Abstract
It is estimated that ∼1% of the world's population has intellectual disability, with males affected more often than females. OGT is an X-linked gene encoding for the enzyme O-GlcNAc transferase (OGT), which carries out the reversible addition of N-acetylglucosamine (GlcNAc) to Ser/Thr residues of its intracellular substrates. Three missense mutations in the tetratricopeptide (TPR) repeats of OGT have recently been reported to cause X-linked intellectual disability (XLID). Here, we report the discovery of two additional novel missense mutations (c.775 G>A, p.A259T, and c.1016 A>G, p.E339G) in the TPR domain of OGT that segregate with XLID in affected families. Characterization of all five of these XLID missense variants of OGT demonstrates modest declines in thermodynamic stability and/or activities of the variants. We engineered each of the mutations into a male human embryonic stem cell line using CRISPR/Cas9. Investigation of the global O-GlcNAc profile as well as OGT and O-GlcNAc hydrolase levels by Western blotting showed no gross changes in steady-state levels in the engineered lines. However, analyses of the differential transcriptomes of the OGT variant–expressing stem cells revealed shared deregulation of genes involved in cell fate determination and liver X receptor/retinoid X receptor signaling, which has been implicated in neuronal development. Thus, here we reveal two additional mutations encoding residues in the TPR regions of OGT that appear causal for XLID and provide evidence that the relatively stable and active TPR variants may share a common, unelucidated mechanism of altering gene expression profiles in human embryonic stem cells.
Keywords: post-translational modification, glycosyltransferase; O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT); O-linked N-acetylglucosamine (O-GlcNAc), genetic disease, enzyme kinetics, transcriptomics, intellectual disability, RNA-seq, XLID
Introduction
Intellectual disability (ID)2 is defined in the Diagnostic and Statistical Manual-5 (DSM-5) as a childhood-onset neurodevelopmental disorder characterized by limitations in intellectual functioning and adaptive behavior. The estimated worldwide prevalence of ID is ∼1% (1). Although Down's syndrome and Fragile X syndrome are the two most common causes of ID, it is also present as a feature in a number of rare conditions (2). Only a decade ago, ID in up to 80% of analyzed individuals was of unknown etiology, including nongenetic factors (2). The genetic cause of ID is now established in slightly over 70% of affected individuals, an advance made possible by the use of massive parallel sequencing approaches (2). Monogenic defects account for 40% of the identified causes as opposed to those caused by copy-number variants (deletions of duplications) or aneuploidies. 5–10% of all monogenic defects are X-linked disorders (2).
OGT is a gene on the X chromosome encoding the enzyme O-GlcNAc transferase (OGT) (3), which is responsible for the co- and post-translational addition of β-d-N-acetylglucosamine (GlcNAc) to specific serine and threonine residues on a number of nucleocytoplasmic and mitochondrial proteins (4, 5). O-GlcNAc addition is reversed by the action of the enzyme O-GlcNAc hydrolase (OGA) (4). The cycling of O-GlcNAc by these two enzymes on hundreds of proteins in the cell regulates basic cellular processes such as transcription, translation, and proteostasis and influences cell survival, division, signaling, metabolism, and autophagy (4, 6–9). Both OGT and OGA are essential genes in mice (3, 10, 11).
OGT is a multidomain protein with the catalytic center located within its C terminus (12). At its N terminus are multiple tetratricopeptide repeats (TPRs) that mediate protein–protein interactions (7, 13–16). OGT appears to be important for the proper functioning of the brain; it is abundantly expressed in the brain (17), and O-GlcNAc is enriched in both the brain and in synapses (18, 19). OGT is also a regulator of embryonic development and patterning (20–22), a discovery first made using the model organism Drosophila melanogaster (23), in which loss of OGT, also known as super-sex combs (sxc), results in abnormal morphology and death at the pharate adult stage (24, 25).
Although deregulation of O-GlcNAc associated with diseases such as cancer, Alzheimer's disease, diabetes mellitus, and cardiovascular disease has been well established (4), it is only recently, as the result of an X-exome parallel sequencing effort, that a missense mutation in OGT was first directly attributed to causing intellectual disability, accompanied by dysmorphic features and congenital abnormalities (26, 27). Reports have since emerged of at least three other mutations in OGT that segregate with disease in other families with intellectual disability (28, 29).
