Abstract
Alternative end-joining (a-EJ) pathways, which repair DNA double-strand breaks (DSBs), are initiated by end resection that generates 3′ single strands. This reaction is shared, at least in part, with homologous recombination but distinguishes a-EJ from the major nonhomologous end-joining pathway. Although the a-EJ pathways make only a minor and poorly understood contribution to DSB repair in nonmalignant cells, there is growing interest in these pathways, as they generate genomic rearrangements that are hallmarks of cancer cells. Here, we review and discuss the current understanding of the mechanisms and regulation of a-EJ pathways, the role of a-EJ in human disease, and the potential utility of a-EJ as a therapeutic target in cancer.
Keywords: genomic instability, DNA damage, DNA polymerase, DNA endonuclease, chromosomes, deletions, DNA ligation, microhomology, single-strand annealing, translocation, double-strand break, alternative end-joining pathway
Introduction
DNA double-strand breaks (DSB)2 can be generated by exposure to exogenous agents such as ionizing radiation, endogenous agents such as reactive oxygen species–generated aerobic metabolism, or DNA metabolic processes, including DNA replication, meiosis, and rearrangement of genes encoding immunoglobulins and T cell receptors. These are extremely dangerous lesions because the integrity of both strands of the DNA duplex is lost. In a cell with more than one DSB, it is important to rejoin the previously linked DNA ends otherwise a chromosomal translocation will be generated. Surprisingly, the predominant repair pathway in human cells that is often called either classic or canonical nonhomologous end-joining relies upon DNA end-bridging mediated by protein–protein interactions involving DNA-dependent protein kinase molecules to bring together DNA ends (1–3). In this Minireview, we will refer to this pathway simply as nonhomologous end-joining (NHEJ). Although this could potentially join DNA ends from different chromosomes, the repair of DSBs by NHEJ usually results in the rejoining of previously linked DNA ends, possibly because the arrangement of chromatin in loops attached to a scaffold restricts the movement of DNA ends generated by a break in the loop. The majority of DSBs generated by ionizing radiation and oxygen free radicals have damaged and noncomplementary termini that require processing prior to re-joining. This processing frequently results in either the loss or the addition of a few nucleotides at the break site (1, 2). Thus, although NHEJ usually rejoins previously linked DNA ends, the repair of DSBs by this pathway is frequently mutagenic. For more details about the NHEJ pathway, see the accompanying Minireview by Pannunzio et al. (4).
Although DSBs are repaired by NHEJ throughout the cell cycle, a recombinational repair pathway operates during the S and G2 phases of the cell cycle when an intact sister chromatid is available to guide the error-free repair of DSBs (5, 6). The initial steps of this pathway involve resection of the 5′ ends of the DSBs followed by strand invasion into the adjacent intact sister chromatid, generating a D loop structure by strand exchange (6, 7). For more details about the homologous recombination (HR) pathway, see the accompanying Minireview by Wright et al. (8). In this Minireview, we focus on minor DSB repair pathways that are genetically distinct from HR and NHEJ that we will refer to collectively as alternative end-joining (a-EJ) pathways. These pathways do share factors with and/or utilize similar mechanisms to the major DSB repair pathways. All the a-EJ pathways, like HR, are initiated by end resection (Fig. 1) and involve some, if not all, of the factors that constitute the HR end resection machinery (1, 7, 9). The a-EJ pathways also share similarities with NHEJ in that the DNA ends to be joined are juxtaposed without using a homologous template as a guide. They do, however, utilize differing amounts of sequence homology (Fig. 1) to align the DNA molecules (1, 9). Although the a-EJ pathways make only a minor and poorly understood contribution to DSB repair in nonmalignant cells, there is growing interest in these pathways as they generate large deletions, translocations, and end-to-end chromosome fusions, genomic rearrangements that are frequently observed in cancer cells (10–12). Furthermore, they appear to be promising therapeutic targets in cancer cells with defects in either NHEJ or HR (11, 13–16).
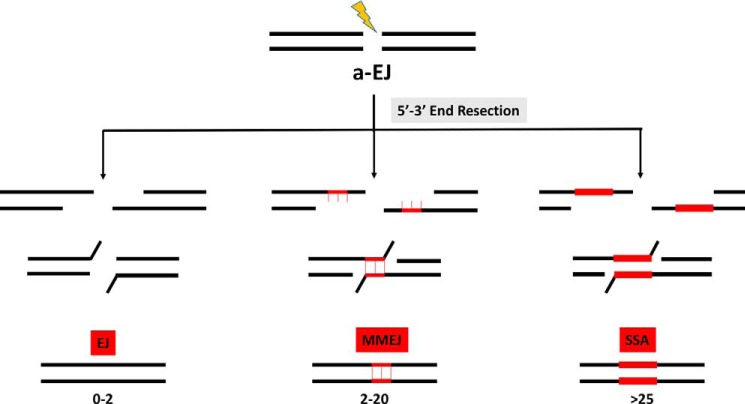
Figure 1.
Role of DNA sequence homology in a-EJ pathways. Resection of the 5′ strand at DSBs is the first common step of all the EJ pathways (a-EJ). Three distinct pathways, single-strand annealing (SSA), microhomology-mediated end-joining (MMEJ), and end-joining (EJ) are distinguished based on the amount of DNA sequence complementarity used to align DNA ends. SSA involves complementary repeat sequences more than 25 nucleotides in length, whereas MMEJ involves shorter tracts of sequence homology, ranging from 2 to 20 nucleotides in length. There is also a third category of DSB repair events that either lack or have very little sequence homology at the repair site generated by a poorly defined EJ pathway.
Overview of a-EJ in humans and its relationship with other pathways of DSB repair
A series of X-ray–sensitive mutants of Chinese hamster ovary cell lines were used to identify human XRCC (X-ray cross-complementing) genes involved in the repair of DSBs both by HR and NHEJ (17, 18). Around the same time, a number of labs described robust DNA end-joining activities in extracts from mammalian cells but did not definitively link these activities to NHEJ factors (19, 20). In a seminal paper, Bauman and West (21) described end joining by a human cell extract that depended upon NHEJ factors but also noted that end-joining activities that were independent of NHEJ could be detected in extracts prepared by different methods.
The initial genetic characterization of a-EJ pathways also occurred around the same time using the yeast Saccharomyces cerevisiae as a model eukaryote. In contrast to mammalian cells, HR is the predominant DSB repair pathway in yeast. Two minor DSB repair pathways, single-strand annealing (SSA) and microhomology-mediated end joining (MMEJ) (22, 23), were identified in HR-deficient yeast strains in addition to the NHEJ pathway (24). Both the SSA and MMEJ pathways are initiated by DNA end resection. In SSA, 5′ to 3′ end resection at both ends exposes single-strand regions with complementary sequences of greater than 25 nucleotides that reside within tandem repeats (Fig. 1). The complementary sequences anneal, generating DNA duplex with noncomplementary 3′ single-strand tails. These tails are removed, followed by gap-filling synthesis and ligation. This pathway usually generates intrachromosomal deletions but may generate translocations through events involving repetitive elements on different chromosomes. In MMEJ, shorter regions of complementary sequence, ranging from 2 to 20 nucleotides that are called microhomologies, are frequently used to align DNA ends prior to gap filling and ligation. Like SSA, this pathway generates deletions, but additional nontemplated nucleotides may be added at the repair site (24).
