components, the activities of other DNA end-joining pathways that typically involve more extensive end resection become apparent. Greater levels of 5′ end resection expose homologous sequences embedded on either side of a DSB, allowing for stable annealing of 3′ single-stranded DNA (ssDNA) that promotes more efficient joining and ligation (9). Although NHEJ usually requires ≤4 bp of microhomology, the alternative end-joining (a-EJ) pathway (also known as Pol θ-mediated end-joining or microhomology-mediated end-joining) (Fig. 1) (10), which utilizes the additional factors of poly(ADP-ribose) polymerase and DNA Pol θ, requires microhomology that ranges between 2 and 20 bp. Although NHEJ dominates DSB repair in most mammalian somatic cells, Pol θ-mediated events appear at an observable frequency in certain cell types (11), for certain repair events (12), and in some organisms (13). Greater levels of resection can further promote the nonconservative homology-directed repair pathway of single-strand annealing (SSA) that requires >25 bp of homology (Fig. 1) (14–17). Therefore, the mechanisms of NHEJ and HR occur on opposite ends of a spectrum with respect to homology usage with a-EJ and SSA occurring between them on a gradient of increasing levels of DNA end resection and homology usage (6).
A key reason for the dominance of NHEJ is that extensive DNA end resection is prevented by Ku binding (18), and the tight affinity and high abundance of Ku in cells increases the likelihood that Ku is the first protein to bind at a broken DNA end (6) (Fig. 1). A small protein called CYREN (cell cycle regulator of NHEJ (69 aa); also called MRI-2, a sub-peptide of C7orf49 (157 aa)) has been proposed to affect Ku DNA binding (not specified how) and thus favor the HR pathway choice in S/G2 (19), although the data on CYREN effects on Ku binding are conflicting (20). Signaling factors appear to be important in controlling resection, as there is evidence that the DNA damage-response protein p53-binding protein 1 (53BP1) is antagonistic to end resection, acting through a number of effector proteins (21, 22). 53BP1 and mediator of DNA damage checkpoint protein 1 (MDC1) are recruited to DSBs through a number of modified histone residues and appear to have distinct roles in DSB repair (8, 23, 24). Further work is required to elucidate specifically how 53BP1 recruitment inhibits extensive end resection. Overcoming this barrier to resection, however, is the first step to enable either a-EJ or SSA.
Following commitment to NHEJ, the nuclease, polymerase, and ligase components act on the DNA ends until repair is complete. Pathway commitment likely is not final until the strands of the break site are ligated, and if the DSB remains unrepaired, the repeated processing of ends may shift repair to another pathway. Below we provide a brief overview of the types of proteins that are involved in NHEJ and their functions, which applies to nearly all vertebrates.
The nucleases of NHEJ
Direct ligation of broken DNA ends is often impeded due to end incompatibility caused by mismatching overhangs or chemical modifications (Fig. 1). Therefore, following commitment to NHEJ, nucleases are required to process mismatched or modified ends to prepare them for ligation. This typically involves removing short regions of the 5′ or 3′ overhangs by either exonucleolytic or endonucleolytic processing to expose short regions of microhomology (≤4 nt) between the strands that can facilitate end joining. Extensive end resection (≥20 nt), which occurs to initiate HR or SSA pathways, is prevented by the presence of Ku, distinguishing the end processing of NHEJ from other DSB repair pathways. When DNA resection is required for NHEJ, DNA-PKcs is recruited in complex with the nuclease Artemis to Ku-bound DNA ends. DNA-PKcs undergoes autophosphorylation and activates Artemis (25, 26). Artemis then gains the ability to cut DNA ends at single-strand–to–double-strand DNA (ss–dsDNA) boundaries, which includes all overhangs and other structures such as gaps, loops, and bubbles that may arise due to mismatches between the two DNA ends being joined (27, 28).
Artemis is a member of the metallo-β-lactamase family of nucleases, containing the conserved metallo-β-lactamase and β-CASP domains. This family of nucleases has the ability to hydrolyze DNA or RNA in various configurations (29). In addition to an intrinsic 5′ exonuclease activity on ssDNA that does not require DNA-PKcs (30, 31), Artemis possesses a DNA–PKcs-dependent endonuclease activity on both 5′ and 3′ DNA overhangs of duplex DNA. Such overhangs often result due to pathological DSBs where breaks on opposite DNA strands occur in very close proximity. Also, Artemis endonuclease activity is essential for the hairpin opening step during V(D)J recombination (following cleavage by recombination activation genes, RAG1 and RAG2), and patients lacking Artemis suffer from severe combined immunodeficiency because of a V(D)J recombination defect in antigen receptor gene assembly (4, 32).
DNA-PKcs interacts with the C terminus of Ku80, which is highly dynamic and flexible (Fig. 2A). The final 12 amino acids of Ku80 are sufficient for interacting with DNA-PKcs (33, 34), but Ku·DNA-PKcs complex formation is very weak unless Ku is bound to a DNA end. The presence of Ku on DNA increases the binding affinity of DNA-PKcs for DNA ends by 100-fold (35). Following binding to the DNA end, DNA-PKcs autophosphorylates, thus activating the endonuclease activity of Artemis (4). This likely occurs when autophosphorylated DNA-PKcs phosphorylates the C-terminal inhibitory region of Artemis (aa 454–458), promoting the dissociation of the inhibitory region from the N-terminal catalytic domain (aa 1–7) (Fig. 2B) (25, 36).
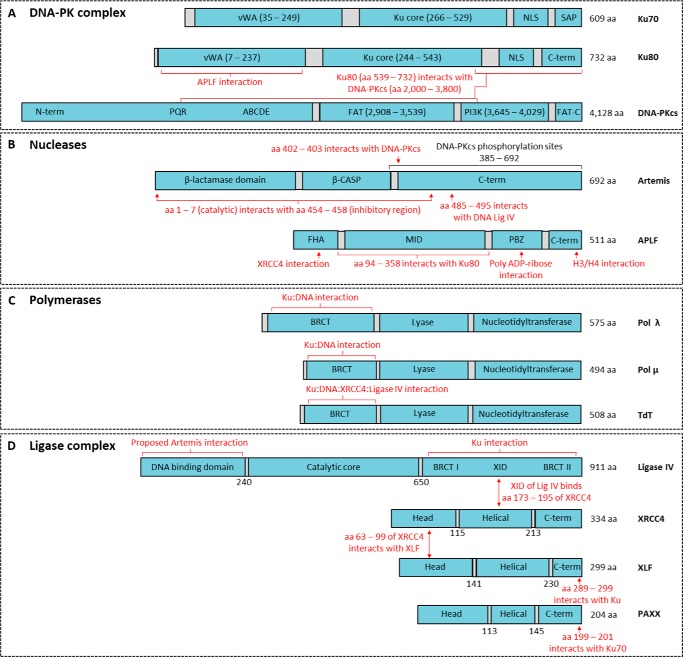
Figure 2.
Nonhomologous end-joining proteins and their known interactions. A, nonhomologous end-joining (NHEJ) DNA-dependent protein kinase (DNA-PK) complex consists of a heterodimer of Ku70 and Ku80 plus DNA-PKcs (catalytic subunit). Ku70 and Ku80 consist of von Willebrand (vWA) domains, the Ku core, and the nuclear localization sequence (NLS). Ku70 also contains a SAF-A/B, Acinus, and PIAS (SAP) domain. DNA-PKcs consists of an N-terminal domain with PQR and ABCDE autophosphorylation clusters implicated in its activation, FAT (FRAP, ATM, TRRAP) domain, followed by the phosphatidylinositol 3-kinase (PI3K) domain, and the FAT-C (C-terminal) domain. B, NHEJ nucleases consist of Artemis and APLF (abbreviation for Aprataxin and PNKP-like factor). Artemis has a catalytic β-lactamase domain, a cleavage and polyadenylation specificity factor (β-CASP) domain, and a disordered C terminus. Amino acids (aa) 454–458 bind aa 1–7 to auto-inhibit Artemis activity (119). APLF consists of a forkhead-associated (FHA) domain, middle (MID) domain, and the poly(ADP-ribose)-binding zinc finger (PBZ) domain (73, 120–122). C, polymerases involved in NHEJ are Pol λ, Pol μ, and terminal deoxynucleotidyltransferase (TdT). They consist of a breast cancer C terminus (BRCT) domain, a lyase domain, and a nucleotidyltransferase domain. D, DNA ligase complex consists of DNA ligase IV, X-ray repair cross-complementing 4 (XRCC4), XRCC4-like factor (XLF), and paralog of XRCC4 and XLF (PAXX). DNA ligase IV consists of an N-terminal DNA-binding domain, a catalytic core, and an XRCC4 interaction domain (XID) flanked by the BRCT I and II domains. XRCC4, XLF, and PAXX are structurally similar with an N-terminal head domain, helical domain, and C terminus. Protein domains are in blue and linker regions in gray.
