Abstract
Glycogen synthase kinase-3 (GSK-3) activity regulates multiple signal transduction pathways and is also a key component of the network responsible for maintaining stem cell pluripotency. Genetic deletion of Gsk-3α and Gsk-3β or inhibition of GSK-3 activity via small molecules promotes stem cell pluripotency, yet the mechanism underlying the role for GSK-3 in this process remains ambiguous. Another cellular process that has been shown to affect stem cell pluripotency is mRNA methylation (m6A). Here, we describe an intersection between these components, the regulation of m6A by GSK-3. We find that protein levels for the RNA demethylase, FTO (fat mass and obesity–associated protein), are elevated in Gsk-3α;Gsk-3β-deficient mouse embryonic stem cells (ESCs). FTO is normally phosphorylated by GSK-3, and MS identified the sites on FTO that are phosphorylated in a GSK-3–dependent fashion. GSK-3 phosphorylation of FTO leads to polyubiquitination, but in Gsk-3 knockout ESCs, that process is impaired, resulting in elevated levels of FTO protein. As a consequence of altered FTO protein levels, mRNAs in Gsk-3 knockout ESCs have 50% less m6A than WT ESCs, and m6A-Seq analysis reveals the specific mRNAs that have reduced m6A modifications. Taken together, we provide the first evidence for how m6A demethylation is regulated in mammalian cells and identify a putative novel mechanism by which GSK-3 activity regulates stem cell pluripotency.
Keywords: RNA methylation, pluripotency, stem cells, protein phosphorylation, ubiquitin
Introduction
Glycogen synthase kinase-3 (GSK-3)5 activity is an important regulator of numerous signal transduction pathways (1). GSK-3 activity is the sum of two largely redundant proteins, GSK-3α and GSK-3β, and in general, GSK-3 is a negative regulator of cellular signaling (2). Rare among kinases, GSK-3 is active at a basal state, whereas pathway activation from upstream signaling cascades results in the inhibition of GSK-3 activity (2). GSK-3α and GSK-3β together regulate signal transduction pathways such as Wnt, protein kinase A, Hedgehog, transforming growth factor-β, nuclear factor of activated T-cells, and phosphatidylinositol 3-kinase (PI3K)-dependent insulin signaling in a variety of biological settings (3–5).
GSK-3 activity can be inhibited through the use of small-molecule inhibitors, such as SB-415,286, CHIR99021 (6–8), and the clinically relevant mood-stabilizer lithium (9, 10); however, a drawback to the use of small molecules to study GSK-3 function is the potential for off-target effects (11). Cells in which Gsk-3α and Gsk-3β have been genetically deleted allow for a more confident assessment of GSK-3–specific functions. Therefore, we utilize mouse embryonic stem cells (ESCs) deficient in both Gsk-3α and Gsk-3β (Gsk-3α−/−;Gsk-3β−/−) (i.e. Gsk-3 double knockout (DKO)) to assess GSK-3–specific functions (12, 13). One prominent phenotype of Gsk-3 DKO ESCs is their persistent pluripotency, assessed by their inability to differentiate into all three germs layers in a teratoma assay as well as by analysis of global gene expression (12, 13). In fact, Gsk-3 DKO ESCs will maintain pluripotency in culture even in the absence of leukemia-inhibitory factor (LIF) (12), which is added to WT mouse ESCs to promote pluripotency (14, 15). Furthermore, the addition of small-molecule inhibitors of both GSK-3 and mitogen-activated protein kinase (extracellular signal-regulated kinase) (termed 2i mixture) to culture media is sufficient to maintain pluripotency of mouse ESCs (16). Finally, the addition of GSK-3 inhibitors has enhanced the efficiency of induced pluripotent stem cell derivation (17, 18) and facilitated the derivation of ESCs from different strains of mice (19, 20) and from different species (21, 22). The mechanism by which GSK-3 inhibition or loss of function promotes pluripotency is not completely resolved (20, 23–35).
Whereas GSK-3 activity clearly plays an important role in stem cell pluripotency, other biological processes are also important for pluripotency. Recent work has demonstrated an important role for the methylation of adenosines (m6A) in mRNA in the regulation of stem cell pluripotency (36–38). m6A is the most common internal modification to mRNA in mammalian cells (39–41), yet little was known about the biology of this modification until a recent flurry of research led to the identification of the enzymes that add and remove m6A (METTL3/METTL14 and FTO/ALKBH5, respectively) (42–44), as well as enrichment-based sequencing experiments to identify mRNAs modified by m6A (45, 46). With respect to stem cell pluripotency, it has been shown that reduced m6A levels promote pluripotency (36–38). One explanation that has been proposed for how m6A levels affect stem cell pluripotency is that mRNAs for several components of the core pluripotency circuitry in mouse ESCs have reduced m6A, thereby increasing the half-life of these factors (36).
Here we describe the regulation of m6A mRNA by GSK-3 in mouse ESCs. We find that FTO (fat mass and obesity–associated) protein levels are elevated in Gsk-3 DKO ESCs compared with WT ESCs, resulting in a concomitant decrease in m6A levels. The mechanism underlying this effect is shown to be the phosphorylation of FTO by GSK-3 and subsequent ubiquitination, a process that is impaired in Gsk-3 DKO ESCs.
Results
FTO protein levels are elevated in Gsk-3 DKO ESCs
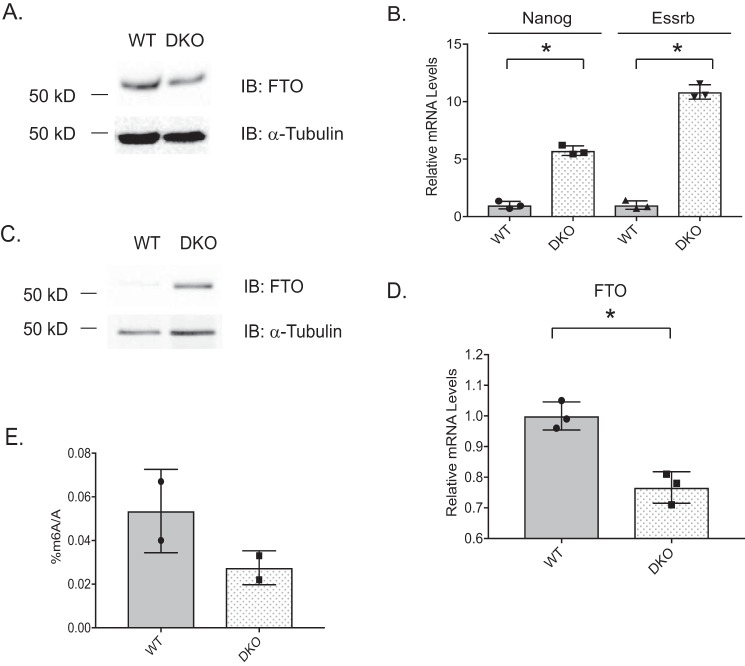
Reduced GSK-3 activity and decreased levels of m6A are both associated with promotion of pluripotency. We therefore investigated whether there was a direct connection between GSK-3 and m6A. Because stem cell pluripotency is enhanced by reduced m6A mRNA levels, an increase in FTO protein would be one route to achieving decreased m6A mRNA levels. Because the stereotypical role for GSK-3 is to negatively regulate target substrates, our hypothesis was that FTO protein levels are kept low by GSK-3 activity, and consequently, we should observe an increase in FTO protein levels in Gsk-3 DKO ESCs. We initiated our study by examining the levels of FTO protein in WT and Gsk-3 DKO ESCs grown under standard conditions (supplemented with LIF). We observed no difference in FTO protein between these cells (Fig. 1A). We then realized that, by using culture conditions containing LIF, both WT and Gsk-3 DKO ESCs would be pluripotent. Thus, adding LIF would not help us to understand how loss of GSK-3 was promoting pluripotency. It had been shown previously that Gsk-3 DKO ESCs would remain pluripotent when grown in the absence of LIF for as long as 14 days (12). Under these same conditions, WT ESCs progressively differentiate. By growing ESCs without LIF, we were able to obtain conditions in which we could now assess how loss of GSK-3 promotes pluripotency. Persistence of pluripotency in Gsk-3 DKO ESCs in the absence of LIF was verified by examining mRNA levels of the pluripotency-associated genes Nanog and estrogen receptor-β (Esrrb) by qPCR. As seen in Fig. 1B, relative levels of Nanog and Esrrb mRNA remain elevated in Gsk-3 DKO ESCs compared with WT ESCs, demonstrating that even without supplementation with LIF, Gsk-3 DKO ESCs remain pluripotent. We then isolated proteins from WT and Gsk-3 DKO ESCs, separated by SDS-PAGE, and examined FTO protein levels by Western blotting. We observed substantially higher FTO protein levels in Gsk-3 DKO ESCs (Fig. 1C). We then asked whether higher FTO protein levels was due to increased levels of mRNA. qPCR analysis of FTO mRNA revealed no increase in Gsk-3 DKO ESCs, and in fact, the mRNA levels were decreased (Fig. 1D). These data suggest that the elevated levels of FTO protein seen in Gsk-3 DKO ESCs are probably due to post-transcriptional effects of GSK-3.
Figure 1.