It should be noted that the NHEJ pathway also utilizes microhomologies, such as those generated by restriction endonucleases, during end joining. Although the complementary single-strand overhangs generated by restriction endonucleases are usually accurately rejoined by the NHEJ pathway, microhomologies less than four nucleotides produced by limited nucleolytic processing and error-prone gap-filling synthesis likely play a role in end alignment during the repair of DSBs with noncomplementary ends by NHEJ, resulting in the characteristic small insertions and deletions (1). The ring-shaped Ku heterodimer initiates the repair of DSBs by NHEJ. This factor binds rapidly and stably to DSBs, preventing end degradation by the HR end resection machinery and limiting resection by other nucleases. It also serves as the platform for the assembly of the other NHEJ factors, including end-processing factors, in a multiprotein complex (1, 2, 25). MMEJ and the other a-EJ pathways are distinct from NHEJ in that they are Ku-independent, require components of HR end-resection machinery, and frequently involve longer tracts of microhomology. The competition between Ku and the end-resection machinery for a DSB end determines whether that end will be repaired by NHEJ or channeled into the HR and occasionally the a-EJ pathways. Our current understanding of the mechanisms that determine DSB repair pathway choice is the focus of the accompanying Minireview by Her and Bunting (26).
The identification of PARP-1 and DNA ligase IIIα (LigIIIα), neither of which are present in S. cerevisiae, as participants in a-EJ in mammalian cells (Table 1), provided the first evidence that there are likely to be differences in the repertoire and mechanisms of a-EJ pathways between yeast and mammalian cells (27–29). The repair of DSBs by a-EJ, in particular MMEJ, is more evident in mammalian cells that are deficient in NHEJ (30–33). For example, class switch recombination that is normally dependent upon NHEJ factors occurs by an a-EJ pathway in the absence of a functional NHEJ pathway (30, 33). Similarly, cells that are deficient in HR are more dependent upon a-EJ pathways for the repair of DSBs (11, 13). Compared with HR and NHEJ, the factors involved in a-EJ and the mechanisms by which they act together to process and repair DSBs are not well-defined. It is possible that the repair of DSBs by a-EJ that occurs in repair-deficient cells may not be carried out by distinct pathways. Instead, the factors required to repair a DSB may be dictated by the nature of the defect in the NHEJ or HR pathway.
Table 1.
Proteins involved in a-EJ pathways
For the proteins shown in boldface type, there is strong evidence identifying them as a key factor in the indicated EJ pathway.

Although it is evident that the MMEJ pathway serves as a back-up pathway in cells that are deficient in either NHEJ or HR, the joining of DSBs by MMEJ can be detected in cells that are proficient for both NHEJ and HR (34). The role(s) of the a-EJ pathways when the major DSB repair pathways are functional is poorly understood. A recent study showing that MMEJ activity is induced by ionizing radiation suggests that this pathway may be responsible for the repair of a subset of DSBs with damaged termini that render them refractory to repair by NHEJ (35). Because the end resection machinery is activated in S phase cells, it is possible that the SSA pathway acts to repair DSBs in S phase cells that occur in unreplicated DNA (9). In the following sections, we provide a brief overview of the mechanisms of the a-EJ pathways, SSA and MMEJ, that are shown schematically in Figs. 2 and 3, respectively.
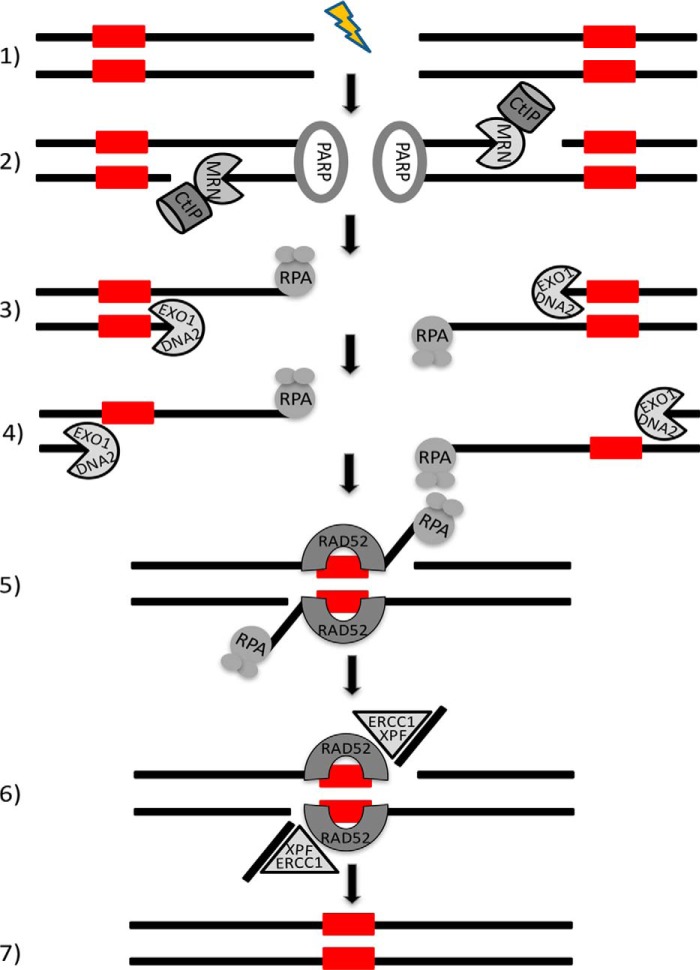
Figure 2.
Repair of DSBs by the single-strand annealing pathway. 1) Introduction of a DSB break. 2) PARP-1 (PARP) mediates the rapid recruitment of MRN and CtIP to the DSB end. CtIP enhances the MRN endonuclease activity resulting in an internal single-strand break within the 5′ strand. The short single-strand fragment at the DSB end is then degraded by the MRN exonuclease activity. 3) The resultant single-strand region, which is rapidly bound by RPA, serves as the binding site for one of the processive 5′ to 3′ exonucleases, either Exo1 or DNA2. 4) The resultant long range resection by Exo1 or DNA2 exposes complementary single-strand regions, greater than 25 nucleotides in length. 5) Rad52 interacts with the RPA-coated single strands and anneals the complementary regions, aligning the DNA ends and exposing nonhomologous 3′ single-strand tails. 6) The single-strand tails are removed by ERCC1/XPF, a DNA structure-specific endonuclease that cleaves the 3′ strand at duplex/single-strand junctions. 7) After any gaps are filled, both strands are ligated to generate an intact duplex that is missing one of the repeats and the DNA region between the repeats. The DNA polymerases and DNA ligases involved in the last steps of SSA have not been identified.
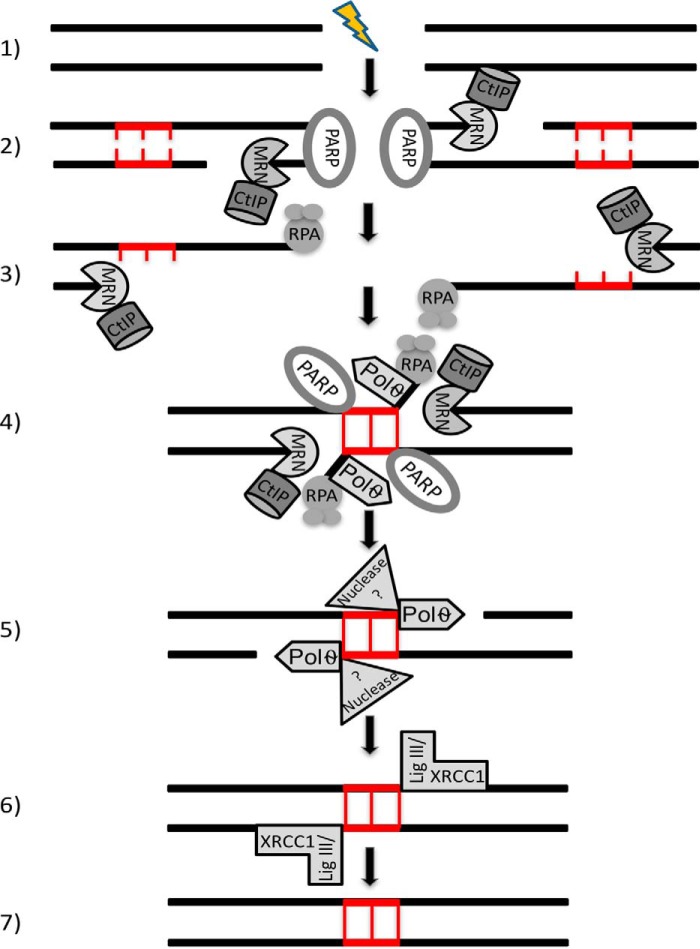
Figure 3.