It has been estimated that 20–50% of ionizing radiation-induced DSBs require Artemis for repair (37, 38). One possibility is that the remaining DSBs can be joined without the need of any nuclease (Fig. S2), but considering the number of nucleases present in the cell, it seems likely that other nucleases could be employed at incompatible ends, especially when Artemis is not present. Among those suggested to be involved in DSB repair include APLF, which is an abbreviation for Aprataxin and PNKP-like factor (also known as PALF) (39–41), flap structure-specific endonuclease 1 (FEN1), DNA replication helicase/nuclease 2 (DNA2), and exonuclease 1 (EXO1). In addition to nucleases, the Werner syndrome ATP-dependent helicase/nuclease (WRN) and the Bloom syndrome RecQ-like helicase (BLM) may also be involved in processing of DSB ends by creating a cleavage substrate for several of the aforementioned nucleases (42–44).
Another possible factor is the MRN complex (consisting of MRE11, RAD50, and NBS1), which is important for the resection step of the HR and SSA pathways to generate extensive 3′-terminated ssDNA overhangs. The intrinsic 3′→5′ exonuclease activity of the MRE11 component cannot generate these 3′-terminated overhangs by acting directly at a DNA end and relies on the C-terminal-binding protein interacting protein (CtIP) to stimulate MRN endonuclease activity to incise distal from the break. Next, the 3′ exonuclease activity can degrade DNA from the incision back toward the DNA end, thus creating the 3′-terminated ssDNA overhangs that can further undergo long range resection (e.g. by EXO1 or DNA2-BLM) (45, 46). This processing may have implications for the binding of Ku to DNA ends because MRE11 endonuclease activity occurs upstream of the Ku-bound DNA end.
CtIP is an important regulator of end processing as it not only stimulates MRN but also the long range resection by BLM and DNA2 (44). Importantly, CtIP is phosphorylated and active in S and G2 (47), indicating that cell cycle is another factor that dictates nuclease involvement. Furthermore, the abundance and localization of these nucleases at DSB sites will determine which nucleases are responsible for the most resection at DSBs. Because Artemis is recruited to breaks by DNA-PKcs at the early stages of NHEJ, and because only limited resection occurs, Artemis appears to the primary nuclease for most NHEJ repair events (27).
The polymerases of NHEJ
Members of the Pol X family of polymerases participate in DSB repair by NHEJ. DNA Pol μ and Pol λ are the two members involved in NHEJ in the majority of human cells (48, 49). Each of these polymerases has an N-terminal BRCA1 C terminus (BRCT) domain that allows them to interact with Ku (Fig. 2) (50). Primary cells derived from mice with genetic knockouts of both Pol μ and Pol λ exhibit little or no sensitivity to ionizing radiation, although knockouts in cell lines can have some deficit in DSB repair in some assays (51, 52). Pol μ and Pol λ can incorporate both dNTPs and rNTPs (48, 49), with any incorporated ribonucleotides subsequently removed by base excision repair (53). Importantly, both Pol μ and Pol λ can incorporate nucleotides in a template-dependent or template-independent manner (51), although template-independent insertion by Pol μ appears stronger than that of Pol λ (54, 55). Both of these polymerases appear to be able to use an unstable primer-template junction, such as would exist during intermediate stages of NHEJ. The activity of these polymerases further explains the high level of diversity that can occur at NHEJ junctions and demonstrates that although resection is one way of generating short stretches of homology between broken DNA ends, template-independent nucleotide addition of one or both broken DNA ends is another.
DNA polymerase β (Pol β) is another member of the Pol X family, but it lacks a BRCT domain (56), which is a likely reason why it is not involved in NHEJ. The final known member of the Pol X family is terminal deoxynucleotidyltransferase (TdT). TdT is only expressed in early B- and T-lymphocytes, making it most relevant to the NHEJ repair that occurs during V(D)J recombination, where it has a major role in promoting immunoglobulin diversity. DNA polymerases outside the Pol X family are able to incorporate nucleotides during NHEJ, but only in a template-dependent manner (16, 17, 57–59).
The ligase complex of NHEJ
DNA ligase IV (Lig4) functions exclusively in NHEJ, making it a central component of the repair process. Lig4 acts in complex with the X-ray repair cross-complementing 4 (XRCC4) enzyme (9), which stimulates Lig4 enzyme activity in biochemical assays (60). Loss of either Lig4 or XRCC4 severely compromises NHEJ. Several other factors have also been implicated for efficient ligation. A screen for XRCC4-interacting factors yielded the XRCC4-like factor (XLF; also known as Cernunnos), a 33-kDa protein with weak sequence homology and structural similarity to XRCC4 (61–63). The N-terminal head domain of XLF interacts with the N-terminal head domain of XRCC4 (62) allowing XLF to complex with XRCC4·Lig4 (Fig. 2). This interaction would presumably stabilize the juxtaposition of the DNA ends prior to covalent ligation, but this is still an area of active investigation. Another protein found to have structural similarity to XRCC4 is the 22-kDa protein PAXX (paralog of XRCC4 and XLF) (64, 65). The C terminus of PAXX (aa 199–201) interacts with Ku, and similar to XLF mutants, PAXX mutants are more sensitive to ionizing radiation and DSB-inducing agents (Fig. 2) (64, 66, 67).
Accessory proteins of NHEJ: Tyrosyl DNA phosphodiesterase 1, polynucleotide kinase, and aprataxin
Although the above proteins can carry out a majority of the NHEJ reactions, some circumstances require the activity of other proteins to chemically modify DNA ends to make them suitable for repair. For example, tyrosyl DNA phosphodiesterase 1 (TDP1) is the only identified enzyme that can specifically process the 3′-phosphoglycolates (3′-PG) that can form as a by-product of up to 10% of ionizing radiation-induced DSBs (68, 69). Ends with 3′-PG adducts are unligatable and must be removed for NHEJ to proceed.
Polynucleotide kinase (PNK) and aprataxin are two more factors that may be enlisted in DSB repair by NHEJ. Human PNK possesses both kinase and phosphatase activity. Phosphorylation by PNK is necessary when a 5′ end lacks a phosphate group, and the phosphatase activity is important for removing 3′ phosphates that can arise following some types of oxidative damage (70). Aprataxin is employed when Lig4 initiates but does not complete a covalent join, resulting in an aborted ligation product where the AMP group remains covalently bound to the 5′ strand of one of the DNA ends. In this case, the deadenylation reaction catalyzed by aprataxin is required to remove the AMP group (71). Following phosphorylation of XRCC4 by CK2, both PNK and aprataxin can bind to XRCC4 via their forkhead-associated domain (72). Therefore, although PNK and aprataxin may not initially localize to the DSB site, they can be recruited if necessary. This may occur if the DSB remains unrepaired after a certain length of time, indicating that the first set of NHEJ proteins that responded to the site were unable to complete repair.
Optimal NHEJ component utilization is influenced by DNA end configuration
NHEJ is a single pathway, but the DNA end configurations at a given DSB determine which NHEJ components are most important for efficient ligation. In other words, NHEJ has several enzymes at its disposal, but it does not need to engage all of them unless presented with certain DNA end configurations (5). Even the core NHEJ components may load and act in various combinations, highlighting the flexibility of NHEJ and explaining the diversity of repair products generated for the very same DSB configuration and DNA end sequence. This model is supported by many structural and biochemical studies demonstrating the different routes DNA end processing can take to reach a ligatable joint (Fig. 3). The stability of this ligatable joint is greatly enhanced when base pairing of ssDNA from either side of the break can occur via microhomology, although for NHEJ this microhomology need not be extensive, as even a single base pairing (even a non-Watson-Crick base pairing) will increase the stability enough to improve ligation efficiency a few fold over what is observed for NHEJ at blunt ends (74). In some cases, simple breathing of the DNA ends that exposes a complementary base pair between two broken ends may be adequate for repair, whereas in other cases more extensive processing by nucleases and polymerases may be required (16, 17).