GSK-3 activity regulates FTO protein levels. A, Western blot analysis of FTO protein in WT and Gsk-3 DKO ESCs grown under standard conditions in the presence of LIF. Blots were then stripped and reprobed with α-tubulin antibody to ensure equal loading of protein lysates. B, qPCR data showing the expression of pluripotency-related genes when grown in the absence of LIF. WT and Gsk-3 DKO ESCs were grown in the absence of LIF for 14 days before collecting RNA. mRNA levels of pluripotency markers Nanog and Esrrb are shown as relative with respect to Gapdh mRNA levels. Each experiment shown was performed in triplicate. Error bars, S.D. *, statistical significance between the indicated groups (p < 0.05, unpaired t test, two-tailed). C, Western blot analysis of FTO protein in WT and Gsk-3 DKO ESCs grown in the absence of LIF for 14 days. Blots were then stripped and reprobed with α-tubulin antibody to ensure equal loading of protein lysates. D, qPCR analysis of FTO mRNA levels in WT and Gsk-3 DKO ESCs in the absence of LIF. Relative mRNA levels are with respect to Gapdh mRNA. Each experiment shown was performed in triplicate. Error bars, S.D. *, statistical significance between the indicated groups (p < 0.05, unpaired t test, two-tailed). E, m6A quantification. mRNA was isolated from WT and Gsk-3 DKO ESCs grown in the absence of LIF for 14 days. mRNA was digested into single nucleotides and quantified by UHPLC. Data depict the ratio of m6A with respect to the total number of adenosines in each sample and are the average of three technical replicates for each sample. Results are from two biological replicates.
m6A quantification
Given that FTO is an RNA demethylase and FTO protein is elevated in Gsk-3 DKO ESCs relative to WT ESCs, we sought to quantitatively measure the amounts of m6A mRNA in ESCs by UHPLC-MS. Total RNA was isolated from WT and Gsk-3 DKO ESCs grown in the absence of LIF for 14 days. We then enriched for mRNA, which was then digested into single nucleotides. Targeted UHPLC-MS detection of individual nucleotides allowed us to calculate the relative amount of m6A, which was normalized against the total adenosine pool to correct for any differences in the amount of RNA analyzed in each experiment. Analysis revealed a 49% reduction in m6A nucleotides in Gsk-3 DKO ESCs compared with WT ESCs (Fig. 1E), consistent with the higher levels of FTO protein observed in Gsk-3 DKO ESCs.
Small-molecule inhibitors of GSK-3 increase FTO protein
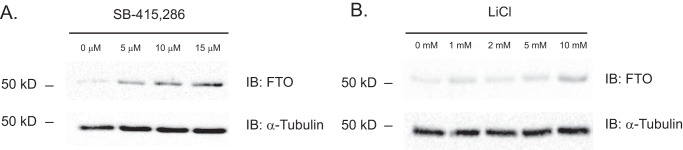
The elevation of FTO protein was not limited to Gsk-3 DKO ESCs. We extended our initial observation to WT ESCs treated with small-molecule inhibitors of GSK-3. WT ESCs were grown in the absence of LIF for 14 days and then treated with increasing doses of the GSK-3 inhibitors SB-415,286 or lithium chloride for 48 h. Examination of FTO protein by Western blotting revealed a dose-dependent increase in FTO protein (Fig. 2, A and B). These results strongly support the notion that GSK-3 inhibition, even for a short time, is sufficient to result in elevated levels of FTO protein in ESCs.
Figure 2.
Small-molecule Gsk-3 inhibitors stabilize FTO protein in WT ESCs. A and B, Western blots showing FTO protein levels in WT ESCs grown in the absence of LIF for 14 days and then treated with the indicated final concentrations of the GSK-3 inhibitors SB-415,286 (A) or lithium chloride (LiCl) for 2 days (B). Blots were then stripped and reprobed with α-tubulin antibody to ensure equal loading of protein lysates.
FTO phosphorylation
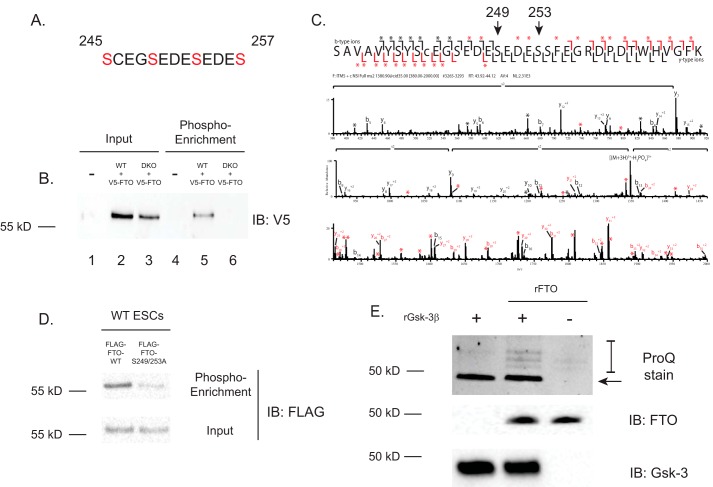
Because FTO protein levels are modulated by GSK-3 activity, we hypothesized that FTO is phosphorylated by GSK-3, and a repeated GSK-3 consensus motif was found within mouse FTO (amino acids 245–257) (Fig. 3A). To test whether FTO phosphorylation occurs in mouse ESCs, we overexpressed murine V5-tagged FTO in both WT and Gsk-3 DKO ESCs. Phosphorylated proteins were enriched using a Pro-Q Diamond kit, and proteins were analyzed by Western blotting for the presence of FTO. Examination of total protein lysates revealed that FTO-V5 was expressed in both WT and Gsk-3 DKO ESCs, indicating successful transfection of cells. Analysis of phospho-enriched proteins revealed FTO-V5 in WT ESCs, but not in Gsk-3 DKO ESCs (Fig. 3B). From this experiment, we conclude that FTO is normally phosphorylated in WT ESCs, and this phosphorylation is impaired in Gsk-3 DKO ESCs.
Figure 3.
Analysis of FTO phosphorylation in ESCs. A, consensus GSK-3 phosphorylation motif in mouse FTO. The numbering indicates amino acid position within FTO protein; serines are highlighted in red. B, Western blotting of overexpressed V5-tagged FTO (V5-FTO) in WT and Gsk-3 DKO ESCs before (lanes 1–3) and after enrichment for phosphorylated proteins (lanes 4–6). C, identification of FTO phosphorylation sites. Mass spectrometry was performed on FLAG-FTO overexpressed in WT ESCs. Black bars located within the sequence represent singly and doubly charged nonphosphorylated b- or y-ions. Red bars represent singly and doubly charged phosphorylated b- or y-ions. Asterisks represent neutral water losses from nonphosphorylated fragment ions (black, −18 Da) and neutral phosphate losses from phosphorylated fragment ions (red, −98 Da). c, a carbamidomethylated Cys residue. FTO is phosphorylated on serines 249 and 253 in mouse ESCs, within the GSK-3 consensus sequence. D, FLAG Western blotting of overexpressed WT FLAG-tagged FTO (FLAG-FTO-WT) and FLAG-FTO mutated at the GSK-3 phosphorylation site (FLAG-FTO-S249A/S253A) in WT ESCs after enrichment for phosphorylated proteins. Protein lysates before phospho-enrichment (Input) show total expression of FLAG-FTO proteins. E, in vitro kinase assay. Recombinant GSK-3β (rGsk-3β) was incubated with recombinant mouse FTO (rFTO) for 1 h at 30 °C. Proteins were then separated on 7.5% Tris/Tricine acrylamide gels and stained with Pro-Q Diamond stain to detect phosphorylated proteins (top). The arrow shows that the recombinant GSK-3β is phosphorylated. Phosphorylated FTO is denoted by the bracket. Middle panel, Western blotting showing the amount of recombinant FTO added to each reaction; bottom panel, Western blotting showing the amount of recombinant GSK-3β added to each reaction.
Mapping of FTO phosphorylation in vivo
We next sought to identify the precise amino acids within FTO that are phosphorylated in WT ESCs and not in Gsk-3 DKO ESCs. ESCs were transfected with murine FLAG-tagged FTO and then immunoprecipitated, trypsinized, and analyzed by LC-MS/MS. These results definitively showed the presence of phosphorylation in the region of FTO containing the GSK-3 consensus sequences, specifically on Ser-249 and Ser-253. Collision-induced dissociation of the singly phosphorylated peptide and carbamidomethylated (M + 3H)3+ ion (1380.2174 m/z, sequence SAVAVYSYSCEGSEDESEDESSFEGRDPDTWHVGFK) generates fragment ions that suggest a mixed population of isobaric peptides that harbor phosphorylated species on at least two residues (Fig. 3C). The observed fragment ions primarily suggest a mix of phosphorylation sites on both serine residues found in the sequence ∦ EG(SEDES)ED ∦ (where the parentheses denote the amino acid motif that was found to be phosphorylated, where both serines were phosphorylated). No doubly or triply phosphorylated forms of this sequence were observed.
To confirm that phosphorylation of FTO within the GSK-3 consensus motif accounts for the phosphorylation identified by phospho-enrichment, we created murine FTO in which serines 249 and 253, found to be phosphorylated by LC-MS/MS, are mutated to alanines (pCAGEN-FLAG-FTO-S249A/S253A). When WT FLAG-FTO and FLAG-FTO S249A/S253A were transfected into WT ESCs and phosphorylated proteins were enriched using the Pro-Q Diamond kit, we recovered less of the serine mutant FTO compared with WT FTO (Fig. 3D). This suggests that changing the GSK-3 consensus motif in FTO is sufficient to reduce its phosphorylation in vivo, further strengthening the argument that GSK-3 is the kinase responsible for FTO phosphorylation.