In this study, we report the clinical features/phenotypes of three separate patients, two of whom are brothers, with what appears to be syndromic intellectual disability. Using exome sequencing to determine the etiology of the disease, we discover two separate novel missense mutations in the TPR domain of OGT that segregate with X-linked ID (XLID) in these patients. We undertake the in vitro biochemical characterization of these and other currently known XLID missense variants of OGT using recombinant proteins, revealing that the stability and activity of these OGT variants is only modestly reduced. To use as an ex vivo human isogenic model to study the effects of the XLID mutations on early development and disease pathogenesis, we engineered a human male embryonic stem cell line using CRISPR/Cas9 to express these OGT variants. We investigated and found no significant changes in the steady-state global O-GlcNAc profile as well as OGT and OGA levels in these cells by Western blotting. Analysis of the transcriptome of these cells, however, reveals shared dysregulation of genes and pathways involved in early development, providing a starting point for dissecting the impact of OGT variants on common phenotypes in the XLID-affected individuals.
Results
Case reports of three patients with intellectual disability
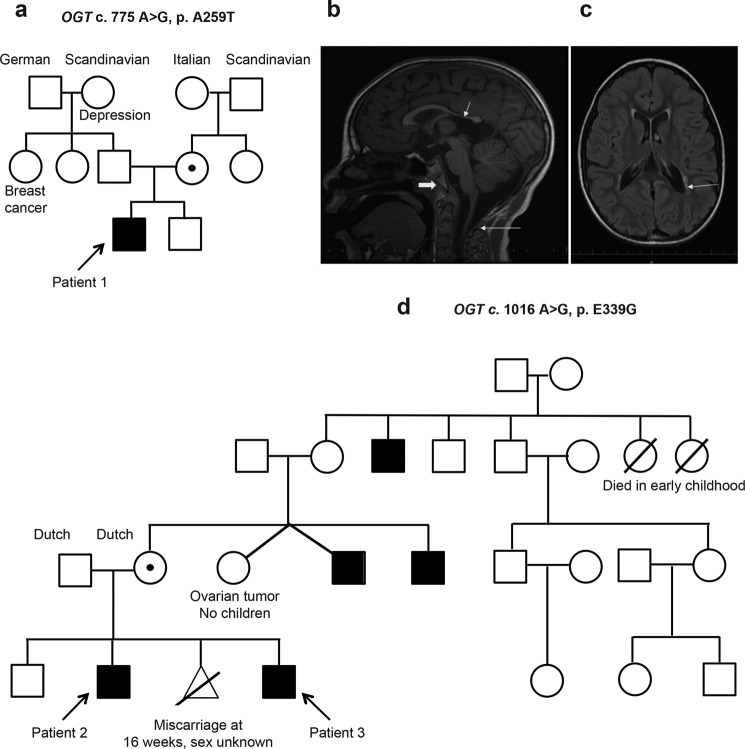
At the time of diagnosis, Patient 1 was a 7-year-old boy with moderate ID, microcephaly, hypothyroidism, abnormal sleep pattern, nystagmus, and epilepsy. No other family member was similarly affected (Fig. 1a). During pregnancy, there was concern for intrauterine growth retardation. Patient 1 was born at 39 weeks' gestation with APGAR scores of 8 at 1 min and 9 at 5 min, weighing 6 pounds 8 ounces (>10th centile). Postnatal course was significant for failure to thrive, with short stature and delayed milestones. The height and weight since early infancy were always below the 3rd centile. Patient 1 had microcephaly since birth with head size consistently below 2nd centile. He walked at 3 years of age and was nonverbal until 5 years of age. At 7 years of age, it was estimated that the patient functioned at the level of an 18–24-month-old. The patient was diagnosed with hypothyroidism at infancy and continued to receive thyroid hormones at diagnosis. He had mild conductive hearing loss despite use of myringotomy tubes. At the age of 4 years, the patient began to experience episodes of full body stiffening, cyanosis, and unresponsiveness. His seizures included atonic seizures, tonic seizures, generalized tonic clonic seizures, and staring spells. His seizures were treated with oxcarbazepine, lamotrigine, and clobazam and were well-controlled at the time of diagnosis. His EEG at onset of seizures showed multifocal spikes. The patient also had insomnia, and his EEG revealed disorganized background during sleep with no sleep spindles and vertex waves during sleep. A brain MRI at 8 months of age demonstrated thin corpus callosum. At age 7 years, his brain MRI revealed extremely thin corpus callosum particularly the posterior body and splenium, mild ventriculomegaly, and abnormal periventricular T2 and FLAIR signal (Fig. 1b). In addition, the clivus appeared short, and the posterior arch of the first cervical vertebra was relatively hypoplastic (Fig. 1c). An MRI of the spine at 7 years of age revealed diminished vertebral body heights at multiple levels, especially the thoracic and lumbar levels. Mild disc bulge was also seen at multiple levels in the lumbar region. Patient 1's karyotype, comparative genomic hybridization, and DNA test for Fragile X did not reveal any abnormalities. Several random glucose levels were mildly elevated. Routine metabolic testing, including serum lactate and ammonia, serum amino acids, urine organic acids, serum acylcarnitine profile, plasma very long chain fatty acids, lysosomal enzyme assays, and transferrin testing for congenital disorders of glycosylation were normal.