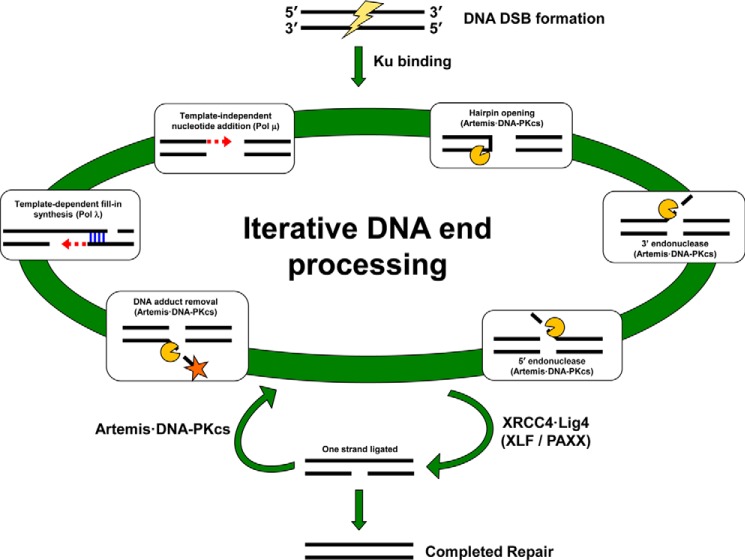
Figure 3.
DNA ends undergo iterative processing during NHEJ. NHEJ is single pathway with multiple components available to process the diversity of DNA end configurations at any given DSB. The first major step following formation of either a pathological or physiological DSB is binding of the Ku70·Ku80 complex (Ku) to protect DNA ends. The Ku·DNA complex is able to efficiently bind and thereby recruit other NHEJ components. An iterative processing occurs to make the two broken DNA ends optimal for ligation. Several types of processing performed by the Artemis·DNA-PKcs complex or DNA polymerases are shown in the white boxes along the large green circle. It would be difficult to represent all the possible DNA end configurations and every type of enzymatic processing in one figure; therefore, this depiction is not meant to be comprehensive but is merely to highlight some of the possibilities with the key components for each process indicated in parentheses. Any of these processes can occur to either end of a break in any order and multiple times. Once XRCC4·Lig4 is able to successfully ligate across a break, an intermediate with one strand ligated can form. Ligation of the second strand will complete repair. Alternatively, the gapped intermediate generated by ligating one strand has two ss–dsDNA boundaries, and Artemis·DNA-PKcs can cut at either boundary to generate a new DSB, thereby returning the ends to the iterative processing step where they can undergo further alterations.
The iterative nature of NHEJ means that multiple components can act on a single DSB during multiple rounds of processing (Fig. 3). Nucleases can remove nucleotides from a DNA end, with Pol μ subsequently adding nucleotides to that very same DNA end. Similarly, XRCC4·Lig4 can successfully ligate one DNA strand of a DSB only to have Artemis·DNA-PKcs reverse this by cleaving the newly ligated strand at the DNA gap generated by the ligation. Therefore, use of one set of components is not mutually exclusive to the use of other components, and all are active and in play as long as a DSB remains incompletely repaired.
Blunt-end ligation by Ku-XRCC4·Lig4
Biochemical studies have demonstrated that Ku is required for the efficient joining of blunt DNA ends lacking microhomology by NHEJ. When a ligatable joint is formed using exposed microhomology, however, Ku may not be necessary, indicating that Ku becomes less important as ends are able to form a thermodynamically stable joint through terminal base pairing (55). Ku is highly abundant in cells and has a high affinity for DNA ends (KD = 6 × 10−10 m), allowing it to quickly respond to a break and promote the binding of XRCC4·Lig4 to the DNA ends (75). The C terminus of Lig4 contains two BRCT domains that allow it to bind to two Ku complexes, conceivably one attached to each of the DSB ends (76). The region between these two BRCT domains of Lig4 carries the interaction domain that binds a homodimer of XRCC4 (Fig. 2D) where the 2 to 1 ratio of XRCC4 to Lig4 further stabilizes the bridging between the two DNA ends (77–79). The further activity of DNA-PKcs, Artemis, or Pol μ is not required, as efficient ligation is achieved with the Ku-XRCC4·Lig4 complex alone in reconstitution assays using human proteins (80). Therefore, at least for blunt DNA ends, direct ligation is preferred over extensive processing. This contrasts with results from Saccharomyces cerevisiae where blunt end ligation was found to be inefficient (81, 82), but this may be due to greater DNA end resection that occurs prior to repair by HR, which is the more dominant repair mechanism in yeast.
Previous cryo-EM studies have shown interaction between two DNA-PK complexes (83). The recent 4.3 Å crystal structure of DNA-PKcs also raises the possibility that dimerization of DNA-PKcs contributes to bridging of DNA ends (84); however, this particular observation of a dimeric arrangement may be due to crystal packing. DNA is not present in this crystal structure; thus one can only speculate about this interaction. The ligation of ends with only Ku and XRCC4·Lig4 provides biochemical evidence that DNA end-bridging is not reliant on DNA-PKcs or NHEJ factors other than Ku and XRCC4·Lig4 (5). It is clear that the joining of the blunt ends (signal ends) during V(D)J recombination also does not require any NHEJ proteins other than Ku and XRCC4·Lig4 (9), and this is consistent with the biochemistry of blunt end ligation.
The nucleases of NHEJ can process multiple DNA end configurations
Artemis has been implicated as the major nuclease involved in NHEJ when such activity is required. Although the role Artemis plays in DNA hairpin opening during V(D)J recombination is well-characterized, its role in NHEJ is now beginning to be understood. Recent biochemical studies have revealed that the ligation of incompatible overhangs is strongly stimulated in the presence of the Artemis·DNA-PKcs complex. Therefore, Artemis is recruited to process various DNA overhangs at broken DNA ends to promote formation of a stable ligatable joint. This makes sense when one considers that DNA hairpins are structurally similar to DNA overhangs, due to a sterically-constrained hairpin tip that results in only transient base pairing of the terminal base pairs (4 nt), thus creating a ss–dsDNA boundary (85). This ability of Artemis to act at ss–dsDNA boundaries gives it the flexibility to process a number of DNA end configurations.
The endonuclease activity of the Artemis·DNA-PKcs complex can remove both 5′ and 3′ DNA overhangs to create DNA end structures that can be ligated by the XRCC4·Lig4 complex (50, 86) (Fig. 3). At 5′ overhangs, Artemis cuts directly at the ss–dsDNA boundary, but when processing 3′ overhangs and DNA hairpins, Artemis preferentially leaves a 4-nt 3′ overhang. Long 5′ and 3′ overhangs can also be endonucleolytically processed by Artemis, and this may be useful to make microhomology embedded within the overhang available for annealing to create a stable ligatable joint (5). These observations suggest a model in which Artemis·DNA-PKcs binds to the ss–dsDNA boundary to occupy 4 nts along the single-stranded segment at the boundary followed by nicking on the 3′ side of the 4 nts (27).
In addition to overhangs, evidence also shows that when blunt DNA ends breathe between a closed, fully hydrogen-bonded state to an open, partially hydrogen-bonded state, they form ss–dsDNA boundaries upon which Artemis can act (27). Repair of such ends is relevant as blunt DNA ends may be generated by chemotherapeutic agents, reactive oxygen species, or ionizing radiation (87). Furthermore, breathing allows the Artemis·DNA-PKcs complex to resect into the duplex to generate short overhangs that can form microhomology (5), explaining why even NHEJ of blunt ends can display nucleotide loss at repair junctions. Still, the fact that Artemis·DNA-PKcs does not strongly stimulate the ligation of blunt-ended DNA suggests that even though Artemis·DNA-PKcs is able to resect at blunt ends, these ends are usually joined directly without resection (5, 27).
The versatility of Artemis to act at many different types of ends leads to a unifying model explaining the essential structural features of all DNA substrates at which Artemis functions. Although it may appear that Artemis has the ability to recognize a number of different structures, in fact it is one structure, an ss–dsDNA boundary, that is recognized in a variety of different forms. 5′ and 3′ overhangs, hairpins, and blunt ends in an open state all have potential regions of ss–dsDNA that can act as contact points for Artemis (28). The Artemis active site can then act within the single-stranded portion of the overhang or the hairpin to achieve hydrolysis of the phosphodiester backbone. Although this model must await the elucidation of a DNA-Artemis structure, it explains the diversity of cutting patterns of Artemis.
Besides the role in processing DNA overhangs, Artemis appears to be necessary for removing damaged DNA from broken ends (Fig. 3). When ionizing radiation-induced DSBs bear a 3′-PG terminus (88–90), for example, these DNA ends are unable to undergo ligation because this step requires a 3′-hydroxyl on one end and a 5′-phosphate on the other. TDP1 is able to remove these 3′ modifications; however, TDP1 mutant cells are only marginally radiosensitive compared with Artemis mutants, and it has been demonstrated biochemically that the Artemis·DNA-PKcs complex is able to process these ends (91, 92). This suggests that Artemis can work with or in place of TDP1 to repair the large number of DSBs that can occur following radiation exposure.