FTO is a substrate for GSK-3β
Because the role for GSK-3 in controlling FTO protein levels could be indirect, we sought to ask whether GSK-3 could directly phosphorylate FTO. We performed in vitro kinase assays using recombinant GSK-3β and recombinant mouse FTO protein. Using conditions that resulted in the phosphorylation of recombinant 2N4R Tau (47) by GSK-3β (data not shown), incubation of FTO with recombinant GSK-3β in the presence of fresh ATP resulted in phosphorylation of FTO (Fig. 3E). From this experiment, we conclude that FTO is probably a direct substrate for GSK-3β. In addition, the relatively weak phosphorylation of FTO by GSK-3β points to the likely requirement of a priming kinase for optimal FTO phosphorylation by GSK-3β.
FTO ubiquitination
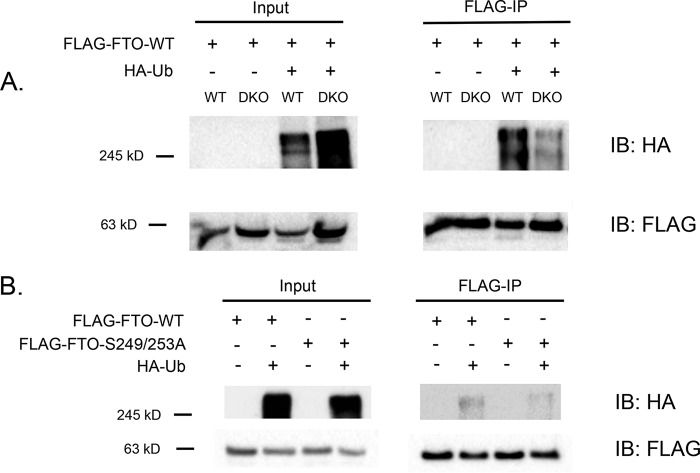
Because FTO protein levels are increased in Gsk-3 DKO ESCs compared with WT ESCs, we speculated that this could be due to a reduction in FTO ubiquitination and subsequent degradation. To assess this possibility, we examined the status of FTO ubiquitination in WT and Gsk-3 DKO ESCs. Cells were co-transfected with FLAG-FTO and HA-ubiquitin, then treated with lactacystin to inhibit the proteasome, allowing for the accumulation of ubiquitinated proteins. We then immunoprecipitated FTO using anti-FLAG antibody and performed Western blotting for ubiquitin using an anti-HA antibody. We found that FTO is modified by ubiquitin in WT ESCs, but this modification is significantly impaired in Gsk-3 DKO ESCs (Fig. 4A).
Figure 4.
Analysis of FTO ubiquitination in ESCs. A, Western blotting of WT and Gsk-3 DKO ESCs transfected with FLAG-FTO-WT alone or with HA-ubiquitin (HA-Ub). FTO was immunoprecipitated with anti-FLAG (FLAG-IP). Input and FLAG-IP lysates were then separated by SDS-PAGE. Top and middle panels, HA Western blots. Bottom panel, FLAG Western blotting on the same lysates. B, Western blotting of WT ESCs transfected with FLAG-FTO-WT or FLAG-FTO-S249A/S253A, with and without HA-Ub. FTO was immunoprecipitated with anti-FLAG (FLAG-IP). Input and FLAG-IP lysates were then separated by SDS-PAGE. Top, HA Western blotting. Bottom, FLAG Western blotting on the same lysates.
Next, we examined whether preventing FTO phosphorylation by GSK-3 was sufficient to affect FTO ubiquitination. WT ESCs were transfected with HA-ubiquitin and either FLAG-FTO or FLAG-FTO S249A/S253A. Using the same procedure described above for Fig. 4A, we observed that FTO modification by ubiquitin is reduced in the FTO S249A/S253A mutant compared with WT FTO (Fig. 4B). Therefore, polyubiquitination associated with proteasome-mediated protein degradation is impaired with respect to FTO in Gsk-3 DKO ESCs, providing a likely explanation for the increased FTO protein levels seen in Gsk-3 DKO ESCs.
GSK-3β rescues m6A levels and pluripotency gene expression in Gsk-3 DKO ESCs
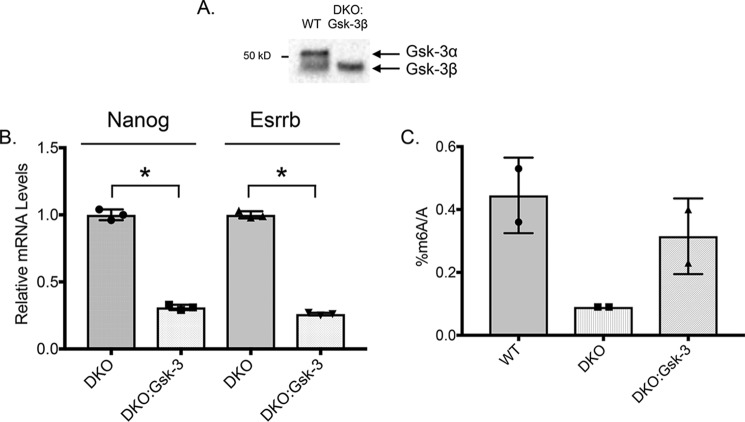
Reducing GSK-3 activity via genetic deletion or small-molecule inhibitors promotes stem cell pluripotency, as well as regulating m6A mRNA levels. Re-expressing GSK-3β in Gsk-3 DKO ESCs would therefore be predicted to reverse these effects. Gsk-3 DKO ESCs stably expressing GSK-3β (DKO:GSK-3β) were created (Fig. 5A) and analyzed. WT, Gsk-3 DKO, and DKO:GSK-3β ESCs were grown in the absence of LIF for 14 days and then analyzed for gene expression and m6A levels. Compared with Gsk-3 DKO ESCs, DKO:GSK-3β ESCs had significantly reduced levels of expression of both pluripotency-associated genes Nanog and Esrrb, as assessed by qPCR (Fig. 5B). Furthermore, the reduced levels of m6A seen in Gsk-3 DKO ESCs were almost completely restored to WT levels in DKO:GSK-3β ESCs (Fig. 5C). These data, along with the in vitro kinase assay results, help to reinforce the notion that GSK-3 is indeed regulating FTO in vivo, and this regulation is intimately connected to controlling stem cell pluripotency.
Figure 5.
Expression of GSK-3β in Gsk-3 DKO ESCs rescues m6A. A, Western blotting of WT and Gsk-3 DKO ESCs stably expressing GSK-3β. ESCs were grown in the absence of LIF for 14 days. GSK-3α/β Western blotting demonstrates the expression of GSK-3β in Gsk-3 DKO ESCs (DKO:GSK-3β). Note the absence of GSK-3α protein. B, qPCR analysis of the expression of pluripotency-related genes in DKO:GSK-3β ESCs after 14 days in culture in the absence of LIF. Relative mRNA levels of pluripotency markers Nanog and Esrrb are shown with respect to Gapdh mRNA. Each experiment shown was performed in triplicate. Error bars, S.D. *, statistical significance between the indicated groups (p < 0.05, unpaired t test, two-tailed). C, m6A quantification. Representative experiment showing the relative quantification of m6A levels in RNA isolated from WT, Gsk-3 DKO, and DKO:GSK-3β ESCs grown in the absence of LIF for 14 days. Each experiment shown was performed in duplicate. Error bars, S.D.
m6A-Seq
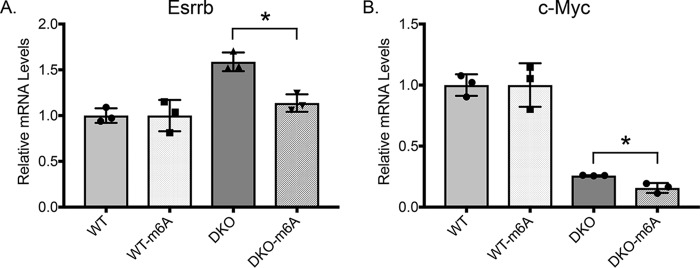
Whereas m6A mRNA levels in Gsk-3 DKO ESCs were approximately half of that detected in WT ESCs, the LC-MS/MS analysis gave no information as to which specific mRNAs had reduced m6A modifications. To obtain this information, we performed m6A-Seq, making modifications to the approach taken by Meyer et al. (45) and Dominissini et al. (46). We first isolated total RNA from WT and Gsk-3 DKO ESCs, enriched for mRNA using oligo(dT) beads, and then immunoprecipitated m6A mRNA using an anti-m6A antibody coupled to magnetic beads. The resulting m6A mRNA was then chemically fragmented, cDNA was created using random primers, adaptors were ligated to the ends, and next-generation sequencing was performed. m6A-Seq analysis was performed to identify transcripts that had at least 2-fold fewer m6A modifications in Gsk-3 DKO ESCs compared with WT ESCs and relative to input reads for WT and Gsk-3 DKO ESCs. 899 mRNAs were identified that met these criteria (Fig. S1). To verify the results obtained from the m6A-Seq experiment, we performed meRIP-qPCR on the mRNA from WT and Gsk-3 DKO ESCs that was immunoprecipitated with the m6A antibody, specifically examining mRNAs that were found to be hypomethylated in Gsk-3 DKO ESCs. We found that m6A levels for the pluripotency-associated mRNAs Esrrb (Fig. 6A) and c-myc (Fig. 6B) were both reduced by ∼40% with respect to input mRNA in Gsk-3 DKO ESCs, providing independent validation of our m6A-Seq results.