Figure 1.
Pedigrees and phenotypes of the patients. a, three-generation pedigree of Patient 1 (with OGT c.775 G>A, p.A259T). Proband is identified by the arrow. Circles denote females; circles with dots, carrier females confirmed using exome sequencing or Sanger sequencing; open squares, unaffected males; and closed squares, males with XLID. b, MRI of brain of Patient 1 at 7 years of age. Sagittal T1 image reveals extremely thin corpus callosum, particularly the posterior body and splenium (short thin arrow), short appearing clivus (thick arrow), and relatively hypoplastic posterior arch of first cervical vertebra (long thin arrow). c, MRI of brain at of Patient 1 at 7 years of age. Axial T2 FLAIR image reveals mild ventriculomegaly (arrow). d, four-generation pedigree of Patients 2 and 3 (with OGT c.1016 A>G, p.E339G). Probands are identified by arrows, and diagram has same coding as in a.
Patients 2 and 3 are siblings and unrelated to Patient 1 (Fig. 1d). There was a maternal family history of intellectual impairment with the mother's two male siblings and a maternal great uncle being affected (Fig. 1d). The parents' first child is healthy with no history of developmental or learning difficulties (Fig. 1d). Patient 2 was born at 39 weeks after an uncomplicated pregnancy. His birth weight was 2820 g (10th centile). In early childhood his motor development was within normal limits, but he was noted to have language delay. He was later diagnosed with mild to moderate ID. He also had behavioral difficulties. He experienced recurrent middle ear infections and required multiple tympanostomy tube insertions. Examination findings in Patient 2 at age 8 years 4 months were weight 22.4 kg (10–25th centile), height 127.4 cm (50th centile), and head circumference 49.5 cm (2nd centile). He had a dolicocephalic face shape with a high arched palate and malalignment of the teeth (with absence of two upper lateral incisors. His ears were long (length >97th centile). On neurological examination, he had generalized mild hypotonia and subtle asymmetry of deep tendon reflexes. Patient 3 (Fig. 1d) was born after an uncomplicated pregnancy. He also experienced recurrent middle ear infections. His early childhood development was within normal limits. He was noted to have learning difficulties when he reached primary school age. He was diagnosed with dyspraxia and attention-deficit hyperactivity disorder (ADHD), which was treated with methylphenidate. Examination findings in Patient 3 at age 11 years 10 months were weight 32.2 kg (10-25th centile), height 142.5 cm (50th centile), and head circumference 52.2 cm (2nd to 50th centile). He had a dolicocephalic face shape with broad nasal root and downslanted palpebral fissures. His philtrum was long and smooth, and his upper lip was thin. His palate was highly arched. He had bilateral fifth finger clinodactyly. G-banded and molecular karyotypes and Fragile X testing were normal in both Patients 2 and 3.