The finding that the C-terminal region of Artemis (aa 485–495) interacts with the N-terminal head domain of Lig4 (Fig. 2B) (93–95) adds a further dimension to the role Artemis may play in NHEJ. Although the DNA-PKcs–independent 5′ exonuclease activity has been described, recent data show that Artemis has a DNA-PKcs–independent 3′ endonuclease activity stimulated by XRCC4·Lig4 (96). The interaction between Lig4 and the C-terminal regulatory region of Artemis may recruit Artemis and alter the protein conformation, permitting endonuclease activity without the need for activation by DNA-PKcs. In addition to its crucial role in ligating a stable joint intermediate, the extreme radiosensitivity of Lig4 mutants may be due to its ability to stimulate or recruit various NHEJ components to a DSB.
The DNA polymerases of NHEJ work to create a stable ligatable joint
DNA polymerases can serve two important roles in NHEJ: fill-in synthesis of gaps and nucleotide addition to broken DNA ends. Both processes can enhance formation of a stable intermediate for ligation by XRCC4·Lig4. The DNA polymerases Pol μ and Pol λ are recruited to the DNA end by interaction of their N-terminal BRCT domain with the Ku·DNA complex (Fig. 2C) (50). Pol μ primarily adds nucleotides in a template-independent manner, whereas Pol λ primarily has template-dependent polymerase activity, although limited template-independent activity has been reported (54). Pol μ, and also TdT, carries a protein domain, loop 1, that affects association with a DNA template through hydrogen bonding and allows for template-independent nucleotide addition (56, 97).
The template-dependent activity of Pol λ is mostly required when long ssDNA ends are annealed with terminal microhomology, leaving a gap. Fill-in synthesis of this gap will further stabilize the annealed intermediate and promote the ligation (52, 55). When the 3′ overhangs are mismatched and therefore unable to form an annealed intermediate, Pol λ has little effect on NHEJ because there is no DNA template to act upon (5).
Pol μ strongly promotes the ligation of incompatible 3′ overhangs in reactions containing only the Ku-XRCC4·DNA ligase 4 complex (55). By adding nucleotides to the ends of these overhangs in both template-dependent and template-independent mechanisms, Pol μ generates regions of microhomology for subsequent annealing and ligation (55). Nucleotide addition can occur on 3′ overhangs as short as 1 to 2 nts (52). In biochemical reactions containing Artemis, the joining of two mismatched 3′ overhangs is strongly stimulated by Pol μ, promoting the formation of terminal microhomology with limited processing by Artemis (Fig. 3) (5). Interestingly, sequencing of NHEJ junctions reveals that if the ends are compatible, meaning they already share microhomology, nucleotide addition by Pol μ does not occur or is limited (5). This illustrates once again that ends capable of forming a thermodynamically stable intermediate are ligated efficiently without having to recruit additional factors.
XLF and PAXX stimulate ligation by the XRCC4·Lig4 complex
XLF and PAXX are the most recently characterized NHEJ factors shown to support ligation by the Lig4 complex. Both XLF and PAXX share structural similarity with XRCC4 (62, 64). Individual XLF and PAXX mutants display only a mild phenotype, but XLF PAXX double mutants are synthetically lethal in mice and reduce V(D)J recombination in human B-lymphocytes (98–101), suggesting that although they may be redundant, at least one is necessary for efficient repair by NHEJ. The main purpose of XLF and PAXX appears to be in providing additional structural support to stabilize two DNA ends, thereby enhancing the ability of XRCC4·Lig4. This likely occurs in a subset of NHEJ repair reactions where the two broken ends are incompatible and lack the thermodynamic stability provided by annealed microhomology.
Homodimers of XLF bind directly to XRCC4 via an N-terminal head domain (102). This head domain also allows XLF to interact with the Ku·DNA complex (103). In biochemical reactions containing only Ku and the XRCC4·Lig4 complex, XLF was shown to only stimulate the ligation of short, incompatible 3′ overhangs (55). In another study, however, XLF was shown to promote the ligation of all mismatched and noncohesive overhangs in the presence of Ku, DNA-PKcs, and XRCC4·Lig4 (104). Although it is possible that DNA-PKcs could affect XLF interactions, it is also possible that differences in the DNA substrates used in each study affect the outcome because the study involving DNA-PKcs used >3 kb of linearized plasmids and the other used fragments of ∼70 bp. Although further study is required to fully understand the major role of XLF in NHEJ, it seems that XLF promotes annealing of at least some incompatible substrates.
Genetic studies in mice complement these biochemical findings as it was found that an XLF DNA-PKcs double knockout is synthetically lethal. Interestingly, a Ku70 knockout rescues this synthetic lethality (105). Similar to a study showing that a Ku80 deletion rescues the lethality of a Lig4 knockout (106), this demonstrates that several NHEJ factors are epistatic to Ku. Loss of both XLF and DNA-PKcs must severely impair the ability to repair a DSB by NHEJ. Further genetic studies and analysis of DSB repair junctions in these deficient mice will provide more information as to the critical role of XLF.
Like XLF, PAXX also forms homodimers, and its C terminus has been found to associate with Ku (Fig. 2D) (64, 65). In reactions containing only Ku and the XRCC4·Lig4 complex, PAXX was shown to promote the ligation of two blunt ends (64). In some cases, XLF and PAXX may work together to stabilize DNA ends. In reactions containing Ku, XRCC4·Lig4, and XLF, PAXX promoted the ligation of a blunt end to a 3′ overhang (65). Interestingly, a more recent biochemical study showed that if Artemis and Pol μ are included, PAXX does not stimulate NHEJ for 3′ overhangs, but it does for 5′ overhangs (5), indicating that the role of PAXX may be to stabilize substrates that cannot generate microhomology by end processing or nucleotide addition.
Structural biology of NHEJ
There has been remarkable progress in determining progressively higher resolution structures of some of the NHEJ proteins or, at least, portions of them (Fig. 4) (48, 49, 64, 84, 94, 107–112). Readers should also refer to detailed reviews about the structural aspects of the interactions of ligase IV with XRCC4, XLF, and Artemis (113, 114). However, we still lack a convincing comprehensive view of how the enzymatic components are positioned at a single DNA end or at a pair of DNA ends. We also do not know the relative position of each component relative to most of the others in a large multiprotein complex during NHEJ.
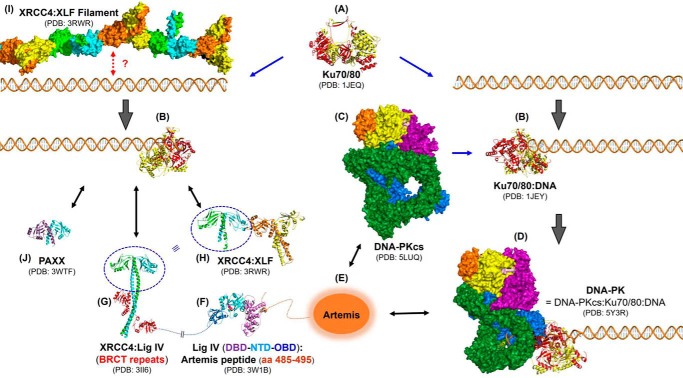
Figure 4.