Figure 6.
m6A-Seq validation. A and B, meRIP-qPCR analysis of input (pre-meRIP) and m6A levels of Esrrb (A) and c-myc (B) mRNAs from WT and Gsk-3 DKO ESCs. Bars labeled WT or DKO, mRNA levels before meRIP. Bars labeled WT-m6A and DKO-m6A, mRNA levels after Me-RIP. All meRIP-qPCR analyses are relative to Gapdh mRNA levels. Each experiment shown was performed in triplicate. Error bars, S.D. *, statistical significance between the indicated groups (p < 0.05, unpaired t test, two-tailed).
Our approach for detecting m6A relied on an antibody against m6A, but this antibody was recently shown to detect both m6A and N6,2′-O-dimethyladenosine (m6Am) (48), making it impossible for us to discriminate between these two RNA modifications using this technique (49). However, because m6Am is only found directly adjacent to the 7-methylguanosine cap on mRNA (50), we are confident that most of the changes we observe are m6A, but we cannot discount the possibility of changes in m6Am as well.
Because we were interested in whether there is a direct connection between mRNAs with GSK-3–dependent reduction and mouse ESC pluripotency, we wanted to examine whether any of the core pluripotency factors described by Young (51) had reduced m6A modifications in Gsk-3 DKO ESCs. We found that only a handful of the mRNAs for these pluripotency factors (Esrrb, c-myc, Sox2, and Smad1) had reductions in m6A. Furthermore, because Mettl3−/− ESCs also have reduced m6A mRNA levels as well as enhanced pluripotency (36), we looked for concordance between mRNAs that show reduced m6A modifications in both Gsk-3 DKO ESCs and Mettl3−/− ESCs. With respect to the mRNAs described as being part of the pluripotency circuit, we found reduced m6A levels on Lin28, c-myc, and Max in Gsk-3 DKO ESCs. Therefore, whereas there are modest effects on known regulators of pluripotency, the overall effect on these regulators could help to explain the persistent pluripotency seen in Gsk-3 DKO ESCs.
Discussion
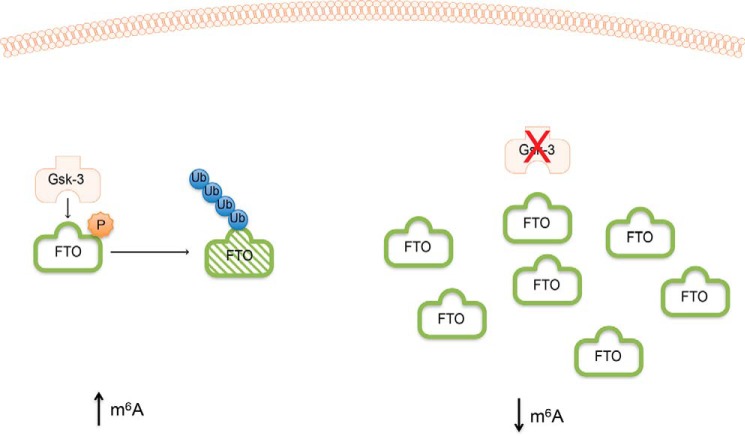
The epitranscriptome landscape in eukaryotic cells has recently been revealed in detail. Besides m6A-Seq studies that have precisely identified the location of methylated adenosines in RNA molecules (45, 46), details are also emerging regarding the reversible nature of epigenetic marks on RNA (52). This reversibility implies that RNA modifications are regulated, but the nature of this regulation is just beginning to be understood. Here, we describe a compelling role for GSK-3 in the regulation of m6A levels via FTO (Fig. 7).
Figure 7.
Proposed model. Left, when GSK-3 is active, it phosphorylates FTO, which targets it for ubiquitin-mediated degradation, keeping FTO levels low and resulting in elevated levels of m6A mRNA. Right, in the absence of GSK-3 activity, FTO protein accumulates, resulting in decreased levels of m6A mRNA.
We provide evidence that GSK-3 plays a role in modulating mRNA adenosine methylation in mouse embryonic stem cells through controlling the protein levels of the RNA demethylase FTO. Whereas GSK-3 inhibition or deletion has been known to regulate embryonic stem cell pluripotency, the precise role for GSK-3 has been difficult to pin down. Several different aspects of GSK-3 function have been investigated, but a great deal of focus has been on examining the GSK-3–dependent signaling pathways that promote pluripotency. Most of these studies have focused on Wnt and PI3K signaling because activation of each pathway results in inhibition of GSK-3, but the conclusions have not been clear-cut. For example, with respect to Wnt signaling, it has been proposed that pluripotency via GSK-3 inhibition works through both Tcf3-dependent (26, 53, 54) and Tcf3-independent (24) mechanisms. In addition, several lines of evidence also support a role for PI3K in promoting stem cell pluripotency (55). Detailed mechanistic studies have determined that GSK-3 activity was regulated in part by PI3K signaling, yet the precise mechanism by which inhibition of GSK-3 was promoting stem cell pluripotency remained elusive (8, 29, 34, 56). Our findings raise the possibility that Wnt signaling and/or PI3K signaling via GSK-3 could be regulating mRNA methylation, thereby controlling stem cell pluripotency. Further studies will be required to determine whether specific signaling pathways have an effect on FTO phosphorylation and mRNA methylation.
The phosphorylation of proteins has been shown to be a common prerequisite for subsequent polyubiquitination (57), and GSK-3 activity has been shown to precede substrate ubiquitination (58). A prime example is β-catenin, the main effector of canonical Wnt signaling. β-Catenin is ubiquitinated and degraded only after phosphorylation by GSK-3 on N-terminal serines and threonines (59). Activation of Wnt signaling by the addition of Wnt ligand (60), genetic deletion of GSK-3 (12), and small-molecule inhibition of GSK-3 (10, 11, 61) all result in the accumulation of β-catenin protein. We observe a similar paradigm for GSK-3 with respect to FTO. Although we provide evidence that FTO is a direct target of GSK-3, the in vitro kinase assay for FTO lacked the robustness of phosphorylation seen in the positive control (Tau). This result probably reflects that GSK-3 often phosphorylates substrates that have been primed by another kinase (62); Tau does not have a requirement for priming with respect to GSK-3 (63). Therefore, it is quite possible that phosphorylation of FTO by another kinase would result in enhanced phosphorylation by GSK-3. Because our in vitro kinase assay only examined the effect of GSK-3 on FTO phosphorylation, the next reasonable step would be to identify kinases that work in tandem with GSK-3 in the phosphorylation of FTO.
With our data describing a novel role for GSK-3 activity in modulating post-transcriptional modifications of mRNA, one could envision how signaling pathways that converge on GSK-3 via distinct routes (e.g. Wnt and PI3K) could synergize with respect to gene expression. For example, whereas one GSK-3–dependent pathway could promote the transcription of a specific mRNA, a different GSK-3–dependent pathway could elevate FTO protein levels, thereby reducing methylation of the same mRNA. An exciting possibility raised by our work is that transcription and RNA modifications are occurring concurrently, resulting in a fine-tuning of the net transcriptional response to signaling events.
These data may also be relevant in further understanding how signaling pathways have the capacity to directly alter the m6A modifications of specific mRNAs, as well as providing insights into the mechanisms of GSK-3–mediated signaling in other cell types and disease states. FTO has been shown to be expressed at high levels in the brain (64–66), implicating an important role in regulating mRNA methylation. Although details on the relationship between RNA modifications and disease are just beginning to be examined in depth (67), it is worth noting that GSK-3 is inhibited by the mood-stabilizing drug lithium (9, 10). A recent study showed that genes involved in m6A modification are associated with major depressive disorder in a Han Chinese population (68). The data presented here raise the possibility that toggling the levels of methylated adenosine on mRNA in the brain could play a role in mood stabilization.
The FTO locus in humans has been shown unequivocally to be associated with predisposition to obesity (69–72), and genetic deletion of FTO in mice was shown to protect animals from becoming obese. Despite the connection between FTO and obesity, the precise role that FTO plays in this process is not well-understood, although regulation of the expression of nearby genes has been proposed (73). However, this explanation is independent of the biological function of the FTO protein as a 2-oxoglutarate–dependent RNA demethylase (64, 74). GSK-3 is a major regulator of cellular metabolism and has been linked to obesity via its role in regulating the insulin/PI3K pathway. Interestingly, it was recently found that transgenic mice expressing a form of GSK-3 that is insensitive to inhibition by PI3K signaling were protected from developing metabolic syndrome, which includes obesity (75). Although it is not yet known if the role of FTO in obesity is connected to GSK-3 activity and mRNA methylation levels, it is a tantalizing possibility that should be explored further.
Although we identify a new mechanism that results in changes in mRNA methylation, the biological effects of these changes are likely to be complicated. Since the discovery that m6A tags on mRNA are reversible (43), it has become widely accepted that this modification results in decreased mRNA half-life. This is due to the recognition of m6A mRNA by the reader protein YTH domain family 2 (YTHDF2) (76). Yet numerous studies have shown that the m6A modification of mRNA affects nearly every step in its life cycle, and the resulting effect on m6A-modified mRNA depends on the specific YTH domain family reader protein that binds to the mRNA (77). Further complicating the fate of m6A-modified mRNA is the recent discovery that insulin-like growth factor-2 mRNA-binding proteins (IGFBP1–3) also bind to m6A-containing transcripts and have the opposite effect as YTHDF2, resulting in stabilization of mRNA (78). Taken together, this means that we are unable to speculate what effect reducing m6A marks has on the mRNAs affected in Gsk-3 DKO ESCs.