Identification of two distinct missense mutations in the TPR region of OGT causal for XLID
To investigate the etiology of ID in Patient 1, full exome sequencing was performed using genomic DNA isolated from the patient, his parents, and a healthy brother. Initially, 20 unique alterations in 20 genes were identified. Manual review to rule out sequencing artifacts and polymorphisms as well as genes lacking clinical overlap with patient's clinical course resulted in a short list of four genes with four unique alterations. Further analysis considering likely autosomal and X-linked inheritance models and familial segregation revealed a hemizygous threonine to alanine (p.A259T) variant in exon 7 of the OGT gene as the causal mutation (Fig. 2, a–c). Based on data from the Exome Aggregation Consortium (30), the OGT c.775G>A alteration was not observed among 60,706 individuals tested. Allele frequency data for this nucleotide position are not currently available from the 1000 Genomes Project (31), and the alteration is not currently listed in the database of single nucleotide polymorphisms (dbSNP) (32). The p.Ala-259 amino acid is invariable across available vertebrate species. The p.A259T alteration is predicted to be probably damaging by Polyphen (33) (score 1.00) and deleterious by SIFT (34) (score 0.01) in silico analyses. Co-segregation analysis of the c.775G>A (p.A259T) alteration revealed that it is present in the patient's mother and absent in his father and brother (Fig. 1a).
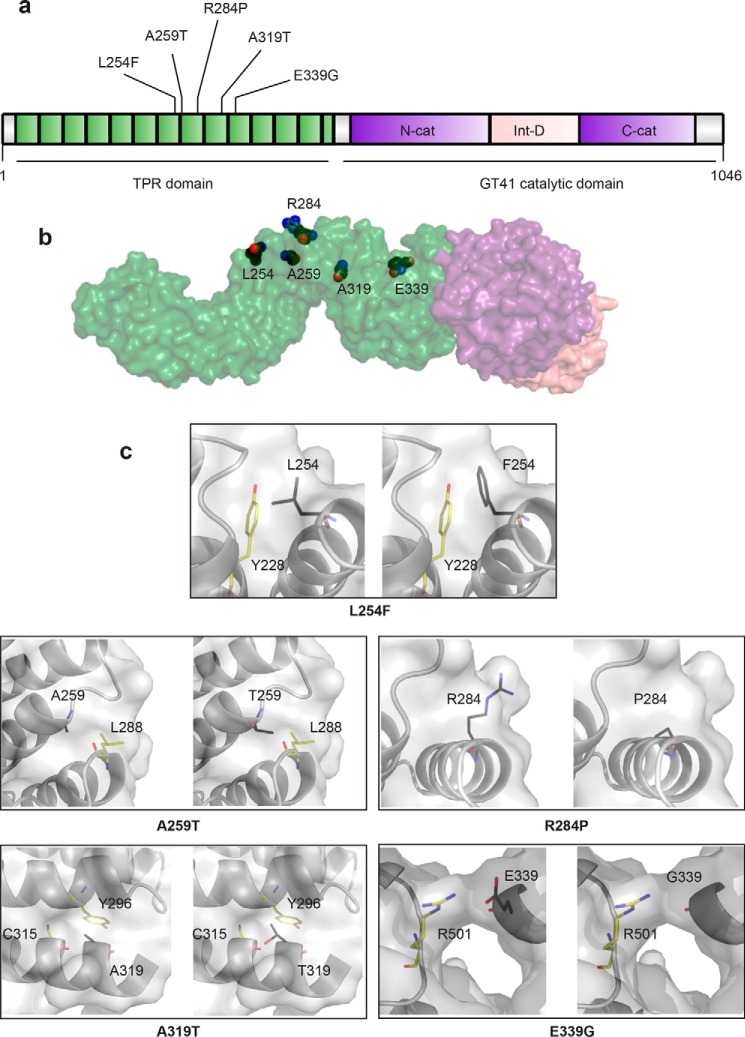
Figure 2.
All known missense mutations in the TPR domain that are causal for XLID. a, schematic of the domain organization of OGT. The N-terminal TPR domain is depicted in green with XLID variants noted, the intervening domain in pink, and the lobes of the catalytic domain in purple. b, model of full-length OGT based on the structure of the first 11.5 TPRs (PDB code 1W3B (14)) and the structure of the catalytic domain with the last 2.5 TPRs (PDB code 4AY5 (52)). The domains in the model are colored as in the schematic, and the side chains of the residues in the TPR that are mutated in XLID are depicted as spheres with black carbons. c, close-up of residues that are mutated in XLID. Panels on left show the WT residue in the amino acid position indicated, and panels on right show the replacing residue modeled into the structure (side chains are shown as sticks with black carbons). Surface representation shown in both panels is from OGT-WT. Side chains of neighboring residues that could clash are shown as sticks with yellow carbons.