Repair of DSBs by the microhomology-mediated end-joining pathway. 1) Introduction of a DSB break. 2) PARP-1 (PARP) mediates the rapid recruitment of MRN and CtIP to the DSB end. CtIP enhances the MRN endonuclease activity resulting in an internal single-strand break within the 5′ strand. The short single-strand fragment at the DSB end is then degraded by the MRN exonuclease activity. 3) Short regions of sequence complementarity, ranging from 2 to 20 nucleotides, are exposed within the RPA-coated single-strand regions. 4) The DNA ends are transiently aligned via the short microhomologies. PARP-1, MRN, and Pol θ have each been implicated in the end alignment. It is likely that a similar, possibly MRN independent process of end alignment also occurs between ends that are being resected by the long-range exonucleases, Exo1 and DNA2 (see Fig. 2, step 3). 5) Nonhomologous 3′ tails are removed prior to error-prone gap-filling DNA synthesis by Pol θ. It is assumed that several functionally redundant nucleases will participate in end processing. 6) Both strands are ligated by the LigIIIα–XRCC1 complex (LigIII/XRCC1). 7) The DNA duplexes generated by MMEJ are characterized by deletions and the presence of sequence microhomologies at the repair site.
End resection (MRN/CtlP)
As mentioned above, the a-EJ pathways are similar to HR in that they are initiated by end resection (Fig. 1). For HR and a-EJ (Figs. 2 and 3), end resection is initiated by the Mre11/Rad50/Nbs1 (MRN) complex and CtIP (Table 1). Studies with the functionally homologous yeast proteins suggest that an initial 3′ single-strand region is generated by CtIP-enhanced MRN endonuclease activity followed by 3′ to 5′ exonucleolytic digestion by MRN (7, 36, 37). This allows the loading of the more processive nucleases, either Exo1 or DNA2, that then generate long stretches of single-strand DNA (7). Because PARP-1 is required for the rapid recruitment of MRN to DSBs (38), it is possible that the involvement of PARP-1 in a-EJ is due to its role in recruiting MRN for the initial phase of end resection. This, however, seems unlikely, because PARP inhibitors do not reduce the viability of cells with a functional HR pathway, whereas if they disrupted MRN recruitment and end resection then they would also be expected to impact the repair of DSBs by HR.
Although genetic studies have implicated MRN and CtIP in both SSA and MMEJ (32, 34, 39–43), the role of Exo1 and DNA2 is not clear. If there are defects in HR at the strand exchange or later stages, it seems likely that Exo1 and/or DNA2 will have been involved in generating the single-strand ends. Functional redundancy between Exo1 and DNA2 may explain why DNA2, Exo1, and also BLM, which interacts with both Exo1 and DNA2, appear to be dispensable for MMEJ (7). Because more extensive DNA end resection is likely to be needed to expose the homologous repeats required for SSA, DSB repair by this pathway probably involves Exo1 and/or DNA2 (Fig. 2), whereas the more limited end resection by CtIP and MRN may be sufficient for at least some of the DSB repair events catalyzed by the other a-EJ pathways involving either a few nucleotides of microhomology (MMEJ) (Fig. 3) or no homology (EJ) (34).
At the present time, little is known about how the extent of end resection is controlled either during HR or a-EJ. Notably, the 3′ to 5′ exonuclease of Mre11, which functions as a dimer within the MRN complex, is sensitive to the presence of other DNA ends with noncomplementary ends enhancing degradation and complementary ends inhibiting degradation (44, 45). When degradation by Mre11 exposes a short tract of sequence that is complementary to another DNA end, the Mre11 exonuclease appears to pause, presumably transiently stabilizing DNA end alignment via the microhomologies (44, 45). The involvement of the endo- and, in particular, the exonuclease activity of Mre11 in MMEJ is supported by results from a recent study showing that the repair of a plasmid substrate by an MMEJ-proficient XRCC1 immunoprecipitate that contains MRN and CtIP was blocked by small molecule inhibitors of either the endo- or the exonuclease activities of Mre11 (35). Interestingly, the loss of WRN, a member of the RecQ helicase family that is defective in the prototypic premature aging syndrome, Werner's syndrome, increased the size of deletions generated during MMEJ event suggesting that WRN functions to suppress resection in MMEJ even though WRN also possesses nuclease activity (46).
Given the role of end resection in a-EJ, it was generally assumed that DSB repair via SSA, MMEJ, and the other EJ pathway occurs predominantly in S and G2 phase cells when the end-resection machineries are active. Despite the negative regulation of end resection in G1 cells, it is evident that the repair of DSBs by MMEJ also occurs in this phase of the cell cycle (47). Both CtIP and MRN appear to be responsible for the end resection in G1 cells with 53BP1 enhancing both end resection and MMEJ (47). Knockdown of 53BP1 in G1 cells resulted in about a 2-fold increase in ionizing radiation-induced γ-H2AX foci remaining 8 and 24 h after radiation, indicating that a significant fraction of ionizing radiation-induced DSBs are repaired by MMEJ in G1 cells (47). Furthermore, because inactivation of NHEJ resulted in increased repair of DSBs by MMEJ in G1 cells, it appears that NHEJ suppresses DSB repair by MMEJ in G1 cells (47). Although phosphorylation of CtIP by cyclin-dependent protein kinases is critical for its role in initiating resection for HR in G2 cells, CtIP is phosphorylated in a DNA damage-inducible manner by Polo-like kinase 3 to activate CtIP/MRN-dependent resection in G1 cells (48, 49). In accord with the evidence that there is competition between NHEJ and MMEJ in G1 cells (47), resection by CtIP/MRN in G1 cells inhibits DSB repair by NHEJ (48). It should be noted that there is also evidence that DSB repair in G1 cells occurs via a resection-dependent version of NHEJ rather than MMEJ in addition to resection-independent NHEJ (50). Further studies are needed to resolve these apparently contradictory findings.
DNA end-bridging and alignment
A critical step in all the a-EJ pathways is the juxtaposing of DNA ends. Because SSA likely requires extensive resection by Exo1 and/or DNA2 to expose the homologous repeats (Fig. 2 and Table 1), this repair pathway is probably most active during the S and G2 phases of the cell cycle. For post-replication breaks, the ends are likely held in the same vicinity as a consequence of sister chromatid cohesion with the repair event generating intrachromosomal deletions, whereas resection at DSBs in nonreplicated DNA in cells could lead to translocations involving homologous repeats on different chromosomes (9). Initial genetic studies in yeast identified Rad52 as a key component of the SSA pathway with similar studies implicating mammalian Rad52 protein (22, 51). Notably, Rad52 protein has a robust single annealing activity and is able to anneal complementary single strands that are coated with RPA (52, 53). Together, these studies suggest that, during SSA, Rad52 anneals complementary RPA coated-sequences exposed by end resection (Fig. 2 and Table 1).