While this manuscript was in preparation, data from Jaffrey and colleagues (48) were published showing that FTO does not specifically demethylate m6A RNA, but instead prefers to demethylate m6Am. Our approach for detecting m6A relied on an antibody against m6A, but this antibody was shown to detect both m6A and m6Am (49). In addition, whereas the approach we used for m6A-Seq did not provide us with the precise locations of demethylated adenosines in RNA from Gsk-3 DKO ESCs, we nonetheless observed that hundreds of mRNAs had regions largely devoid of methylated adenosines. This strongly suggests that demethylation of m6Am by FTO is not the only RNA demethylation occurring in Gsk-3 DKO ESCs. Mauer et al. (48) also provided evidence that m6A demethylation is mediated by Alkbh5. Our m6A-Seq data raise the possibility that GSK-3 activity could be directly or indirectly regulating Alkbh5, in addition to FTO. We do not observe an increase in Alkbh5 protein or mRNA in Gsk-3 DKO ESCs (data not shown), but it is possible that either GSK-3 regulates Alkbh5 activity or that there is coordination between the demethylation of m6Am and m6A. Further investigation will be required to determine whether either of these possibilities is indeed true.
In summary, we have uncovered the first example of a mechanism by which mRNA demethylation can be regulated. This finding dovetails nicely with the recent discovery from Vallier and colleagues (79) that Activin/Nodal signaling via Smad2/Smad3 regulates the activity of the Mettl3 mRNA methyltransferase complex. Interestingly, it has been shown that simultaneous inhibition of GSK-3 and Activin/Nodal signaling in mouse ESCs promotes pluripotency (80), yet at the time of publication, neither of these molecules were known to regulate mRNA methylation. In light of both the findings from Bertero et al. (79) and those described here by our group on the regulation of mRNA methylation, an intriguing possibility is that these experiments could be reinterpreted as attaining pluripotency via the simultaneous inhibition of mRNA methylation and promotion of mRNA demethylation. Further experiments will be able to directly test this hypothesis. Taken together, we provide strong evidence demonstrating that the effects of GSK-3 are not limited to canonical signaling pathways but can affect epigenetic processes as well.
Experimental procedures
Plasmids
The mammalian expression vector pCAGEN (from Connie Cepko, Addgene plasmid 11160) was linearized with EcoRI, and synthetic gBlocks (IDT) containing V5-FTO, FLAG-FTO, or FLAG-FTO-S249A/S253A were then cloned into pCAGEN using Gibson Assembly master mix (New England Biolabs). Plasmids containing inserts were identified by restriction digests and confirmed by DNA sequencing (Eton Biosciences).
Cell culture and transfection
Feeder-free WT mouse (E14K), Gsk-3 DKO ESCs, and Gsk-3 DKO/Gsk-3β rescue ESCs were grown on 0.1% gelatin-coated plates with high-glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 15% fetal bovine serum (HyClone), 1% nonessential amino acids, 1% sodium pyruvate, 1% l-glutamine, 1% penicillin/streptomycin (Gibco) 55 μm 2-mercaptoethanol, and 1000 units/ml recombinant LIF or ESGRO (Millipore). Medium was replenished every other day. WT and Gsk-3 DKO ESCs were transfected using PEI (81). 1 × 106 ESCs/well were resuspended in Opti-MEM (Gibco) along with either 1800 ng of pCAGEN-V5-FTO, pCAGEN-FLAG-FTO, or pCAGEN-FLAG-FTO-S249A/S253A, along with 200 ng of pMax-GFP. After incubation for 30 min at room temperature, cells were added to gelatin-coated 6-well plates containing complete medium. Transfection efficiencies were confirmed after 18 h via fluorescence microscopy (Evos FL, Thermo Fisher Scientific). For identification of phosphorylation sites on FTO, we plated 6 × 106 WT and Gsk-3 DKO ESCs per 10-cm plate and used 10 plates of each cell line. Cells on each plate were transfected with 11 μg of pCAGEN-FLAG-FTO and 1 μg of pMax-GFP using the PEI method.
Immunoprecipitation and Western blotting
For protein extraction, ESCs were resuspended in lysis buffer (137 mm NaCl, 10 mm Tris, pH 7.4, 1% IGEPAL) containing protease inhibitor mixture (1:100; Sigma) and lysed by vigorous vortexing. For V5 immunoprecipitations, 10% of the lysate was set aside, whereas the remaining protein lysate was added to V5-agarose beads (Bethyl Laboratories). Lysates were incubated with beads for 2 h at room temperature while rotating and then centrifuged at 3000 × g for 1 min at 4 °C to pellet beads. Pellet was washed four times with lysis buffer containing protease inhibitor mixture (1:100; Sigma). Protein was eluted from the beads by adding 40 μl of 1× Tris/Tricine sample buffer and incubating for 5 min at 100 °C. Beads were pelleted by centrifugation at 14,000 rpm for 1 min, and the supernatant was transferred to a fresh microcentrifuge tube and stored at −20 °C until use. For FLAG immunoprecipitations, 10% of the lysate was set aside, whereas the remaining protein lysate was added to FLAG-magnetic agarose beads (Sigma). Lysates were incubated with beads for 4 h at room temperature while rotating, placed on the magnet, and supernatant was saved. Beads were washed three times in lysis buffer and then stored at 4 °C.
Western blotting was performed after electrophoresis on 7.5% Tris/Tricine acrylamide gels and transfer of protein onto nitrocellulose membranes at 100 V for 1 h. Blots were blocked for 1 h with either 5% milk/TBST (150 mm NaCl, 50 mm Tris, pH 7.4, 0.1% Tween) (for α-FTO, α-V5, and α-FLAG antibodies) or 5% BSA/TBST (for α-tubulin antibody). Primary antibodies (α-FTO mAb (1:1000; PhosphoSolutions, catalog no. 597-FTO, lot CH7166), α-V5 mouse mAb (1:5000; Bethyl Laboratories, catalog no. A190-120A, lot A190-120A-6), α-FLAG M2 mouse mAb (1:1000; Sigma, catalog no. F3165, lot SLBQ7119V), α-HA rabbit mAb (1:1000; Cell Signaling Technologies; catalog no. 3724, lot 8), and α-tubulin rabbit mAb (1:1000; Cell Signaling Technologies, catalog no. 2125, lot 11)) were diluted in blocking solutions overnight at 4 °C. Blots were washed and incubated for 30 min in α-mouse or α-rabbit IgG horseradish peroxidase secondary antibody (GE Healthcare) diluted 1:20,000 in 5% milk/TBS or 5% BSA/TBST. Proteins were visualized using ECL Plus detection reagents (GE Healthcare).
RNA isolation, cDNA synthesis, and quantitative PCR
1 × 106 ESCs/well were plated on a gelatin-coated 6-well plate. RNA was isolated using TRIzol reagent (Thermo Fisher Scientific) and extracted using Direct-zol RNA Miniprep columns (Zymo) following the manufacturer's protocol. RNA was quantified using a Nanodrop 2000 (Thermo Fisher Scientific).
2 μg of total RNA was used to synthesize cDNA. cDNA was synthesized using the High Capacity Reverse Transcriptase kit (Applied Biosystems) according to the manufacturer's protocol. The amount of input RNA used was kept constant for each reverse transcription reaction. Reactions were run on a StepOne Real-Time PCR System (Applied Biosystems) using PrimeTime gene expression master mix (IDT) and PrimeTime qPCR assays for FTO (Mm.PT.58.32888407), Nanog (Mm.PT.58.23510265), and Esrrb (Mm.PT.58.14246772) (IDT). Three biological replicates and three technical replicates were used for WT, Gsk-3 DKO, and Gsk-3 DKO/Gsk-3β ESCs. All threshold cycle (Ct) values were normalized to a mouse Gapdh endogenous control (Mm.PT.39a.1) (IDT), and relative quantification was calculated from the median Ct value.
Enrichment of phosphoproteins
Total protein was isolated from WT and Gsk-3 DKO mESCs that were transfected with pCAGEN-FTO-V5, pCAGEN-FTO-FLAG, or pCAGEN-FTO-S249A/S253A-FLAG. Samples were enriched for phosphoproteins using the Pro-Q Diamond phosphoprotein enrichment kit (Molecular Probes) according to the manufacturer's protocol for nondenatured protein lysates. Lysates were then separated on 7.5% Tris/Tricine acrylamide gels, and Western blotting for V5 or FLAG was performed as described above.
In vitro kinase assay
Recombinant human GSK-3β (Sigma-Aldrich) was incubated with recombinant mouse FTO (Sigma-Aldrich) or recombinant 2N4R His-tagged Tau in the presence of fresh ATP for 1 h at 30 °C. Reactions were then run on 7.5% Tris/Tricine acrylamide gels, and then the gels were stained with Pro-Q Diamond for 1 h followed by destaining for 1 h to detect phosphorylated proteins. Gels were imaged using a Gel Doc XRS+ (Bio-Rad) using the 560DF50 filter.
Protein mass spectrometry
Immunoprecipitated FLAG-FTO from WT and Gsk-3 DKO ESCs washed with 100 μl of 100 mm ammonium bicarbonate (pH 8.0) three times while retained on FLAG magnetic beads. Cysteine residues were reduced and alkylated at 22 °C using 10 mm DTT (Sigma-Aldrich) for 1.5 h followed by 25 mm iodoacetamide (Sigma-Aldrich) for 45 min in the dark. Proteins were digested on beads (1:20 enzyme/protein) with either trypsin or endoproteinase LysC (Promega) for 6 h at room temperature on a shaker. Peptides were removed with the supernatant and acidified to pH 2.5 using formic acid (Fisher).