To find the cause of ID in Patients 2 and 3, the genomic DNA of Patient 2 was subjected to X-chromosome exome sequencing, following which confirmation of variants and segregation analysis in further family members, including Patient 3, was performed by Sanger sequencing (see under “Experimental procedures”). Resultant output variants (eight unique alterations in seven genes were found, with one of these genes harboring a single silent mutation in addition to a missense mutation) were then assessed based on the presence in known SNP databases, including the Genome Aggregation Database (gnomAD) (30), predicted functional impact, nucleotide and amino acid conservation, and biochemical nature of amino acid substitutions. Only one variant remained after the filtering process; a hemizygous mutation c.1016A>G (p.E339G) located in exon 8 of the OGT gene (Fig. 2, a–c). This variant is not present in Exome Aggregation Consortium or gnomAD, nor is it listed in dbSNP (32). The amino acid p.E339 is invariable across available vertebrate species and is predicted to be probably damaging by Polyphen (33) (score 1.00) and deleterious by SIFT (34) (score 0). Confirmation of OGT c.1016A>G and segregation analysis in further family members by sequencing confirmed that this variant was present in the proband (Patient 2), as well as the patient's mother and affected brother (Patient 3) (Fig. 1d). It was absent in his father and unaffected brother (Fig. 1d).
The identification of these two novel mutations in OGT now results in a total of five known missense mutations in the TPR domain of OGT causal for XLID (Fig. 2, a and b) (26–29). Patient phenotypes resulting from each of these mutations are summarized in Table 1. Modeling of the XLID–TPR mutations within the structure of OGT (Fig. 2c) shows that substitutions at these residues would likely result in steric clashes with side chains of neighboring residues (as in the case of L254F, A259T, R284P, and A319T) and the loss of secondary structure elements (in the case of R284P) or of stabilizing interactions (in the case of E339G), which could lead to changes in the structure and thermodynamic stability of OGT.
Table 1.
Patient phenotypes caused by the XLID-associated variants of OGT
NA indicates not available. Numbers indicate number of affected patients over number of patients examined for the phenotype.
| Vaidyanathan et al. (27) | This study | Willems et al. (29) | Bouazzi et al. (28) | This study | Total | |
|---|---|---|---|---|---|---|
| Genotype | ||||||
| cDNA (NM_181672.2) | 762 G>T | 775 G>A | 851 G>C | 955 G>A, cosegregates with MED12 c.5922 G>T | 1016 A>G | |
| Skewing in X-chromosome inactivation in carrier | + | ND | NR | − | ND | |
| Protein | L254F | A259T | R284P | A319T, cosegregates with MED12 p. Q1974H | E339G | |
| Phenotype | ||||||
| Affected males | 3 | 1 | 1 | 3 | 2 | 10 |
| Intellectual disability | 3/3 | 1 | 1 | 3/3 | 2/2 | 10/10 |
| Developmental delay | 3/3 | 1 | 1 | 3/3 | 2/2 | 10/10 |
| Behavioral problems | 0/3 | 1 | 1 | 2/3 | 1/2 | 5/10 |
| Brain abnormalities | NA | 1 | 1 | 0/1 | 0/2 | 2/5 |
| Cardiac symptoms | 1/3 | 0 | 1 | NA | 0/2 | 2/7 |
| Dysmorphic features | 3/3 | 1 | 1 | 2/3 | 2/2 | 9/10 |
| Clinodactyly | 3/3 | 0 | 1 | 1/3 | 1/2 | 6/10 |
| Eye abnormalities | 3/3 | 1 | 1 | 2/2 | 0/2 | 7/9 |