Structural aspects of NHEJ. Select reported three-dimensional structures of NHEJ components are shown. Protein structures are shown in ribbon representation except for DNA-PKcs and an XRCC4·XLF filament, which are shown in surface representations. A, crystal structure of Ku70/80 heterodimer alone at 2.7 Å (PDB code 1JEQ) is shown in the middle (109). Ku70 and Ku80 are shown in red and yellow, respectively. B, these toroidal Ku70/80 proteins bind to broken dsDNA ends to form a Ku70/80·DNA complex (solved at 2.5 Å) (PDB code 1JEY) (109). C, crystal structure of DNA-PKcs at 4.3 Å is shown in the center (PDB code 5LUQ) (84). The DNA-PKcs is color-coded as follows: N terminus (blue); circular cradle (green); head comprising FAT region (purple); kinase (yellow); FRB (orange); FATC (light pink). D, DNA-PKcs binds the Ku70/80·DNA to form a DNA-PK complex. A 6.6-Å cryo-EM structure of DNA-PK holoenzyme is shown (PDB code 5Y3R) (111). E, structure of Artemis has not been reported yet. F, crystal structure of the catalytic region of DNA ligase IV (DBD-NTD-OBD) in complex with an Artemis fragment (aa 485–495) was solved at 2.4 Å (PDB code 3W1B) (94). The Artemis fragment is shown in orange and interacts with the DNA-binding domain (DBD), which is shown in violet. The nucleotidyltransferase domain (NTD) is shown in cyan. The catalytic lysine (Lys-273), which forms a covalent AMP-lysine intermediate, is shown as a sphere, and a possible αPO4 is attached to the lysine. The OB-fold domain (OBD) is shown in blue. G, crystal structure of the complex of XRCC4 homodimer and the BRCT repeats of ligase IV (Lig IV) at 2.4 Å is shown (PDB code 3II6) (107). Each XRCC4 molecule is shown in cyan and green. Two BRCT domains are shown in red. H, crystal structure of XRCC4(1–224)·XLF(1–157) complex (both are homodimers) at 3.94 Å is shown (PDB code 3RWR) (110). The XRCC4 homodimer is shown in cyan and green. The XLF homodimer is shown in yellow and orange. I, this XRCC4·XLF complex can form filaments, shown in the same color scheme at the left top corner, which might bridge DNA ends. J, crystal structure of PAXX homodimer at 3.45 Å is shown in cyan and purple (PDB code 3WTF) (64). Note that Ku70/80 bound on the DNA end can recruit XRCC4·ligase IV complex, and Ku70/80 also directly interacts with and recruits XLF and PAXX through their C termini. Also note that structures of the Pol X family polymerases are not shown here due to a space limitation. The figure was created using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Recently, a cryo-EM structure of DNA-PK was reported at 6.6 Å by docking the available crystal structures of DNA-PKcs (at 4.3 Å) (84) and Ku70/80·DNA complex (at 2.5 Å) (109). This finally allowed positioning of DNA-PKcs relative to the Ku70/80·DNA complex. However, statistics on the structural analysis must be much improved, and the position of quite a number of side chains of DNA-PKcs (>500 amino acids) is still questionable (84, 111, 115). Moreover, we still do not know how the C-terminal domain of Ku80 interacts with DNA-PKcs, and a structure of Artemis has not yet been reported. Furthermore, because some of the reported structures lack their C-terminal portions (e.g. Ku, XRCC4, XLF, and PAXX), an understanding on how these flexible regions work as full-length molecules will be critical for understanding the function of these complexes (64, 109–111).
Higher order structures have also been proposed for some NHEJ components. For example, the Lig4 complex, including XRCC4, XLF, and sometimes PAXX, has been proposed to form a sleeve around the DNA duplex (116–118), but the precise geometry is still not clear. It will be interesting to determine how such models will include Ku, DNA-PKcs, Artemis, and the polymerases μ and λ.
In many ways, the major future questions will require increasing reliance on structural insights.
Concluding comments
DNA DSBs are potentially lethal events that must be repaired in a manner that does not compromise genome integrity. NHEJ is the major pathway that repairs DSBs in mammalian cells. DSBs can occur due to various pathological or physiological events; however, the configuration of the DNA ends at breaks is not uniform. Therefore, NHEJ must be highly flexible so that it can deploy multiple enzymes to process the various types of DNA ends it may encounter. Biochemical and genetic studies have provided mechanistic insight into which NHEJ proteins are utilized, depending on the DNA end configuration. Two blunt DNA ends may only require Ku and XRCC4·Lig4 for joining, whereas incompatible 3′ ends may require processing by Artemis·DNA-PKcs, and incompatible 5′ ends may require XLF or PAXX for additional structural support. Time is likely a critical factor as the longer a break remains, the more accessory NHEJ factors may be recruited to a break in an attempt to repair it.
Many attempts have been made to subdivide the NHEJ pathway based upon the diversity of joining products that occur. However, this diversity of products highlights the flexibility of the NHEJ pathway. Repair by NHEJ does not mean a precise join because the activity of Artemis can lead to nucleotide loss, and the activity of Pol μ can lead to nucleotide gain. Also, the term “nonhomologous” was not meant to imply a total lack of homology usage in repair, as up to 4 nts of microhomology is typical for NHEJ repair. Instead, it was only meant to distinguish NHEJ from HR, which can use several hundred base pairs of homology during repair. Still, NHEJ is far from being completely understood, as evidenced by the discovery of new factors (PAXX) and new activities of known factors (Artemis). Continued research in this area will help elucidate why NHEJ is the dominant repair pathway in mammals and reveal more factors that contribute to DSB repair.
Supplementary Material
Acknowledgment
We apologize for any work that we have overlooked.
This work was supported by National Institutes of Health grants (to M. R. L.). This is the second article in the Thematic Minireview series “DNA double-strand break repair and pathway choice.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
- DSB
- double-strand break
- PDB
- Protein Data Bank
- NHEJ
- nonhomologous DNA end joining
- Pol
- polymerase
- HR
- homologous recombination
- aa
- amino acid
- SSA
- single-strand annealing
- TdT
- terminal deoxynucleotidyltransferase
- 3′-PG
- ′-phosphoglycolate
- PNK
- polynucleotide kinase
- CtIP
- C-terminal-binding protein interacting protein
- XLF
- XRCC4-like factor
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- a-EJ
- alternative end-joining
- nt
- nucleotide.
References
- 1. Lieber M. R. (2016) Mechanisms of human lymphoid chromosomal translocations. Nat. Rev. Cancer 16, 387–398 10.1038/nrc.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moore J. K., and Haber J. E. (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in S. cerevisiae. Mol. Cell. Biol. 16, 2164–2173 10.1128/MCB.16.5.2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meek K., Dang V., and Lees-Miller S. P. (2008) DNA-PK: the means to justify the ends? Adv. Immunol. 99, 33–58 10.1016/S0065-2776(08)00602-0 [DOI] [PubMed] [Google Scholar]
- 4. Ma Y., Pannicke U., Schwarz K., and Lieber M. R. (2002) Hairpin opening and overhang processing by an Artemis:DNA-PKcs complex in V(D)J recombination and in nonhomologous end joining. Cell 108, 781–794 10.1016/S0092-8674(02)00671-2 [DOI] [PubMed] [Google Scholar]
- 5. Chang H. H., Watanabe G., Gerodimos C. A., Ochi T., Blundell T. L., Jackson S. P., and Lieber M. R. (2016) Different DNA end configurations dictate which NHEJ components are most important for joining efficiency. J. Biol. Chem. 291, 24377–24389 10.1074/jbc.M116.752329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang H. H. Y., Pannunzio N. R., Adachi N., and Lieber M. R. (2017) Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 10.1038/nrm.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beucher A., Birraux J., Tchouandong L., Barton O., Shibata A., Conrad S., Goodarzi A. A., Krempler A., Jeggo P. A., and Löbrich M. (2009) ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 28, 3413–3427 10.1038/emboj.2009.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lisby M., and Rothstein R. (2015) Cell biology of mitotic recombination. Cold Spring Harb. Perspect. Biol. 7, a016535 10.1101/cshperspect.a016535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lieber M. R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saito S., Kurosawa A., and Adachi N. (2016) Mutations in XRCC4 cause primordial dwarfism without causing immunodeficiency. J. Hum. Genet. 61, 679–685 10.1038/jhg.2016.46 [DOI] [PubMed] [Google Scholar]