Peptides were loaded directly onto a Waters nanoAcquity UPLC for separation on a 75 μm × 250-mm C18 BEH UPLC column (1.7-μm particle size; Waters) coupled to a Thermo LTQ-Orbitrap Velos mass spectrometer. A linear 70 min, 0.3 μl/min flow UPLC gradient was performed using 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B) with the following conditions: 2% solvent B hold for 17 min, 2–7% solvent B in 3 min, 7–32% solvent B in 25 min, 32–95% solvent B in 5 min, 95% solvent B hold for 4 min, 95 to 2% B in 1 min, 15-min hold at 2% B. MS scans were acquired at 60,000 resolution in the Orbitrap, and a top 10 data-dependent CID tandem MS method was used to acquire MS/MS scans in the ion trap. All data were searched against a Uniprot Mus musculus protein database using Mascot (Matrix Science) with the following parameters: 50-ppm peptide mass tolerance; 0.5-Da fragment ion tolerance; variable modifications including Gln to pyro-Glu conversion on N-terminal Gln, oxidation of Met, and phosphorylation of Ser/Thr/Tyr; fixed modification carbamidomethyl Cys; 1% false discovery rate; and two missed cleavages with trypsin enzyme specificity. All expected proteolyzed peptides were investigated by hand for the presence of any singly or doubly phosphorylated forms in the MS and MS/MS spectra. Phosphorylated peptides were sequenced by hand and confirmed by manual inspection of the MS/MS spectra. An additional LC-MS/MS experiment was conducted using the same parameters as above, plus a targeted MS/MS scan event at 1380.9 m/z to sequence the (M + 3H)3+ carbamidomethylated and singly phosphorylated peptide sequence SAVAVYSYSCEGSEDESEDESSFEGRDPDTWHVGFK.
Detection of ubiquitinated FTO
For detection of ubiquitinated FTO, we plated 6 × 106 WT and Gsk-3 DKO ESCs per 10-cm plate. Cells on each plate were transfected with 5.5 μg of pCAGEN-FLAG-FTO, 5.5 μg of HA-ubiquitin, and 1 μg of pMax-GFP using the PEI method. 24 h later, medium was changed, and cells were treated with 10 μm lactacystin for 6 h. Cells were then collected in lysis buffer containing deubiquitination enzyme inhibitors (N-ethylmaleimide and TPEN (N,N,N′,N′-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine)) to ensure recovery of ubiquitinated proteins. Lysates were separated on 7.5% Tris/Tricine acrylamide gels, and Western blotting for HA and FLAG was performed as described above.
m6A quantification
To quantify the amount m6A-modified mRNA, we first enriched for mRNA by isolating the population of poly(A)+ RNA from total RNA. 5 μg of total RNA was isolated from WT and Gsk-3 DKO ESCs as described above and then incubated with oligo(dT)25 magnetic beads (New England Biolabs) on a rotator overnight at 4 °C. Tubes were then placed on a magnetic plate for 1 min, and supernatant was saved as “oligo(dT) other” for further analysis. Beads were washed with 100 μl of RNase-free H2O three times. To elute RNA from beads, 20 μl of cold 10 mm Tris-HCl, pH 7.5, was added to beads and incubated at 80 °C for 2 min. Tubes were returned to the magnet, and eluted RNA was immediately transferred to a new tube and placed on ice. Poly(A)+ RNA was quantified using the Qubit HS RNA assay kit (Thermo Fisher Scientific). For m6A quantification, we used the method described by Jia et al. (43). 1 μg of poly(A)+ mRNA was digested by 4 μl of nuclease P1 (2 units) in 40 μl of LC-MS/MS buffer (25 mm NaCl and 2.5 mm ZnCl2) for 1 h at 37 °C, followed by the addition of 1 m NH4HCO3 and 0.5 units of alkaline phosphatase and incubation at 37 °C for another 1 h. To confirm digestion of RNA, samples were run on a 1% agarose gel, with an undigested RNA sample as a control. Samples were then analyzed by UHPLC-MS (Vanquish UHPLC coupled online with a Q Exactive quadrupole-Orbitrap mass spectrometer, operated in positive-ion mode; Thermo Fisher Scientific) at the University of Colorado School of Medicine Proteomics Core Facility, and the ratios of methyladenosine to adenosine were reported. Retention times and linearity ranges were determined against in-house libraries of light and deuterated standards available in the core for adenosine and methyladenosine (Sigma-Aldrich).
m6A levels were also quantified using the EpiQuik m6A RNA methylation quantification kit (Epigentek) following the manufacturer's instructions. Briefly, 200 ng of total RNA from WT, Gsk-3 DKO, and Gsk-3 DKO/Gsk-3β rescue ESCs was bound to wells in a clear 96-well plate via RNA high-binding solution and then incubated with m6A capture and detection antibodies, followed by colorimetric detection of signal (A450 nm) using an EnSpire plate reader (PerkinElmer Life Sciences). The readings were then used to calculate the relative quantification using a formula provided by the manufacturer.
m6A-Seq
RNA was isolated from WT and Gsk-3 DKO ESCs using TRIzol (Thermo Fisher Scientific) as described above. 15 μg of total RNA was used for mRNA isolation with oligo(dT)25 magnetic beads (New England Biolabs). 7 μg of poly(A)+ RNA from each sample was then incubated with Protein G magnetic beads (New England Biolabs) coupled with anti-m6A antibody (New England Biolabs, catalog no. E1611A, lot 0011602) rotating overnight at 4 °C. Beads were washed at room temperature, two times with reaction buffer (150 mm NaCl, 10 mm Tris HCl, pH 7.5, and 0.1% Nonidet P-40), two times with low-salt reaction buffer (50 mm NaCl, 10 mm Tris HCl, pH 7.5, and 0.1% Nonidet P-40), and two times with high-salt reaction buffer (500 mm NaCl, 10 mm Tris HCl, pH 7.5, and 0.1% Nonidet P-40). Beads were then resuspended in Buffer RLT (Qiagen) and placed on a magnet, and then m6A mRNA-containing solution was removed and placed in new tubes. Samples were concentrated further using MyOne Silane Dynabeads (Thermo Fisher Scientific). m6A mRNA values obtained using Qubit were: as follows 122 ng (WT) and 163 ng (DKO). m6A mRNA was chemically fragmented by incubating in 100 mm Tris-HCl, pH 7.0, and 100 mm ZnCl2 at 94 °C for 5 min, and the reaction was stopped with 0.5 m EDTA (82). Fragmented RNA was cleaned and concentrated using a commercial kit (Zymo). m6A-Seq was performed using the IonProton sequencing platform (Thermo Fisher Scientific) at PrimBio (Exton, PA).
meRIP-qPCR
50 ng of WT and Gsk-3 DKO mRNA from cells grown without LIF before and after immunoprecipitation with anti-m6A antibody was used for meRIP-qPCR. Briefly, SuperScript IV VILO cDNA kit (Thermo Fisher Scientific) was used to synthesize cDNA using the manufacturer's protocol. cDNA was diluted to 0.5 ng/μl for use in qPCR. Reactions were run on a StepOne real-time PCR system (Applied Biosystems) using PrimeTime gene expression master mix (IDT) and PrimeTime qPCR assays (IDT) for c-myc (Mm.PT.58.13590978) and Esrrb (Mm.PT.58.14246772). All Ct values were normalized to a mouse Gapdh endogenous control (Mm.PT.39a.1) (IDT), and relative quantification was calculated from the median Ct value.
Author contributions
J. N. E. initiated the study. K. J. F., J. N. E., and L. J. S. performed Western blotting, qPCR, and m6A quantification experiments. K. J. F. created FTO expression constructs and performed phospho-enrichment experiments. L. J. S. also performed in vitro kinase assays, ubiquitination assays, and Me-RIP qPCR. A. D., K. C. H., and J. L. B. performed and analyzed mass spectrometry experiments. M. K. M., S. G., and H. K. are undergraduates who provided technical assistance with a variety of experiments. C. J. P. conceived and coordinated the study, analyzed m6A-Seq data, and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Jeff Kuret, Xiaojun Ren, and Manabu Kurokawa for critical reading of the manuscript and helpful discussions. We also thank Jeff Kuret for the gift of recombinant Tau protein and Bradley Doble for the Gsk-3 DKO ESCs.
This work was supported in part by National Institutes of Health Grant R15GM119103-01 (to C. J. P.) and funding from the University of Colorado Denver. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1.
m6A-Seq data have been deposited to the GEO database and are available under accession ID GSE108327.
- GSK-3
- glycogen synthase kinase-3
- FTO
- fat mass and obesity–associated
- m6A
- methylated adenosine on the 6-carbon
- m6Am
- N6,2′-O-dimethyladenosine
- ESC
- embryonic stem cell
- DKO
- double knockout
- LIF
- leukemia-inhibitory factor
- qPCR
- quantitative PCR
- UHPLC
- ultra-high pressure liquid chromatography
- Esrrb
- estrogen receptor-β
- meRIP
- methylated RNA immunoprecipitation
- YTHDF2
- YTH domain family 2
- IGFBP
- insulin-like growth factor mRNA-binding protein
- PI3K
- phosphatidylinositol 3-kinase
- PEI
- polyethyleneimine
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- IB
- immunoblotting.