For the other a-EJ pathways, different proteins have been suggested to play roles in the bridging of DNA ends and alignment via microhomologies (Table 1). PARP-1, which competes with Ku for binding to DSBs, appears to have end-bridging activity (27), although the mechanism by which PARP-1 mediates end-synapsis, in particular the contribution of poly(ADP-ribosylation), has not been elucidated. As noted previously, PARP-1 is involved in the rapid recruitment of the MRN complex to DNA ends (38). Both the yeast and human versions of this complex have robust end-bridging activity (54, 55), suggesting that the MRN complex can simultaneously engage two DNA ends, compare sequences, and transiently align the DNA ends via exposed microhomologies during endo- and exonucleolytic digestion (44, 45).
More recently, an A-family DNA polymerase, Pol θ, has been identified as a key factor in a-EJ (11, 13). Pol θ has a helicase-like domain at its N terminus that is separated from the C-terminal polymerase domain by a long, unstructured central region. Notably, Pol θ displaces RPA from single-strand DNA and also interacts with Rad51 and inhibits Rad51-dependent HR (11, 13, 56). Thus, Pol θ appears to actively compete with the HR machinery for resected DNA ends. Furthermore, Pol θ is capable of searching for and aligning microhomologies, thereby contributing to end bridging (57, 58).
Removal of nonhomologous 3′ tails
During the repair of DSBs by SSA, the long 3′ tails are coated with RPA (9). Following Rad52-mediated annealing of homologous repeat sequences (Fig. 2), the noncomplementary 3′ tails are removed by the DNA structure-specific endonuclease ERCC1/XPF (Table 1) that interacts with and is stimulated by Rad52 (59). Although it is likely the end-joining events catalyzed by the other a-EJ pathways will also involve removal of noncomplementary 3′ tails (Fig. 1), the identity of the nuclease(s) involved has not been definitively established (Table 1), presumably reflecting redundancy among these enzymes.
Gap filling DNA synthesis
It is likely that, after the removal of the nonhomologous tails, the aligned DNA duplexes will contain gaps. The identity of the DNA polymerase(s) involved in gap filling during the repair of DSBs by SSA (9) and microhomology-independent EJ pathway (Fig. 1) has not been definitively established (Table 1). In contrast, there is compelling evidence that Pol θ participates in the majority of DSBs repaired by MMEJ (11, 13, 58). Notably, Pol θ has robust terminal transferase activity in addition to template-directed synthesis activity and so generates insertions with significant sequence diversity at repair sites (58, 60, 61). Although Pol θ is clearly a key MMEJ factor, there is currently no evidence of functional interactions between Pol θ and other MMEJ factors.
Ligation
The repair of DSBs by the a-EJ pathways is completed by a DNA ligase once ligatable termini have been generated by end processing. Among the DNA ligases encoded by the three mammalian LIG genes, DNA ligase IV appears to only function in NHEJ, leaving the DNA ligases encoded by the LIG1 and LIG3 genes as the candidate enzymes for a-EJ (2, 62, 63). Because S. cerevisiae lacks a homolog of the mammalian LIG3 gene (64), it is likely that Cdc9 DNA ligase, the functional homolog of human LigI, is the predominant DNA ligase in yeast a-EJ. Although the contribution of the DNA ligases encoded by the mammalian LIG1 and LIG3 genes to SSA has not been established (Table 1), there is substantial evidence indicating that LigIIIα is the major DNA ligase in the MMEJ pathway (27, 28, 65, 66). In the nucleus, LigIIIα forms a stable complex with XRCC1, a DNA repair protein that is often referred to as a scaffold protein because of its interactions with a large number of DNA repair proteins (67). Both LigIIIα and XRCC1 preferentially interact with poly(ADP-ribosylated) PARP-1, interactions that underlie the recruitment of the LigIIIα/XRCC1 to in vivo DNA single-strand breaks (68, 69). Although PARP inhibitors reduce the repair of DSBs by MMEJ, there was no reduction in the recruitment of XRCC1 to DSBs (14, 35), indicating that the recruitment of LigIIIα/XRCC1 is not dependent upon poly(ADP-ribosylated) PARP-1. Interestingly, LigIIIα/XRCC1 also physically and functionally interacts with the MRN complex with these two complexes acting together to digest and join DNA duplexes with noncomplementary termini utilizing internal sequence microhomologies (35, 70). Thus, it is possible that this interaction directs the recruitment of LigIIIα/XRCC1 to DSBs undergoing repair by MMEJ. In support of this idea, ionizing radiation induces increased association of XRCC1 with Mre11 and CtIP, co-localization of XRCC1 with Mre11, and increased MMEJ activity (35). These changes in the behavior of XRCC1 and MMEJ activity are dependent upon phosphorylation of XRCC1 by casein kinase 2 in response to ionizing radiation (35), suggesting that this phosphorylation event(s) enhance the interaction with the MRN complex. Interestingly, the N-terminal zinc finger of LigIIIα, which is required for intermolecular ligation in vitro, is also required for LigIIIα-dependent MMEJ (65, 71–73).
In mouse cells, the NHEJ pathway acts to suppress formation of translocations by LigIIIα-dependent MMEJ in response to DSBs induced either by site-specific nucleases or ionizing radiation (12, 65, 66). This is consistent with studies implicating MMEJ in the formation of translocations due to aberrant class switch recombination in mice deficient in NHEJ (10, 30, 33). The observation in some studies that translocation formation was not dependent upon XRCC1 (66, 74) was surprising given the role of this protein in maintaining the stability and activity of nuclear LigIIIα and other studies indicating that XRCC1 is a key component of the MMEJ pathway (35, 75). Although there may be sufficient residual nuclear LigIIIα for translocation formation in the absence of XRCC1 (75), it is also possible that, when nuclear LigIIIα is absent, translocation formation occurs by a LigI-dependent EJ pathway (65). Notably, these events do not appear to involve end alignment via microhomologies (65). At the present time, it is not known how LigI is recruited to the repair site and whether other LigI-interacting proteins such as proliferating cell nuclear antigen and replication factor C are involved in this EJ pathway (Table 1).
Role of A-EJ in genome instability and human disease
The repair of DSBs by a-EJ is inherently mutagenic, potentially giving rise to chromosomal translocations as well as intra- and interchromosomal deletions and insertions (10, 33). In addition, the MMEJ pathway also contributes to the formation of end-to-end chromosome fusions (11). Given the prevalence of these types of rearrangements in the genomes of cancer cells, there is significant interest in understanding the contribution of a-EJ to cancer formation and progression. Although the majority of studies of genome instability have focused on the nuclear genome, deletions within the circular mitochondrial genome that have been implicated in a wide variety of human diseases frequently occur between repeats and/or involve microhomologies at the repaired site (76–80). These observations suggest that a-EJ pathways may contribute to deleterious changes in mitochondrial DNA, but our understanding of the repertoire and mechanisms of DNA repair pathways, in particular DSB repair pathways, operating in mitochondria is still very limited. Although there is compelling evidence that LigIIIα is the only DNA ligase in mitochondria (81–86) and mitochondrial extracts are capable of utilizing internal microhomologies to join DNA molecules with noncomplementary ends (86), further work is needed to definitively establish the mitochondrial localization of other a-EJ factors, such as Pol θ and Mre11, and their participation in the repair of DSBs by a-EJ in mitochondria (86, 87).