- 11. Schimmel J., Kool H., van Schendel R., and Tijsterman M. (2017) Mutational signatures of non-homologous and polymerase θ-mediated end-joining in embryonic stem cells. EMBO J. 36, 3634–3649 10.15252/embj.201796948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saito S., Maeda R., and Adachi N. (2017) Dual loss of human POLQ and LIG4 abolishes random integration. Nat. Commun. 8, 16112 10.1038/ncomms16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan S. H., Yu A. M., and McVey M. (2010) Dual roles for DNA polymerase θ in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6, e1001005 10.1371/journal.pgen.1001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gauss G. H., and Lieber M. R. (1996) Mechanistic constraints on diversity in human V(D)J recombination. Mol. Cell. Biol. 16, 258–269 10.1128/MCB.16.1.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bhargava R., Onyango D. O., and Stark J. M. (2016) Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32, 566–575 10.1016/j.tig.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Daley J. M., Laan R. L., Suresh A., and Wilson T. E. (2005) DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 280, 29030–29037 10.1074/jbc.M505277200 [DOI] [PubMed] [Google Scholar]
- 17. Daley J. M., Palmbos P. L., Wu D., and Wilson T. E. (2005) Nonhomologous end joining in yeast. Annu. Rev. Genet. 39, 431–451 10.1146/annurev.genet.39.073003.113340 [DOI] [PubMed] [Google Scholar]
- 18. Mimitou E. P., and Symington L. S. (2010) Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29, 3358–3369 10.1038/emboj.2010.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arnoult N., Correia A., Ma J., Merlo A., Garcia-Gomez S., Maric M., Tognetti M., Benner C. W., Boulton S. J., Saghatelian A., and Karlseder J. (2017) Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552 10.1038/nature24023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Slavoff S. A., Heo J., Budnik B. A., Hanakahi L. A., and Saghatelian A. (2014) A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J. Biol. Chem. 289, 10950–10957 10.1074/jbc.C113.533968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J. T., Tkáč J., Cook M. A., Rosebrock A. P., Munro M., Canny M. D., Xu D., and Durocher D. (2013) A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 22. Symington L. S. (2016) Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 51, 195–212 10.3109/10409238.2016.1172552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Daley J. M., Niu H., Miller A. S., and Sung P. (2015) Biochemical mechanism of DSB end resection and its regulation. DNA Repair 32, 66–74 10.1016/j.dnarep.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F. W., Chen J., Jackson S. P., and Scully R. (2007) Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell 28, 1045–1057 10.1016/j.molcel.2007.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goodarzi A. A., Yu Y., Riballo E., Douglas P., Walker S. A., Ye R., Härer C., Marchetti C., Morrice N., Jeggo P. A., and Lees-Miller S. P. (2006) DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 25, 3880–3889 10.1038/sj.emboj.7601255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu J., Li S., Zhang X., Wang L. C., Niewolik D., Schwarz K., Legerski R. J., Zandi E., and Lieber M. R. (2010) DNA-PKcs regulates a single-stranded DNA endonuclease activity of Artemis. DNA Repair 9, 429–437 10.1016/j.dnarep.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang H. H., Watanabe G., and Lieber M. R. (2015) Unifying the DNA end-processing roles of the artemis nuclease: Ku-dependent artemis resection at blunt DNA ends. J. Biol. Chem. 290, 24036–24050 10.1074/jbc.M115.680900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang H. H., and Lieber M. R. (2016) Structure-specific nuclease activities of Artemis and the Artemis: DNA-PKcs complex. Nucleic Acids Res. 44, 4991–4997 10.1093/nar/gkw456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dominski Z. (2007) Nucleases of the metallo-β-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 42, 67–93 10.1080/10409230701279118 [DOI] [PubMed] [Google Scholar]
- 30. Li S., Chang H. H., Niewolik D., Hedrick M. P., Pinkerton A. B., Hassig C. A., Schwarz K., and Lieber M. R. (2014) Evidence that the DNA endonuclease ARTEMIS also has intrinsic 5′-exonuclease activity. J. Biol. Chem. 289, 7825–7834 10.1074/jbc.M113.544874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pawelczak K. S., and Turchi J. J. (2010) Purification and characterization of exonuclease-free Artemis: implications for DNA-PK-dependent processing of DNA termini in NHEJ-catalyzed DSB repair. DNA Repair 9, 670–677 10.1016/j.dnarep.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Diest F. L., Tezcan I., Sanal O., Bertrand Y., Philippe N., Fischer A., and de Villartay J.-P. (2001) Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105, 177–186 10.1016/S0092-8674(01)00309-9 [DOI] [PubMed] [Google Scholar]
- 33. Gell D., and Jackson S. P. (1999) Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 27, 3494–3502 10.1093/nar/27.17.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ding Q., Reddy Y. V., Wang W., Woods T., Douglas P., Ramsden D. A., Lees-Miller S. P., and Meek K. (2003) Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 23, 5836–5848 10.1128/MCB.23.16.5836-5848.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. West R. B., Yaneva M., and Lieber M. R. (1998) Productive and nonproductive complexes of Ku and DNA-PK at DNA termini. Mol. Cell. Biol. 18, 5908–5920 10.1128/MCB.18.10.5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma Y., Pannicke U., Lu H., Niewolik D., Schwarz K., and Lieber M. R. (2005) The DNA-PKcs phosphorylation sites of human artemis. J. Biol. Chem. 280, 33839–33846 10.1074/jbc.M507113200 [DOI] [PubMed] [Google Scholar]
- 37. Riballo E., Kühne M., Rief N., Doherty A., Smith G. C., Recio M.-J., Reis C., Dahm K., Fricke A., Krempler A., Parker A. R., Jackson S. P., Gennery A., Jeggo P. A., and Löbrich M. (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol. Cell 16, 715–724 10.1016/j.molcel.2004.10.029 [DOI] [PubMed] [Google Scholar]
- 38. Kurosawa A., Koyama H., Takayama S., Miki K., Ayusawa D., Fujii M., Iiizumi S., and Adachi N. (2008) The requirement of Artemis in double-strand break repair depends on the type of DNA damage. DNA Cell Biol. 27, 55–61 10.1089/dna.2007.0649 [DOI] [PubMed] [Google Scholar]
- 39. Kanno S., Kuzuoka H., Sasao S., Hong Z., Lan L., Nakajima S., and Yasui A. (2007) A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 26, 2094–2103 10.1038/sj.emboj.7601663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li S., Kanno S., Watanabe R., Ogiwara H., Kohno T., Watanabe G., Yasui A., and Lieber M. R. (2011) Polynucleotide kinase and aprataxin-like forkhead-associated protein (PALF) acts as both a single-stranded DNA endonuclease and a single-stranded DNA 3′ exonuclease and can participate in DNA end joining in a biochemical system. J. Biol. Chem. 286, 36368–36377 10.1074/jbc.M111.287797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grundy G. J., Rulten S. L., Zeng Z., Arribas-Bosacoma R., Iles N., Manley K., Oliver A., and Caldecott K. W. (2013) APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 32, 112–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mimitou E. P., and Symington L. S. (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu Z., Chung W. H., Shim E. Y., Lee S. E., and Ira G. (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 10.1016/j.cell.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Daley J. M., Jimenez-Sainz J., Wang W., Miller A. S., Xue X., Nguyen K. A., Jensen R. B., and Sung P. (2017) Enhancement of BLM-DNA2-mediated long-range DNA end resection by CtIP. Cell Rep. 21, 324–332 10.1016/j.celrep.2017.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cannavo E., and Cejka P. (2014) Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122–125 10.1038/nature13771 [DOI] [PubMed] [Google Scholar]
- 46. Anand R., Ranjha L., Cannavo E., and Cejka P. (2016) Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell 64, 940–950 10.1016/j.molcel.2016.10.017 [DOI] [PubMed] [Google Scholar]
- 47. Huertas P., and Jackson S. P. (2009) Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bebenek K., Pedersen L. C., and Kunkel T. A. (2014) Structure-function studies of DNA polymerase λ. Biochemistry 53, 2781–2792 10.1021/bi4017236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moon A. F., Pryor J. M., Ramsden D. A., Kunkel T. A., Bebenek K., and Pedersen L. C. (2014) Sustained active site rigidity during synthesis by human DNA polymerase μ. Nat. Struct. Mol. Biol. 21, 253–260 10.1038/nsmb.2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ma Y., Lu H., Tippin B., Goodman M. F., Shimazaki N., Koiwai O., Hsieh C.-L., Schwarz K., and Lieber M. R. (2004) A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell 16, 701–713 10.1016/j.molcel.2004.11.017 [DOI] [PubMed] [Google Scholar]