References
- 1. Force T., and Woodgett J. R. (2009) Unique and overlapping functions of GSK-3 isoforms in cell differentiation and proliferation and cardiovascular development. J. Biol. Chem. 284, 9643–9647 10.1074/jbc.R800077200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kockeritz L., Doble B., Patel S., and Woodgett J. R. (2006) Glycogen synthase kinase-3: an overview of an over-achieving protein kinase. Curr. Drug Targets 7, 1377–1388 10.2174/1389450110607011377 [DOI] [PubMed] [Google Scholar]
- 3. Grimes C. A., and Jope R. S. (2001) The multifaceted roles of glycogen synthase kinase 3β in cellular signaling. Prog. Neurobiol. 65, 391–426 10.1016/S0301-0082(01)00011-9 [DOI] [PubMed] [Google Scholar]
- 4. Frame S., and Cohen P. (2001) GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359, 1–16 10.1042/0264-6021:3590001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doble B. W., and Woodgett J. R. (2003) GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175–1186 10.1242/jcs.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coghlan M. P., Culbert A. A., Cross D. A., Corcoran S. L., Yates J. W., Pearce N. J., Rausch O. L., Murphy G. J., Carter P. S., Roxbee Cox L., Mills D., Brown M. J., Haigh D., Ward R. W., Smith D. G., et al. (2000) Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem. Biol. 7, 793–803 10.1016/S1074-5521(00)00025-9 [DOI] [PubMed] [Google Scholar]
- 7. Meijer L., Flajolet M., and Greengard P. (2004) Pharmacological inhibitors of glycogen synthase kinase 3. Trends Pharmacol. Sci. 25, 471–480 10.1016/j.tips.2004.07.006 [DOI] [PubMed] [Google Scholar]
- 8. Bone H. K., Damiano T., Bartlett S., Perry A., Letchford J., Ripoll Y. S., Nelson A. S., and Welham M. J. (2009) Involvement of GSK-3 in regulation of murine embryonic stem cell self-renewal revealed by a series of bisindolylmaleimides. Chem. Biol. 16, 15–27 10.1016/j.chembiol.2008.11.003 [DOI] [PubMed] [Google Scholar]
- 9. Klein P. S., and Melton D. A. (1996) A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. U.S.A. 93, 8455–8459 10.1073/pnas.93.16.8455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stambolic V., Ruel L., and Woodgett J. (1996) Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr. Biol. 6, 1664–1668 10.1016/S0960-9822(02)70790-2 [DOI] [PubMed] [Google Scholar]
- 11. Phiel C. J., and Klein P. S. (2001) Molecular targets of lithium action. Annu. Rev. Pharmacol. Toxicol. 41, 789–813 10.1146/annurev.pharmtox.41.1.789 [DOI] [PubMed] [Google Scholar]
- 12. Doble B. W., Patel S., Wood G. A., Kockeritz L. K., and Woodgett J. R. (2007) Functional redundancy of GSK-3α and GSK-3β in Wnt/β-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell 12, 957–971 10.1016/j.devcel.2007.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bartman C. M., Egelston J., Kattula S., Zeidner L. C., D'Ippolito A., Doble B. W., and Phiel C. J. (2014) Gene expression profiling in mouse embryonic stem cells reveals glycogen synthase kinase-3-dependent targets of phosphotidylinositol 3-kinase and Wnt/β-catenin signaling pathways. Front. Endocrinol. 5, 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., and Gough N. M. (1988) Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684–687 10.1038/336684a0 [DOI] [PubMed] [Google Scholar]
- 15. Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., and Rogers D. (1988) Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688–690 10.1038/336688a0 [DOI] [PubMed] [Google Scholar]
- 16. Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., and Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen J., Liu J., Chen Y., Yang J., Chen J., Liu H., Zhao X., Mo K., Song H., Guo L., Chu S., Wang D., Ding K., and Pei D. (2011) Rational optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells with ultra-high efficiency and fast kinetics. Cell Res. 21, 884–894 10.1038/cr.2011.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bar-Nur O., Brumbaugh J., Verheul C., Apostolou E., Pruteanu-Malinici I., Walsh R. M., Ramaswamy S., and Hochedlinger K. (2014) Small molecules facilitate rapid and synchronous iPSC generation. Nat. Methods 11, 1170–1176 10.1038/nmeth.3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Umehara H., Kimura T., Ohtsuka S., Nakamura T., Kitajima K., Ikawa M., Okabe M., Niwa H., and Nakano T. (2007) Efficient derivation of embryonic stem cells by inhibition of glycogen synthase kinase-3. Stem Cells 25, 2705–2711 10.1634/stemcells.2007-0086 [DOI] [PubMed] [Google Scholar]
- 20. Ye S., Tan L., Yang R., Fang B., Qu S., Schulze E. N., Song H., Ying Q., and Li P. (2012) Pleiotropy of glycogen synthase kinase-3 inhibition by CHIR99021 promotes self-renewal of embryonic stem cells from refractory mouse strains. PLoS One 7, e35892 10.1371/journal.pone.0035892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q. L., and Smith A. (2008) Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298 10.1016/j.cell.2008.12.007 [DOI] [PubMed] [Google Scholar]
- 22. Li P., Tong C., Mehrian-Shai R., Jia L., Wu N., Yan Y., Maxson R. E., Schulze E. N., Song H., Hsieh C. L., Pera M. F., and Ying Q. L. (2008) Germline competent embryonic stem cells derived from rat blastocysts. Cell 135, 1299–1310 10.1016/j.cell.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hnisz D., Schuijers J., Lin C. Y., Weintraub A. S., Abraham B. J., Lee T. I., Bradner J. E., and Young R. A. (2015) Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell 58, 362–370 10.1016/j.molcel.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kelly K. F., Ng D. Y., Jayakumaran G., Wood G. A., Koide H., and Doble B. W. (2011) β-Catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell 8, 214–227 10.1016/j.stem.2010.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lyashenko N., Winter M., Migliorini D., Biechele T., Moon R. T., and Hartmann C. (2011) Differential requirement for the dual functions of β-catenin in embryonic stem cell self-renewal and germ layer formation. Nat. Cell Biol. 13, 753–761 10.1038/ncb2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., and Smith A. (2011) Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 13, 838–845 10.1038/ncb2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hao J., Li T. G., Qi X., Zhao D. F., and Zhao G. Q. (2006) WNT/β-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev. Biol. 290, 81–91 10.1016/j.ydbio.2005.11.011 [DOI] [PubMed] [Google Scholar]
- 28. Wagner R. T., Xu X., Yi F., Merrill B. J., and Cooney A. J. (2010) Canonical Wnt/β-catenin regulation of liver receptor homolog-1 mediates pluripotency gene expression. Stem Cells 28, 1794–1804 10.1002/stem.502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sanchez-Ripoll Y., Bone H. K., Owen T., Guedes A. M., Abranches E., Kumpfmueller B., Spriggs R. V., Henrique D., and Welham M. J. (2013) Glycogen synthase kinase-3 inhibition enhances translation of pluripotency-associated transcription factors to contribute to maintenance of mouse embryonic stem cell self-renewal. PLoS One 8, e60148 10.1371/journal.pone.0060148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kingham E., and Welham M. (2009) Distinct roles for isoforms of the catalytic subunit of class-IA PI3K in the regulation of behaviour of murine embryonic stem cells. J. Cell Sci. 122, 2311–2321 10.1242/jcs.046557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Campbell J. M., Nottle M. B., Vassiliev I., Mitchell M., and Lane M. (2012) Insulin increases epiblast cell number of in vitro cultured mouse embryos via the PI3K/GSK3/p53 pathway. Stem Cells Dev. 21, 2430–2441 10.1089/scd.2011.0598 [DOI] [PubMed] [Google Scholar]
- 32. Hishida T., Nakachi Y., Mizuno Y., Katano M., Okazaki Y., Ema M., Takahashi S., Hirasaki M., Suzuki A., Ueda A., Nishimoto M., Hishida-Nozaki Y., Vazquez-Ferrer E., Sancho-Martinez I., Izpisua Belmonte J. C., and Okuda A. (2015) Functional compensation between Myc and PI3K signaling supports self-renewal of embryonic stem cells. Stem Cells 33, 713–725 10.1002/stem.1893 [DOI] [PubMed] [Google Scholar]
- 33. Lee M. Y., Lim H. W., Lee S. H., and Han H. J. (2009) Smad, PI3K/Akt, and Wnt-dependent signaling pathways are involved in BMP-4-induced ESC self-renewal. Stem Cells 27, 1858–1868 10.1002/stem.124 [DOI] [PubMed] [Google Scholar]
- 34. Storm M. P., Bone H. K., Beck C. G., Bourillot P. Y., Schreiber V., Damiano T., Nelson A., Savatier P., and Welham M. J. (2007) Regulation of Nanog expression by phosphoinositide 3-kinase-dependent signaling in murine embryonic stem cells. J. Biol. Chem. 282, 6265–6273 10.1074/jbc.M610906200 [DOI] [PubMed] [Google Scholar]
- 35. Storm M. P., Kumpfmueller B., Thompson B., Kolde R., Vilo J., Hummel O., Schulz H., and Welham M. J. (2009) Characterization of the phosphoinositide 3-kinase-dependent transcriptome in murine embryonic stem cells: identification of novel regulators of pluripotency. Stem Cells 27, 764–775 10.1002/stem.3 [DOI] [PubMed] [Google Scholar]
- 36. Batista P. J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D. M., Lujan E., Haddad B., Daneshvar K., Carter A. C., Flynn R. A., Zhou C., Lim K. S., Dedon P., et al. (2014) m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 10.1016/j.stem.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y., Li Y., Toth J. I., Petroski M. D., Zhang Z., and Zhao J. C. (2014) N6-Methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A. A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y. S., Ben-Haim M. S., Eyal E., Yunger S., Pinto Y., Jaitin D. A., et al. (2015) Stem cells: m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- 39. Perry R. P., and Kelley D. E. (1974) Existence of methylated messenger RNA in mouse L cells. Cell 1, 37–42 10.1016/0092-8674(74)90153-6 [DOI] [Google Scholar]