As noted previously, a-EJ is more readily detectable in cells that are deficient in NHEJ (10, 31, 32). In mice, genetic inactivation of NHEJ results in an increase in the frequency of chromosomal translocations with evidence of microhomologies at many of the ligation sites, indicative of joining by MMEJ (12, 65). As expected, mouse cells deficient in PARP-1, CtIP, or LigIIIα exhibit a reduced overall frequency of chromosomal translocations and less use of microhomologies in the translocations that do occur (65). Furthermore, p53-null mice that are also deficient in NHEJ develop pro-B–cell lymphomas arising as a consequence of chromosomal translocations between the IgH and the c-myc loci with the repair junctions characterized by insertions, deletions, and microhomology (88, 89). Together, these results indicate that, in the mouse, the majority of translocations in lymphoid cells likely occurs as a result of the joining of DSBs generated by V(D)J recombination with DSBs generated by activation-induced cytidine deaminase and reactive oxygen species at other chromosomal fragile zones (90) by MMEJ, whereas NHEJ prevents tumor incidence by suppressing translocation formation. Although there is compelling evidence indicating that Pol θ is an important contributor to MMEJ (11, 13, 58), there are, however, contradictory reports whether Pol θ-dependent MMEJ enhances or prevents chromosomal translocations (11, 13, 58, 61). As noted above, end-to-end chromosome fusions are generated by Pol θ-dependent MMEJ (11). Although this activity is normally suppressed by protective protein complexes at telomere ends and NHEJ (11), the end-to-end fusion of chromosomes by Pol θ-dependent MMEJ may occur in response to telomere shortening during cancer development.
In contrast to mouse cells (12, 65), the majority of translocations in human cells appears to arise as a consequence of the repair of DSBs by NHEJ (91). It is possible that differences in the relative contributions of NHEJ and A-EJ pathways to DSB repair between mouse and human cells underlies this discrepancy. For example, there is evidence that NHEJ is much less active in mouse cells (92). It is likely that the frequency of translocation formation versus rejoining of previously linked ends is different for each of the a-EJ pathways and that the contribution of an individual a-EJ pathway to translocation formation will be determined by a combination of the tendency of that pathway to generate translocations and the contribution of that pathway to DSB repair. For example, even if the repair of DSBs by NHEJ results in a low frequency of translocations, the NHEJ pathway will be responsible for most of the translocations produced if it is the predominant DSB repair pathway. In addition, it is possible that the contribution of the individual a-EJ pathways to translocation formation may differ depending on whether the DSBs are generated by class switch recombination, site-specific endonucleases, or other mechanisms.
A-EJ as a therapeutic target in cancer
The rational development of olaparib and other PARP inhibitors to selectively target HR-deficient breast and ovarian cancers in patients with an inherited predisposition for these tumors (93–95) has stimulated efforts to design similar synthetic lethal strategies for other cancers. As back-up pathways for the major DSB repair pathways, the a-EJ pathways are attractive potential therapeutic targets in cancers with defects in either HR and NHEJ because inhibiting a-EJ is unlikely to impact the growth and survival of normal tissues but should sensitize the cancer cells that are more dependent on a-EJ pathways to repair DSBs generated by endogenous and/or exogenous agents. In support of this model, reducing MMEJ by knockdown or knockout of Pol θ reduced the survival of both HR- and NHEJ-deficient cells (11, 13, 58). Furthermore, BRCA2-deficient tumor cells have higher steady-state levels of Pol θ (13), suggesting that MMEJ is up-regulated in these cells to compensate for the HR defect, and knockdown of Pol θ enhanced the killing of HR-deficient cells by PARP inhibitors (13), indicating that MMEJ enhances the survival of HR-deficient cells by repairing replication-induced DSBs. Although MMEJ inhibitors are likely to enhance the efficacy of PARP inhibitors in HR-deficient tumors, there is a more urgent need to develop reliable biomarkers to identify HR-deficient sporadic tumors that are likely to respond to PARP inhibitors. Based on initial studies (13), Pol θ expression levels appear to be a promising indicator of HR status.
In contrast to HR, there is less evidence linking mutation of NHEJ genes with genome instability in tumor samples. There is, however, evidence of reduced expression of the NHEJ factors, Ku, Artemis, and LigIV, and a compensatory increase in MMEJ factors, PARP1 and LigIIIα, in tyrosine kinase-activated leukemias, breast cancer, and neuroblastoma (14–16). Notably, expression of either BCR-ABL1 or FLT3-ITD in nonmalignant myeloid cell lines induces expression of c-MYC that in turn enhances expression of the LIG3 and PARP1 genes by suppressing expression of the microRNAs, miR-150 and miR-22 (96). Furthermore, the extent of the change in the expression levels of the DSB factors increases in imatinib-resistant chronic myeloid leukemia cells (16). An increase in the steady-state levels of PARP1 and LigIIIα was also observed in derivatives of an estrogen-responsive breast cell line that had acquired resistance to either tamoxifen or an aromatase inhibitor (15).
The changes in steady-state levels of the DSB repair proteins in the breast cancer and BCR-ABL1–expressing myeloid cell lines correlated with changes in the relative contribution of NHEJ and MMEJ to the repair of a transfected plasmid substrate (15, 16). As expected, incubation of these cell lines with a PARP inhibitor and an inhibitor of DNA ligases I and III reduced the fraction of plasmids repaired by MMEJ (15, 16). The cancer cell lines with dysregulated expression of the DSB repair proteins exhibited sensitivity to the PARP and DNA ligase inhibitors as single agents and in combination (14–16). Knockdown of LigIIIα expression had similar effects to the LigI/III inhibitor suggesting that the activity of the inhibitor is due to inhibition of LigIIIα rather than LigI (16). The synergistic activity of the PARP and DNA ligase inhibitors observed in some cell lines (15) is difficult to reconcile with the inhibition of two proteins in the same repair pathway. A recent study showing that the DNA ligase inhibitor preferentially targets mitochondrial function in cancer cells (97) suggests that the synergy may be due to effects on both mitochondrial DNA metabolism and nuclear DNA repair.
Analysis of the expression levels of a-EJ and NHEJ genes in neuroblastoma showed that high expression of PARP1, LIG3, and LIG1 and low expression of LIG4 correlated with reduced survival and higher stage disease (14). Furthermore, elevated expression of both LIG3 and PARP1 was detected by RT-PCR in bone marrow mononuclear cells from chronic myeloid leukemia patients with imatinib-resistant and imatinib-sensitive disease (16). Notably, increased sensitivity to the combination of PARP-1 and DNA ligase inhibitors was observed in cells with elevated expression of both LIG3 and PARP1 (16). Taken together, these results indicate that MMEJ is a promising therapeutic target in cancers with elevated expression of genes encoding key MMEJ factors and/or reduced expression of genes encoding NHEJ factors.
Concluding comments
In contrast to the two major DSB repair pathways, HR and NHEJ, the protein participants in and the molecular mechanisms of the minor DSB pathways, known collectively as a-EJ, are poorly defined. It had been suggested that a-EJ events did not reflect the activity of distinct DSB repair pathways but instead represented the action of a group of factors whose participation was dictated by the nature of the defect in the NHEJ or HR pathway. This view was based upon the observations that the repair of DSBs by a-EJ was more evident in cells that are deficient in either of the major DSB repair pathways and that all of the factors initially implicated in a-EJ had major roles in other DNA repair pathways. The recent demonstration that the major cellular function of Pol θ is in a-EJ indicates that there are distinct a-EJ pathways. This is further supported by emerging evidence that the a-EJ pathways contribute to DSB repair, even when the major DSB repair pathways are active.