- 51. Bertocci B., De Smet A., Weill J.-C., and Reynaud C. A. (2006) Non-overlapping functions of polX family DNA polymerases, pol m, pol l, and TdT, during immunoglobulin V(D)J recombination in vivo. Immunity 25, 31–41 10.1016/j.immuni.2006.04.013 [DOI] [PubMed] [Google Scholar]
- 52. Pryor J. M., Waters C. A., Aza A., Asagoshi K., Strom C., Mieczkowski P. A., Blanco L., and Ramsden D. A. (2015) Essential role for polymerase specialization in cellular nonhomologous end joining. Proc. Natl. Acad. Sci. U.S.A. 112, E4537–E4545 10.1073/pnas.1505805112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nick McElhinny S. A., and Ramsden D. A. (2003) Polymerase μ is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 23, 2309–2315 10.1128/MCB.23.7.2309-2315.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nick McElhinny S. A., Havener J. M., Garcia-Diaz M., Juárez R., Bebenek K., Kee B. L., Blanco L., Kunkel T. A., and Ramsden D. A. (2005) A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell 19, 357–366 10.1016/j.molcel.2005.06.012 [DOI] [PubMed] [Google Scholar]
- 55. Gu J., Lu H., Tippin B., Shimazaki N., Goodman M. F., and Lieber M. R. (2007) XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 26, 1010–1023 10.1038/sj.emboj.7601559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lange S. S., Takata K., and Wood R. D. (2011) DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110 10.1038/nrc2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lieber M. R. (2006) The polymerases for V(D)J recombination. Immunity 25, 7–9 10.1016/j.immuni.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 58. Daley J. M., and Wilson T. E. (2005) Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell. Biol. 25, 896–906 10.1128/MCB.25.3.896-906.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Daley J. M., and Wilson T. E. (2008) Evidence that base stacking potential in annealed 3′ overhangs determines polymerase utilization in yeast nonhomologous end joining. DNA Repair 7, 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grawunder U., Wilm M., Wu X., Kulesza P., Wilson T. E., Mann M., and Lieber M. R. (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388, 492–495 10.1038/41358 [DOI] [PubMed] [Google Scholar]
- 61. Dai Y., Kysela B., Hanakahi L. A., Manolis K., Riballo E., Stumm M., Harville T. O., West S. C., Oettinger M. A., and Jeggo P. A. (2003) Nonhomologous end joining and V(D)J recombination require an additional factor. Proc. Natl. Acad. Sci. U.S.A. 100, 2462–2467 10.1073/pnas.0437964100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ahnesorg P., Smith P., and Jackson S. P. (2006) XLF interacts with the XRCC4-DNA ligase IV complex to promote nonhomologous end-joining. Cell 124, 301–313 10.1016/j.cell.2005.12.031 [DOI] [PubMed] [Google Scholar]
- 63. Buck D., Malivert L., de Chasseval R., Barraud A., Fondanèche M.-C., Sanal O., Plebani A., Stéphan J.-L., Hufnagel M., le Diest F., Fischer A., Durandy A., de Villartay J.-P., and Revy P. (2006) Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124, 287–299 10.1016/j.cell.2005.12.030 [DOI] [PubMed] [Google Scholar]
- 64. Ochi T., Blackford A. N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V. M., Robinson C. V., Blundell T. L., and Jackson S. P. (2015) DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347, 185–188 10.1126/science.1261971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xing M., Yang M., Huo W., Feng F., Wei L., Jiang W., Ning S., Yan Z., Li W., Wang Q., Hou M., Dong C., Guo R., Gao G., Ji J., et al. (2015) Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 6, 6233 10.1038/ncomms7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roy S., de Melo A. J., Xu Y., Tadi S. K., Négrel A., Hendrickson E., Modesti M., and Meek K. (2015) XRCC4/XLF interaction is variably required for DNA repair and is not required for ligase IV stimulation. Mol. Cell. Biol. 35, 3017–3028 10.1128/MCB.01503-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tadi S. K., Tellier-Lebègue C., Nemoz C., Drevet P., Audebert S., Roy S., Meek K., Charbonnier J. B., and Modesti M. (2016) PAXX is an accessory c-NHEJ factor that associates with Ku70 and has overlapping functions with XLF. Cell Rep. 17, 541–555 10.1016/j.celrep.2016.09.026 [DOI] [PubMed] [Google Scholar]
- 68. Inamdar K. V., Pouliot J. J., Zhou T., Lees-Miller S. P., Rasouli-Nia A., and Povirk L. F. (2002) Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J. Biol. Chem. 277, 27162–27168 10.1074/jbc.M204688200 [DOI] [PubMed] [Google Scholar]
- 69. Chen B., Zhou X., Taghizadeh K., Chen J., Stubbe J., and Dedon P. C. (2007) GC/MS methods to quantify the 2-deoxypentos-4-ulose and 3′-phosphoglycolate pathways of 4′ oxidation of 2-deoxyribose in DNA: application to DNA damage produced by γ radiation and bleomycin. Chem. Res. Toxicol. 20, 1701–1708 10.1021/tx700164y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bernstein N. K., Williams R. S., Rakovszky M. L., Cui D., Green R., Karimi-Busheri F., Mani R. S., Galicia S., Koch C. A., Cass C. E., Durocher D., Weinfeld M., and Glover J. N. (2005) The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell 17, 657–670 10.1016/j.molcel.2005.02.012 [DOI] [PubMed] [Google Scholar]
- 71. Ahel I., Rass U., El-Khamisy S. F., Katyal S., Clements P. M., McKinnon P. J., Caldecott K. W., and West S. C. (2006) The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 443, 713–716 10.1038/nature05164 [DOI] [PubMed] [Google Scholar]
- 72. Koch C. A., Agyei R., Galicia S., Metalnikov P., O'Donnell P., Starostine A., Weinfeld M., and Durocher D. (2004) Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 23, 3874–3885 10.1038/sj.emboj.7600375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mehrotra P. V., Ahel D., Ryan D. P., Weston R., Wiechens N., Kraehenbuehl R., Owen-Hughes T., and Ahel I. (2011) DNA repair factor APLF is a histone chaperone. Mol. Cell 41, 46–55 10.1016/j.molcel.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sinden R. R., and Wells R. D. (1992) DNA structure, mutations and human genetic diseases. Curr. Opin. Biotechnol. 3, 612–622 10.1016/0958-1669(92)90005-4 [DOI] [PubMed] [Google Scholar]
- 75. Mimori T., and Hardin J. A. (1986) Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 261, 10375–10379 [PubMed] [Google Scholar]
- 76. Costantini S., Woodbine L., Andreoli L., Jeggo P. A., and Vindigni A. (2007) Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair 6, 712–722 10.1016/j.dnarep.2006.12.007 [DOI] [PubMed] [Google Scholar]
- 77. Sibanda B. L., Critchlow S. E., Begun J., Pei X. Y., Jackson S. P., Blundell T. L., and Pellegrini L. (2001) Crystal structure of an Xrcc4-DNA ligase IV complex. Nat. Struct. Biol. 8, 1015–1019 10.1038/nsb725 [DOI] [PubMed] [Google Scholar]
- 78. Grawunder U., Zimmer D., Kulesza P., and Lieber M. R. (1998) Requirement for an interaction of XRCC4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J. Biol. Chem. 273, 24708–24714 10.1074/jbc.273.38.24708 [DOI] [PubMed] [Google Scholar]
- 79. Grawunder U., Zimmer D., and Lieber M. R. (1998) DNA ligase IV binds to XRCC4 via a motif located between rather than within its BRCT domains. Curr. Biol. 8, 873–876 10.1016/S0960-9822(07)00349-1 [DOI] [PubMed] [Google Scholar]
- 80. Nick McElhinny S. A., Snowden C. M., McCarville J., and Ramsden D. A. (2000) Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20, 2996–3003 10.1128/MCB.20.9.2996-3003.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Herrmann G., Lindahl T., and Schär P. (1998) S. cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 17, 4188–4198 10.1093/emboj/17.14.4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Westmoreland J. W., Summers J. A., Holland C. L., Resnick M. A., and Lewis L. K. (2010) Blunt-ended DNA double-strand breaks induced by endonucleases PvuII and EcoRV are poor substrates for repair in Saccharomyces cerevisiae. DNA Repair 9, 617–626 10.1016/j.dnarep.2010.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Spagnolo L., Rivera-Calzada A., Pearl L. H., and Llorca O. (2006) Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell 22, 511–519 10.1016/j.molcel.2006.04.013 [DOI] [PubMed] [Google Scholar]
- 84. Sibanda B. L., Chirgadze D. Y., Ascher D. B., and Blundell T. L. (2017) DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 355, 520–524 10.1126/science.aak9654 [DOI] [PubMed] [Google Scholar]
- 85. Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., and Hilbers C. W. (1989) Effects of base sequence on the loop folding in DNA hairpins. Biochemistry 28, 7491–7498 10.1021/bi00444a049 [DOI] [PubMed] [Google Scholar]
- 86. Lu H., Shimazaki N., Raval P., Gu J., Watanabe G., Schwarz K., Swanson P. C., and Lieber M. R. (2008) A biochemically defined system for coding joint formation in human V(D)J recombination. Mol. Cell 31, 485–497 10.1016/j.molcel.2008.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Povirk L. F. (2012) Processing of damaged DNA ends for double-strand break repair in mammalian cells. ISRN Mol. Biol. 2012, 1–16 10.5402/2012/345805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Henner W. D., Grunberg S. M., and Haseltine W. A. (1983) Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J. Biol. Chem. 258, 15198–15205 [PubMed] [Google Scholar]