- 40. Desrosiers R., Friderici K., and Rottman F. (1974) Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 71, 3971–3975 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wei C. M., Gershowitz A., and Moss B. (1975) Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4, 379–386 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- 42. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., Dai Q., Chen W., and He C. (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y. G., and He C. (2011) N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., Vågbø C. B., Shi Y., Wang W. L., Song S. H., Lu Z., Bosmans R. P., Dai Q., Hao Y. J., Yang X., et al. (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., and Jaffrey S. R. (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., and Rechavi G. (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 47. Schafer K. N., Cisek K., Huseby C. J., Chang E., and Kuret J. (2013) Structural determinants of Tau aggregation inhibitor potency. J. Biol. Chem. 288, 32599–32611 10.1074/jbc.M113.503474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A. V., Patil D. P., Linder B., Pickering B. F., Vasseur J.-J., Chen Q., Gross S. S., Elemento O., Debart F., Kiledjian M., and Jaffrey S. R. (2017) Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 10.1038/nature21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Linder B., Grozhik A. V., Olarerin-George A. O., Meydan C., Mason C. E., and Jaffrey S. R. (2015) Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wei C., Gershowitz A., and Moss B. (1975) N6,O2′-Dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253 10.1038/257251a0 [DOI] [PubMed] [Google Scholar]
- 51. Young R. A. (2011) Control of the embryonic stem cell state. Cell 144, 940–954 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jia G., Fu Y., and He C. (2013) Reversible RNA adenosine methylation in biological regulation. Trends Genet. 29, 108–115 10.1016/j.tig.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cole M. F., Johnstone S. E., Newman J. J., Kagey M. H., and Young R. A. (2008) Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 22, 746–755 10.1101/gad.1642408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Göttgens B., Niwa H., and Smith A. (2012) Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell 11, 491–504 10.1016/j.stem.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Voskas D., Ling L. S., and Woodgett J. R. (2014) Signals controlling un-differentiated states in embryonic stem cells and cancer cells: role of the phosphatidylinositol 3′ kinase pathway. J. Cell. Physiol. 229, 1312–1322 10.1002/jcp.24603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Paling N. R., Wheadon H., Bone H. K., and Welham M. J. (2004) Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J. Biol. Chem. 279, 48063–48070 10.1074/jbc.M406467200 [DOI] [PubMed] [Google Scholar]
- 57. Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 58. Xu C., Kim N. G., and Gumbiner B. M. (2009) Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle 8, 4032–4039 10.4161/cc.8.24.10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aberle H., Bauer A., Stappert J., Kispert A., and Kemler R. (1997) β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16, 3797–3804 10.1093/emboj/16.13.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Willert K., Brown J. D., Danenberg E., Duncan A. W., Weissman I. L., Reya T., Yates J. R. 3rd, and Nusse R. (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 10.1038/nature01611 [DOI] [PubMed] [Google Scholar]
- 61. Hedgepeth C. M., Conrad L. J., Zhang J., Huang H. C., Lee V. M., and Klein P. S. (1997) Activation of the Wnt signaling pathway: a molecular mechanism for lithium action. Dev. Biol. 185, 82–91 10.1006/dbio.1997.8552 [DOI] [PubMed] [Google Scholar]
- 62. Sutherland C. (2011) What are the bona fide GSK3 substrates? Int. J. Alzheimers Dis. 2011, 505607 10.4061/2011/505607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cho J. H., and Johnson G. V. (2003) Glycogen synthase kinase 3beta phosphorylates Tau at both primed and unprimed sites: differential impact on microtubule binding. J. Biol. Chem. 278, 187–193 10.1074/jbc.M206236200 [DOI] [PubMed] [Google Scholar]
- 64. Gerken T., Girard C. A., Tung Y.-C. L., Webby C. J., Saudek V., Hewitson K. S., Yeo G. S. H., McDonough M. A., Cunliffe S., McNeill L. A., Galvanovkis J., Rorsman P., Robins P., Prieur X., Coll A. P., et al. (2007) The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472 10.1126/science.1151710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McTaggart J. S., Lee S., Iberl M., Church C., Cox R. D., and Ashcroft F. M. (2011) FTO is expressed in neurones throughout the brain and its expression is unaltered by fasting. PLoS One 6, e27968 10.1371/journal.pone.0027968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hess M. E., Hess S., Meyer K. D., Verhagen L. A., Koch L., Brönneke H. S., Dietrich M. O., Jordan S. D., Saletore Y., Elemento O., Belgardt B. F., Franz T., Horvath T. L., Rüther U., Jaffrey S. R., et al. (2013) The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048 10.1038/nn.3449 [DOI] [PubMed] [Google Scholar]
- 67. Li Z., Weng H., Su R., Weng X., Zuo Z., Li C., Huang H., Nachtergaele S., Dong L., Hu C., Qin X., Tang L., Wang Y., Hong G. M., Huang H., et al. (2017) FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell 31, 127–141 10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Du T., Rao S., Wu L., Ye N., Liu Z., Hu H., Xiu J., Shen Y., and Xu Q. (2015) An association study of the m6A genes with major depressive disorder in Chinese Han population. J. Affect. Disord. 183, 279–286 10.1016/j.jad.2015.05.025 [DOI] [PubMed] [Google Scholar]
- 69. Frayling T. M., Timpson N. J., Weedon M. N., Zeggini E., Freathy R. M., Lindgren C. M., Perry J. R., Elliott K. S., Lango H., Rayner N. W., Shields B., Harries L. W., Barrett J. C., Ellard S., Groves C. J., et al. (2007) A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dina C., Meyre D., Gallina S., Durand E., Körner A., Jacobson P., Carlsson L. M., Kiess W., Vatin V., Lecoeur C., Delplanque J., Vaillant E., Pattou F., Ruiz J., Weill J., et al. (2007) Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39, 724–726 10.1038/ng2048 [DOI] [PubMed] [Google Scholar]
- 71. Scott L. J., Mohlke K. L., Bonnycastle L. L., Willer C. J., Li Y., Duren W. L., Erdos M. R., Stringham H. M., Chines P. S., Jackson A. U., Prokunina-Olsson L., Ding C. J., Swift A. J., Narisu N., Hu T., et al. (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345 10.1126/science.1142382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Scuteri A., Sanna S., Chen W. M., Uda M., Albai G., Strait J., Najjar S., Nagaraja R., Orrú M., Usala G., Dei M., Lai S., Maschio A., Busonero F., Mulas A., et al. (2007) Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115 10.1371/journal.pgen.0030115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Smemo S., Tena J. J., Kim K. H., Gamazon E. R., Sakabe N. J., Gómez-Marín C., Aneas I., Credidio F. L., Sobreira D. R., Wasserman N. F., Lee J. H., Puviindran V., Tam D., Shen M., Son J. E., et al. (2014) Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375 10.1038/nature13138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sanchez-Pulido L., and Andrade-Navarro M. A. (2007) The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 8, 23 10.1186/1471-2091-8-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen H., Fajol A., Hoene M., Zhang B., Schleicher E. D., Lin Y., Calaminus C., Pichler B. J., Weigert C., Häring H. U., Lang F., and Föller M. (2016) PI3K-resistant GSK3 controls adiponectin formation and protects from metabolic syndrome. Proc. Natl. Acad. Sci. U.S.A. 113, 5754–5759 10.1073/pnas.1601355113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang X., Lu Z., Gomez A., Hon G. C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., Ren B., Pan T., and He C. (2014) N6-Methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Patil D. P., Pickering B. F., and Jaffrey S. R. (2018) Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol. 28, 113–127 10.1016/j.tcb.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B. S., Mesquita A., Liu C., Yuan C. L., Hu Y. C., Hüttelmaier S., Skibbe J. R., Su R., Deng X., et al. (2018) Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bertero A., Brown S., Madrigal P., Osnato A., Ortmann D., Yiangou L., Kadiwala J., Hubner N. C., de Los Mozos I. R., Sadée C., Lenaerts A. S., Nakanoh S., Grandy R., Farnell E., Ule J., et al. (2018) The SMAD2/3 interactome reveals that TGFβ controls m(6)A mRNA methylation in pluripotency. Nature 555, 256–259 10.1038/nature25784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tan F., Qian C., Tang K., Abd-Allah S. M., and Jing N. (2015) Inhibition of transforming growth factor β (TGF-β) signaling can substitute for Oct4 protein in reprogramming and maintain pluripotency. J. Biol. Chem. 290, 4500–4511 10.1074/jbc.M114.609016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bartman C. M., Egelston J., Ren X., Das R., and Phiel C. J. (2015) A simple and efficient method for transfecting mouse embryonic stem cells using polyethylenimine. Exp. Cell Res. 330, 178–185 10.1016/j.yexcr.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., and Rechavi G. (2013) Transcriptome-wide mapping of N6-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 8, 176–189 10.1038/nprot.2012.148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.