The role of the a-EJ pathways in the formation of large genomic rearrangements, in particular translocations, that are characteristic of cancer cells is an active area of investigation. Although it is evident that the a-EJ pathways as well as the NHEJ pathway are capable of contributing to this type of genome instability, there are apparently contradictory published findings. This may reflect differences in the utilization of DSB repair pathways between mice and humans and between different cell types. In addition, it is possible that the different assays used to detect genome rearrangements may produce different results. There is, however, compelling evidence that human cancer cells with defects in either the HR or NHEJ pathways are more dependent upon a-EJ pathways, in particular MMEJ for DSB repair, providing a rationale for the development of therapeutic strategies that target the MMEJ pathway. Furthermore, it appears that elevated expression of MMEJ factors, such as PARP-1, LigIIIα, and Pol θ, may serve as biomarkers for cancers with defects in the two major DSB repair pathways, thereby identifying the patient population whose disease is likely to respond to inhibitors of MMEJ.
Acknowledgments
We apologize to all colleagues whose work has not been cited because of space limitations. The University of New Mexico Cancer Center (NCI-designated Comprehensive Cancer Center) was supported by National Institutes of Health Grant CA118100.
This work was supported by National Institutes of Health Grants GM57479, GM47251, ES012512, and CA92584 (to the Tomkinson laboratory). This is the fourth article in the Thematic Minireview series “DNA double-strand break repair and pathway choice.” A. E. T. is a co-inventor on patents that cover the use of DNA ligase inhibitors as anti-cancer agents, and altered expression of DSB repair proteins as biomarkers of increased dependence upon MMEJ. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DSB
- double-strand break
- a-EJ
- alternative end-joining
- NHEJ
- nonhomologous end-joining
- MRN
- Mre11/Rad50/Nbs1
- HR
- homologous recombination
- SSA
- single-strand annealing
- MMEJ
- microhomology-mediated end joining
- PARP
- poly(ADP-ribose) polymerase
- RPA
- replication protein A.
References
- 1. Chang H. H. Y., Pannunzio N. R., Adachi N., and Lieber M. R. (2017) Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 10.1038/nrm.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lieber M. R. (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mladenov E., and Iliakis G. (2011) Induction and repair of DNA double-strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat. Res. 711, 61–72 10.1016/j.mrfmmm.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 4. Pannunzio N. R., Watanabe G., and Lieber M. R. (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 293, 10512–10523 10.1074/jbc.TM117.000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chapman J. R., Taylor M. R., and Boulton S. J. (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- 6. San Filippo J., Sung P., and Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 10.1146/annurev.biochem.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- 7. Daley J. M., Niu H., Miller A. S., and Sung P. (2015) Biochemical mechanism of DSB end resection and its regulation. DNA Repair 32, 66–74 10.1016/j.dnarep.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wright W. D., Shah S. S., and Heyer W.-D. (2018) Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 293, 10524–10535 10.1074/jbc.TM118.000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bhargava R., Onyango D. O., and Stark J. M. (2016) Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32, 566–575 10.1016/j.tig.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boboila C., Jankovic M., Yan C. T., Wang J. H., Wesemann D. R., Zhang T., Fazeli A., Feldman L., Nussenzweig A., Nussenzweig M., and Alt F. W. (2013) Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl. Acad. Sci. U.S.A. 107, 3034–3039 10.1073/pnas.0915067107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mateos-Gomez P. A., Gong F., Nair N., Miller K. M., Lazzerini-Denchi E., and Sfeir A. (2015) Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 10.1038/nature14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simsek D., and Jasin M. (2010) Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat. Struct. Mol. Biol. 17, 410–416 10.1038/nsmb.1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ceccaldi R., Liu J. C., Amunugama R., Hajdu I., Primack B., Petalcorin M. I., O'Connor K. W., Konstantinopoulos P. A., Elledge S. J., Boulton S. J., Yusufzai T., and D'Andrea A. D. (2015) Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 10.1038/nature14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Newman E. A., Lu F., Bashllari D., Wang L., Opipari A. W., and Castle V. P. (2015) Alternative NHEJ pathway components are therapeutic targets in high-risk neuroblastoma. Mol. Cancer Res. 13, 470–482 10.1158/1541-7786.MCR-14-0337 [DOI] [PubMed] [Google Scholar]
- 15. Tobin L. A., Robert C., Nagaria P., Chumsri S., Twaddell W., Ioffe O. B., Greco G. E., Brodie A. H., Tomkinson A. E., and Rassool F. V. (2012) Targeting abnormal DNA repair in therapy-resistant breast cancers. Mol. Cancer Res. 10, 96–107 10.1158/1541-7786.MCR-11-0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tobin L. A., Robert C., Rapoport A. P., Gojo I., Baer M. R., Tomkinson A. E., and Rassool F. V. (2013) Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias. Oncogene 32, 1784–1793 10.1038/onc.2012.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeggo P. A., Tesmer J., and Chen D. J. (1991) Genetic analysis of ionising radiation sensitive mutants of cultured mammalian cell lines. Mutat. Res. 254, 125–133 10.1016/0921-8777(91)90003-8 [DOI] [PubMed] [Google Scholar]
- 18. Thompson L. H., and Jeggo P. A. (1995) Nomenclature of human genes involved in ionizing radiation sensitivity. Mutat. Res. 337, 131–134 10.1016/0921-8777(95)00018-F [DOI] [PubMed] [Google Scholar]
- 19. Fairman M. P., Johnson A. P., and Thacker J. (1992) Multiple components are involved in the efficient joining of double stranded DNA breaks in human cell extracts. Nucleic Acids Res. 20, 4145–4152 10.1093/nar/20.16.4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mason R. M., Thacker J., and Fairman M. P. (1996) The joining of non-complementary DNA double-strand breaks by mammalian extracts. Nucleic Acids Res. 24, 4946–4953 10.1093/nar/24.24.4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baumann P., and West S. C. (1998) DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. U.S.A. 95, 14066–14070 10.1073/pnas.95.24.14066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ivanov E. L., Sugawara N., Fishman-Lobell J., and Haber J. E. (1996) Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma J. L., Kim E. M., Haber J. E., and Lee S. E. (2003) Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23, 8820–8828 10.1128/MCB.23.23.8820-8828.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seol J. H., Shim E. Y., and Lee S. E. (2017) Microhomology-mediated end joining: good, bad and ugly. Mutat. Res. S0027–5107, 30041–30046 10.1016/j.mrfmmm.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hammel M., Yu Y., Radhakrishnan S. K., Chokshi C., Tsai M. S., Matsumoto Y., Kuzdovich M., Remesh S. G., Fang S., Tomkinson A. E., Lees-Miller S. P., and Tainer J. A. (2016) An intrinsically disordered APLF links Ku, DNA-PKcs, and XRCC4-DNA Ligase IV in an extended flexible non-homologous end joining complex. J. Biol. Chem. 291, 26987–27006 10.1074/jbc.M116.751867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Her J., and Bunting S. F. (2018) How cells ensure correct repair of DNA double-strand breaks. J. Biol. Chem. 293, 10502–10511 10.1074/jbc.TM118.000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Audebert M., Salles B., and Calsou P. (2004) Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 279, 55117–55126 10.1074/jbc.M404524200 [DOI] [PubMed] [Google Scholar]