- 89. Henner W. D., Rodriguez L. O., Hecht S. M., and Haseltine W. A. (1983) Gamma ray induced deoxyribonucleic acid strand breaks. 3′ Glycolate termini. J. Biol. Chem. 258, 711–713 [PubMed] [Google Scholar]
- 90. Valerie K., and Povirk L. F. (2003) Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22, 5792–5812 10.1038/sj.onc.1206679 [DOI] [PubMed] [Google Scholar]
- 91. Povirk L. F., Zhou T., Zhou R., Cowan M. J., and Yannone S. M. (2007) Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J. Biol. Chem. 282, 3547–3558 10.1074/jbc.M60774520 [DOI] [PubMed] [Google Scholar]
- 92. Yannone S. M., Khan I. S., Zhou R. Z., Zhou T., Valerie K., and Povirk L. F. (2008) Coordinate 5′ and 3′ endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by Artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Res. 36, 3354–3365 10.1093/nar/gkn205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Malu S., De Ioannes P., Kozlov M., Greene M., Francis D., Hanna M., Pena J., Escalante C. R., Kurosawa A., Erdjument-Bromage H., Tempst P., Adachi N., Vezzoni P., Villa A., Aggarwal A. K., and Cortes P. (2012) Artemis C-terminal region facilitates V(D)J recombination through its interactions with DNA ligase IV and DNA-PKcs. J. Exp. Med. 209, 955–963 10.1084/jem.20111437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ochi T., Gu X., and Blundell T. L. (2013) Structure of the catalytic region of DNA ligase IV in complex with an Artemis fragment sheds light on double-strand break repair. Structure 21, 672–679 10.1016/j.str.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. De Ioannes P., Malu S., Cortes P., and Aggarwal A. K. (2012) Structural basis of DNA ligase IV-Artemis interaction in nonhomologous end-joining. Cell Rep. 2, 1505–1512 10.1016/j.celrep.2012.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gerodimos C. A., Chang H. H., Watanabe G., and Lieber M. R. (2017) Effects of DNA end configuration on XRCC4:DNA ligase IV and its stimulation of Artemis activity. J. Biol. Chem. 292, 13914–13924 10.1074/jbc.M117.798850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Moon A. F., Garcia-Diaz M., Bebenek K., Davis B. J., Zhong X., Ramsden D. A., Kunkel T. A., and Pedersen L. C. (2007) Structural insight into the substrate specificity of DNA polymerase μ. Nat. Struct. Mol. Biol. 14, 45–53 10.1038/nsmb1180 [DOI] [PubMed] [Google Scholar]
- 98. Liu X., Shao Z., Jiang W., Lee B. J., and Zha S. (2017) PAXX promotes KU accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice. Nat. Commun. 8, 13816 10.1038/ncomms13816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Balmus G., Barros A. C., Wijnhoven P. W., Lescale C., Hasse H. L., Boroviak K., le Sage C., Doe B., Speak A. O., Galli A., Jacobsen M., Deriano L., Adams D. J., Blackford A. N., and Jackson S. P. (2016) Synthetic lethality between PAXX and XLF in mammalian development. Genes Dev. 30, 2152–2157 10.1101/gad.290510.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lescale C., Lenden Hasse H., Blackford A. N., Balmus G., Bianchi J. J., Yu W., Bacoccina L., Jarade A., Clouin C., Sivapalan R., Reina-San-Martin B., Jackson S. P., and Deriano L. (2016) Specific roles of XRCC4 paralogs PAXX and XLF during V(D)J recombination. Cell Rep. 16, 2967–2979 10.1016/j.celrep.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kumar V., Alt F. W., and Frock R. L. (2016) PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1-arrested progenitor B-cell line. Proc. Natl. Acad. Sci. U.S.A. 113, 10619–10624 10.1073/pnas.1611882113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Junop M. S., Modesti M., Guarné A., Ghirlando R., Gellert M., and Yang W. (2000) Crystal structure of the XRCC4 DNA repair protein and implications for end joining. EMBO J. 19, 5962–5970 10.1093/emboj/19.22.5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Andres S. N., Modesti M., Tsai C. J., Chu G., and Junop M. S. (2007) Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell 28, 1093–1101 10.1016/j.molcel.2007.10.024 [DOI] [PubMed] [Google Scholar]
- 104. Tsai C. J., Kim S. A., and Chu G. (2007) Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc. Natl. Acad. Sci. U.S.A. 104, 7851–7856 10.1073/pnas.0702620104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Xing M., Bjorås M., Daniel J. A., Alt F. W., and Oksenych V. (2017) Synthetic lethality between murine DNA repair factors XLF and DNA-PKcs is rescued by inactivation of Ku70. DNA Repair 57, 133–138 10.1016/j.dnarep.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Karanjawala Z. E., Adachi N., Irvine R. A., Oh E. K., Shibata D., Schwarz K., Hsieh C. L., and Lieber M. R. (2002) The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair 1, 1017–1026 10.1016/S1568-7864(02)00151-9 [DOI] [PubMed] [Google Scholar]
- 107. Wu P. Y., Frit P., Meesala S., Dauvillier S., Modesti M., Andres S. N., Huang Y., Sekiguchi J., Calsou P., Salles B., and Junop M. S. (2009) Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol. Cell. Biol. 29, 3163–3172 10.1128/MCB.01895-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Garcia-Diaz M., Bebenek K., Krahn J. M., Kunkel T. A., and Pedersen L. C. (2005) A closed conformation for the pol λ catalytic cycle. Nat. Struct. Mol. Biol. 12, 97–98 10.1038/nsmb876 [DOI] [PubMed] [Google Scholar]
- 109. Walker J. R., Corpina R. A., and Goldberg J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 10.1038/35088000 [DOI] [PubMed] [Google Scholar]
- 110. Andres S. N., Vergnes A., Ristic D., Wyman C., Modesti M., and Junop M. (2012) A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 40, 1868–1878 10.1093/nar/gks022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yin X., Liu M., Tian Y., Wang J., and Xu Y. (2017) Cryo-EM structure of human DNA-PK holoenzyme. Cell Res. 27, 1341–1350 10.1038/cr.2017.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Liu M. S., Tsai H. Y., Liu X. X., Ho M. C., Wu W. J., and Tsai M. D. (2016) Structural mechanism for the fidelity modulation of DNA polymerase lambda. J. Am. Chem. Soc. 138, 2389–2398 10.1021/jacs.5b13368 [DOI] [PubMed] [Google Scholar]
- 113. Ochi T., Wu Q., and Blundell T. L. (2014) The spatial organization of non-homologous end joining: from bridging to end joining. DNA Repair 17, 98–109 10.1016/j.dnarep.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Menon V., and Povirk L. F. (2017) XLF/Cernunnos: an important but puzzling participant in the nonhomologous end joining DNA repair pathway. DNA Repair 58, 29–37 10.1016/j.dnarep.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Watanabe G., Lieber M. R., and Williams D. (2017) Structural step forward for NHEJ. Cell Res. 27, 1304–1306 10.1038/cr.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Brouwer I., Sitters G., Candelli A., Heerema S. J., Heller I., de Melo A. J., Zhang H., Normanno D., Modesti M., Peterman E. J., and Wuite G. J. (2016) Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature 535, 566–569 10.1038/nature18643 [DOI] [PubMed] [Google Scholar]
- 117. Reid D. A., Keegan S., Leo-Macias A., Watanabe G., Strande N. T., Chang H. H., Oksuz B. A., Fenyo D., Lieber M. R., Ramsden D. A., and Rothenberg E. (2015) Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 112, E2575–E2584 10.1073/pnas.1420115112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Reid D. A., Conlin M. P., Yin Y., Chang H. H., Watanabe G., Lieber M. R., Ramsden D. A., and Rothenberg E. (2017) Bridging of double-stranded breaks by the nonhomologous end-joining ligation complex is modulated by DNA end chemistry. Nucleic Acids Res. 45, 1872–1878 10.1093/nar/gkw1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Niewolik D., Peter I., Butscher C., and Schwarz K. (2017) Autoinhibition of the nuclease ARTEMIS is mediated by a physical interaction between its catalytic and C-terminal domains. J. Biol. Chem. 292, 3351–3365 10.1074/jbc.M116.770461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Iles N., Rulten S., El-Khamisy S. F., and Caldecott K. W. (2007) APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 27, 3793–3803 10.1128/MCB.02269-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Eustermann S., Brockmann C., Mehrotra P. V., Yang J. C., Loakes D., West S. C., Ahel I., and Neuhaus D. (2010) Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose). Nat. Struct. Mol. Biol. 17, 241–243 10.1038/nsmb.1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Li G. Y., McCulloch R. D., Fenton A. L., Cheung M., Meng L., Ikura M., and Koch C. A. (2010) Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 107, 9129–9134 10.1073/pnas.1000556107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.