- 28. Wang H., Rosidi B., Perrault R., Wang M., Zhang L., Windhofer F., and Iliakis G. (2005) DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 65, 4020–4030 10.1158/0008-5472.CAN-04-3055 [DOI] [PubMed] [Google Scholar]
- 29. Wang M., Wu W., Wu W., Rosidi B., Zhang L., Wang H., and Iliakis G. (2006) PARP-1 and Ku compete for repair of DNA double-strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182 10.1093/nar/gkl840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boboila C., Yan C., Wesemann D. R., Jankovic M., Wang J. H., Manis J., Nussenzweig A., Nussenzweig M., and Alt F. W.. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J. Exp. Med. 207, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corneo B., Wendland R. L., Deriano L., Cui X., Klein I. A., Wong S. Y., Arnal S., Holub A. J., Weller G. R., Pancake B. A., Shah S., Brandt V. L., Meek K., and Roth D. B. (2007) Rag mutations reveal robust alternative end joining. Nature 449, 483–486 10.1038/nature06168 [DOI] [PubMed] [Google Scholar]
- 32. Xie A., Kwok A., and Scully R. (2009) Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16, 814–818 10.1038/nsmb.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yan C. T., Boboila C., Souza E. K., Franco S., Hickernell T. R., Murphy M., Gumaste S., Geyer M., Zarrin A. A., Manis J. P., Rajewsky K., and Alt F. W. (2007) IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 10.1038/nature06020 [DOI] [PubMed] [Google Scholar]
- 34. Truong L. N., Li Y., Shi L. Z., Hwang P. Y., He J., Wang H., Razavian N., Berns M. W., and Wu X. (2013) Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, 7720–7725 10.1073/pnas.1213431110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dutta A., Eckelmann B., Adhikari S., Ahmed K. M., Sengupta S., Pandey A., Hegde P. M., Tsai M. S., Tainer J. A., Weinfeld M., Hegde M. L., and Mitra S. (2017) Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res. 45, 2585–2599 10.1093/nar/gkw1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cannavo E., and Cejka P. (2014) Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122–125 10.1038/nature13771 [DOI] [PubMed] [Google Scholar]
- 37. Garcia V., Phelps S. E., Gray S., and Neale M. J. (2011) Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479, 241–244 10.1038/nature10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haince J. F., McDonald D., Rodrigue A., Déry U., Masson J. Y., Hendzel M. J., and Poirier G. G. (2008) PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197–1208 10.1074/jbc.M706734200 [DOI] [PubMed] [Google Scholar]
- 39. Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., and Lopez B. S. (2009) Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16, 819–824 10.1038/nsmb.1641 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y., and Jasin M. (2011) An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 18, 80–84 10.1038/nsmb.1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee-Theilen M., Matthews A. J., Kelly D., Zheng S., and Chaudhuri J. (2011) CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol. 18, 75–79 10.1038/nsmb.1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bennardo N., Cheng A., Huang N., and Stark J. M. (2008) Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 10.1371/journal.pgen.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muñoz M. C., Laulier C., Gunn A., Cheng A., Robbiani D. F., Nussenzweig A., and Stark J. M. (2012) RING finger nuclear factor RNF168 is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1. J. Biol. Chem. 287, 40618–40628 10.1074/jbc.M112.410951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paull T. T., and Gellert M. (1998) The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1, 969–979 10.1016/S1097-2765(00)80097-0 [DOI] [PubMed] [Google Scholar]
- 45. Paull T. T., and Gellert M. (2000) A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl. Acad. Sci. U.S.A. 97, 6409–6414 10.1073/pnas.110144297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sallmyr A., Tomkinson A. E., and Rassool F. (2008) Up-regulation of WRN and DNA ligase IIIa in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood 112, 1413–1423 10.1182/blood-2007-07-104257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiong X., Du Z., Wang Y., Feng Z., Fan P., Yan C., Willers H., and Zhang J. (2015) 53BP1 promotes microhomology-mediated end-joining in G1-phase cells. Nucleic Acids Res. 43, 1659–1670 10.1093/nar/gku1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bakr A., Köcher S., Volquardsen J., Petersen C., Borgmann K., Dikomey E., Rothkamm K., and Mansour W. Y. (2016) Impaired 53BP1/RIF1 DSB mediated end-protection stimulates CtIP-dependent end resection and switches the repair to PARP1-dependent end joining in G1. Oncotarget 7, 57679–57693 10.18632/oncotarget.11023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barton O., Naumann S. C., Diemer-Biehs R., Künzel J., Steinlage M., Conrad S., Makharashvili N., Wang J., Feng L., Lopez B. S., Paull T. T., Chen J., Jeggo P. A., and Löbrich M. (2014) Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J. Cell Biol. 206, 877–894 10.1083/jcb.201401146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Biehs R., Steinlage M., Barton O., Juhász S., Künzel J., Spies J., Shibata A., Jeggo P. A., and Löbrich M. (2017) DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol. Cell 65, 671–684 10.1016/j.molcel.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stark J. M., Pierce A. J., Oh J., Pastink A., and Jasin M. (2004) Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 24, 9305–9316 10.1128/MCB.24.21.9305-9316.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rothenberg E., Grimme J. M., Spies M., and Ha T. (2008) Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc. Natl. Acad. Sci. U.S.A. 105, 20274–20279 10.1073/pnas.0810317106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu Y., Sugiyama T., and Kowalczykowski S. C. (2006) DNA annealing mediated by Rad52 and Rad59 proteins. J. Biol. Chem. 281, 15441–15449 10.1074/jbc.M601827200 [DOI] [PubMed] [Google Scholar]
- 54. Chen L., Trujillo K., Ramos W., Sung P., and Tomkinson A. E. (2001) Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8, 1105–1115 10.1016/S1097-2765(01)00388-4 [DOI] [PubMed] [Google Scholar]
- 55. de Jager M., van Noort J., van Gent D. C., Dekker C., Kanaar R., and Wyman C. (2001) Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8, 1129–1135 10.1016/S1097-2765(01)00381-1 [DOI] [PubMed] [Google Scholar]
- 56. Mateos-Gomez P. A., Kent T., Deng S. K., McDevitt S., Kashkina E., Hoang T. M., Pomerantz R. T., and Sfeir A. (2017) The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol. 24, 1116–1123 10.1038/nsmb.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kent T., Chandramouly G., McDevitt S. M., Ozdemir A. Y., and Pomerantz R. T. (2015) Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 22, 230–237 10.1038/nsmb.2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wyatt D. W., Feng W., Conlin M. P., Yousefzadeh M. J., Roberts S. A., Mieczkowski P., Wood R. D., Gupta G. P., and Ramsden D. A. (2016) Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell 63, 662–673 10.1016/j.molcel.2016.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Motycka T. A., Bessho T., Post S. M., Sung P., and Tomkinson A. E. (2004) Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J. Biol. Chem. 279, 13634–13639 10.1074/jbc.M313779200 [DOI] [PubMed] [Google Scholar]
- 60. Kent T., Mateos-Gomez P. A., Sfeir A., and Pomerantz R. T. (2016) Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining. Elife 5, e13740 10.7554/eLife.13740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yousefzadeh M. J., Wyatt D. W., Takata K., Mu Y., Hensley S. C., Tomida J., Bylund G. O., Doublié S., Johansson E., Ramsden D. A., McBride K. M., and Wood R. D. (2014) Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10, e1004654 10.1371/journal.pgen.1004654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grawunder U., Zimmer D., Fugmann S., Schwarz K., and Lieber M. R. (1998) DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell 2, 477–484 10.1016/S1097-2765(00)80147-1 [DOI] [PubMed] [Google Scholar]
- 63. Ellenberger T., and Tomkinson A. E. (2008) Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 77, 313–338 10.1146/annurev.biochem.77.061306.123941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Simsek D., and Jasin M. (2011) DNA ligase III: a spotty presence in eukaryotes, but an essential function where tested. Cell Cycle 10, 3636–3644 10.4161/cc.10.21.18094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Simsek D., Brunet E., Wong S. Y., Katyal S., Gao Y., McKinnon P. J., Lou J., Zhang L., Li J., Rebar E. J., Gregory P. D., Holmes M. C., and Jasin M. (2011) DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 7, e1002080 10.1371/journal.pgen.1002080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Soni A., Siemann M., Grabos M., Murmann T., Pantelias G. E., and Iliakis G. (2014) Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res. 42, 6380–6392 10.1093/nar/gku298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tomkinson A. E., and Sallmyr A. (2013) Structure and function of the DNA ligases encoded by the mammalian LIG3 gene. Gene 531, 150–157 10.1016/j.gene.2013.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Okano S., Lan L., Caldecott K. W., Mori T., and Yasui A. (2003) Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 23, 3974–3981 10.1128/MCB.23.11.3974-3981.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Okano S., Lan L., Tomkinson A. E., and Yasui A. (2005) Translocation of XRCC1 and DNA ligase IIIα from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res. 33, 422–429 10.1093/nar/gki190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Della-Maria J., Zhou Y., Tsai M. S., Kuhnlein J., Carney J. P., Paull T. T., and Tomkinson A. E. (2011) Human Mre11/human Rad50/Nbs1 and DNA ligase IIIα/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J. Biol. Chem. 286, 33845–33853 10.1074/jbc.M111.274159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Taylor R. M., Whitehouse C. J., and Caldecott K. W. (2000) The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res. 28, 3558–3563 10.1093/nar/28.18.3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cotner-Gohara E., Kim I. K., Hammel M., Tainer J. A., Tomkinson A. E., and Ellenberger T. (2010) Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry 49, 6165–6176 10.1021/bi100503w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kukshal V., Kim I. K., Hura G. L., Tomkinson A. E., Tainer J. A., and Ellenberger T. (2015) Human DNA ligase III bridges two DNA ends to promote specific intermolecular DNA end joining. Nucleic Acids Res. 43, 7021–7031 10.1093/nar/gkv652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Boboila C., Oksenych V., Gostissa M., Wang J. H., Zha S., Zhang Y., Chai H., Lee C. S., Jankovic M., Saez L. M., Nussenzweig M. C., McKinnon P. J., Alt F. W., and Schwer B. (2012) Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1). Proc. Natl. Acad. Sci. U.S.A. 109, 2473–2478 10.1073/pnas.1121470109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Caldecott K. W., Tucker J. D., Stanker L. H., and Thompson L. H. (1995) Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 23, 4836–4843 10.1093/nar/23.23.4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tanhauser S. M., and Laipis P. J. (1995) Multiple deletions are detectable in mitochondrial DNA of aging mice. J. Biol. Chem. 270, 24769–24775 10.1074/jbc.270.42.24769 [DOI] [PubMed] [Google Scholar]
- 77. Fukui H., and Moraes C. T. (2009) Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 18, 1028–1036 10.1093/hmg/ddn437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schon E. A., Rizzuto R., Moraes C. T., Nakase H., Zeviani M., and DiMauro S. (1989) A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science 244, 346–349 10.1126/science.2711184 [DOI] [PubMed] [Google Scholar]
- 79. Eshaghian A., Vleugels R. A., Canter J. A., McDonald M. A., Stasko T., and Sligh J. E. (2006) Mitochondrial DNA deletions serve as biomarkers of aging in the skin, but are typically absent in nonmelanoma skin cancers. J. Invest. Dermatol. 126, 336–344 10.1038/sj.jid.5700088 [DOI] [PubMed] [Google Scholar]
- 80. Holt I. J., Harding A. E., and Morgan-Hughes J. A. (1988) Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331, 717–719 10.1038/331717a0 [DOI] [PubMed] [Google Scholar]
- 81. Gao Y., Katyal S., Lee Y., Zhao J., Rehg J. E., Russell H. R., and McKinnon P. J. (2011) DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature 471, 240–244 10.1038/nature09773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lakshmipathy U., and Campbell C. (1999) The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell. Biol. 19, 3869–3876 10.1128/MCB.19.5.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lakshmipathy U., and Campbell C. (2000) Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 28, 3880–3886 10.1093/nar/28.20.3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lakshmipathy U., and Campbell C. (2001) Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res. 29, 668–676 10.1093/nar/29.3.668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Simsek D., Furda A., Gao Y., Artus J., Brunet E., Hadjantonakis A. K., Van Houten B., Shuman S., McKinnon P. J., and Jasin M. (2011) Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 471, 245–248 10.1038/nature09794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tadi S. K., Sebastian R., Dahal S., Babu R. K., Choudhary B., and Raghavan S. C. (2016) Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions. Mol. Biol. Cell 27, 223–235 10.1091/mbc.E15-05-0260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wisnovsky S., Jean S. R., and Kelley S. O. (2016) Mitochondrial DNA repair and replication proteins revealed by targeted chemical probes. Nat. Chem. Biol. 12, 567–573 10.1038/nchembio.2102 [DOI] [PubMed] [Google Scholar]
- 88. Difilippantonio M. J., Petersen S., Chen H. T., Johnson R., Jasin M., Kanaar R., Ried T., and Nussenzweig A. (2002) Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196, 469–480 10.1084/jem.20020851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhu C., Mills K. D., Ferguson D. O., Lee C., Manis J., Fleming J., Gao Y., Morton C. C., and Alt F. W. (2002) Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109, 811–821 10.1016/S0092-8674(02)00770-5 [DOI] [PubMed] [Google Scholar]
- 90. Pannunzio N. R., and Lieber M. R. (2017) AID and reactive oxygen species can induce DNA breaks within human chromosomal translocation fragile zones. Mol. Cell 68, 901–912 10.1016/j.molcel.2017.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ghezraoui H., Piganeau M., Renouf B., Renaud J. B., Sallmyr A., Ruis B., Oh S., Tomkinson A. E., Hendrickson E. A., Giovannangeli C., Jasin M., and Brunet E. (2014) Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell 55, 829–842 10.1016/j.molcel.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Finnie N. J., Gottlieb T. M., Blunt T., Jeggo P. A., and Jackson S. P. (1995) DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 92, 320–324 10.1073/pnas.92.1.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N. J., and Helleday T. (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- 94. Farmer H., McCabe N., Lord C. J., Tutt A. N., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M., Jackson S. P., Smith G. C., and Ashworth A. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- 95. Lord C. J., and Ashworth A. (2008) Targeted therapy for cancer using PARP inhibitors. Curr. Opin. Pharmacol. 8, 363–369 10.1016/j.coph.2008.06.016 [DOI] [PubMed] [Google Scholar]
- 96. Muvarak N., Kelley S., Robert C., Baer M. R., Perrotti D., Gambacorti-Passerini C., Civin C., Scheibner K., and Rassool F. V. (2015) c-MYC generates repair errors via increased transcription of alternative-NHEJ factors, LIG3 and PARP1, in tyrosine kinase-activated leukemias. Mol. Cancer Res. 13, 699–712 10.1158/1541-7786.MCR-14-0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sallmyr A., Matsumoto Y., Roginskaya V., Van Houten B., and Tomkinson A. E. (2016) Inhibiting mitochondrial DNA ligase IIIα activates caspase 1-dependent apoptosis in cancer cells. Cancer Res. 76, 5431–5441 10.1158/0008-5472.CAN-15-3243 [DOI] [PMC free article] [PubMed] [Google Scholar]