| Genital/reproductive abnormalities | 2/3 | 0 | 1 | NA | 0/2 | 3/7 |
| Ear abnormalities/hearing impairment | 0/2 | 1 | 0 | 1/3 | 2/2 | 4/9 |
| Microcephaly | 1/3 | 1 | 1 | 0/3 | 0/2 | 3/10 |
| Hypothyroidism | NA | 1 | NA | NA | NA | 1/1 |
| Epilepsy/seizures | 0/3 | 1 | NA | 0/1 | 0/2 | 1/7 |
OGT XLID–TPR mutants form functional dimers but demonstrate decreased thermal stability
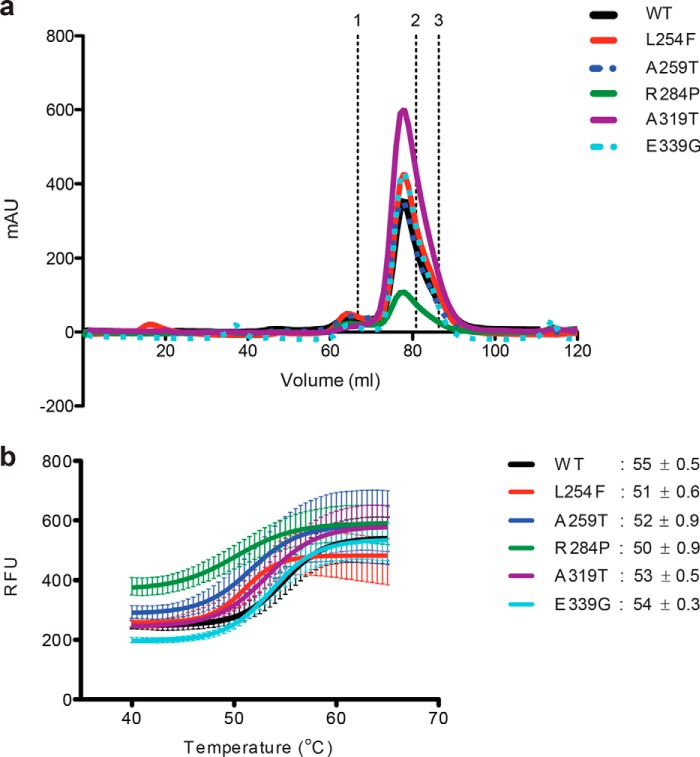
Having identified the mutations causal for ID in our patients, we wanted to biochemically characterize these and other known XLID OGT variants. It is known that OGT forms a functional dimer. Hydrophobic contact between the TPR residues Trp-208 in one monomer and Ile-211 in the second monomer causes OGT to dimerize (14). Given that the XLID mutations lie in the TPR domain of OGT, we wanted to investigate the impact of these amino acid substitutions on OGT dimerization. To do this, we used purified recombinant variant TPR domains alone. Samples of the proteins were run on a Superdex 200 gel-filtration column, and all of the mutants eluted at the same elution volume as the wildtype (WT) TPR domain, indicating that the mutations do not affect the dimerization of OGT (Fig. 3a).
Figure 3.
OGT XLID–TPR mutants form functional dimers but demonstrate decreased thermal stability. a, TPR domain is a constitutive homodimer in solution. Samples of WT and mutant TPR domains were analyzed on a Superdex 200 size-exclusion column. The elution profiles are plotted as A280 against elution volume. Elution time of standards of known molecular weight are marked above; 1, IgG 158 kDa; 2, human albumin 66 kDa; and 3, ovalbumin 44 kDa. The His6-tagged TPR dimer has an expected molecular mass of ∼90 kDa and an apparent molecular mass of ∼120 kDa (14) due to its elongated, nonglobular structure. b, thermal denaturing curves of WT or mutant OGT TPR domains. The melting temperatures (Tm, the temperature at which both the folded and unfolded states of a protein are equally populated at equilibrium) of the proteins are indicated. Fluorescence of Sypro Orange is plotted against temperature. The data were fitted to Boltzmann sigmoidal curve equation using Prism (GraphPad). Experiments were performed in triplicate, and error bars represent mean ± S.E.
Given that the modeling of the mutations into the structure of OGT predicted changes in the structure/stability of the protein, we proceeded to analyze the effect of the mutations on the thermodynamic stability of the TPR domain using differential scanning fluorimetry (Thermofluor assay). Samples of the proteins combined with the Sypro Orange dye were subjected to increasing temperature as a function of time in a real-time PCR machine, which measures the increase in fluorescence of the dye as it binds to hydrophobic regions of the proteins that become exposed as they denature at higher temperatures. This assay revealed that all of the XLID OGT mutant TPR domains are less stable than OGT-WT, with OGT-R284P and OGT-L254F being the most impacted, i.e. least stable. This is in agreement with previously published data for OGT-R284P (Fig. 3b) (29). The high degree of purity of the protein preparations used for this assay are demonstrated by Coomassie G-250 staining following SDS-PAGE separation (Fig. 1a).
XLID–TPR variants of OGT are active enzymes but exhibit impaired glycosyltransferase kinetics
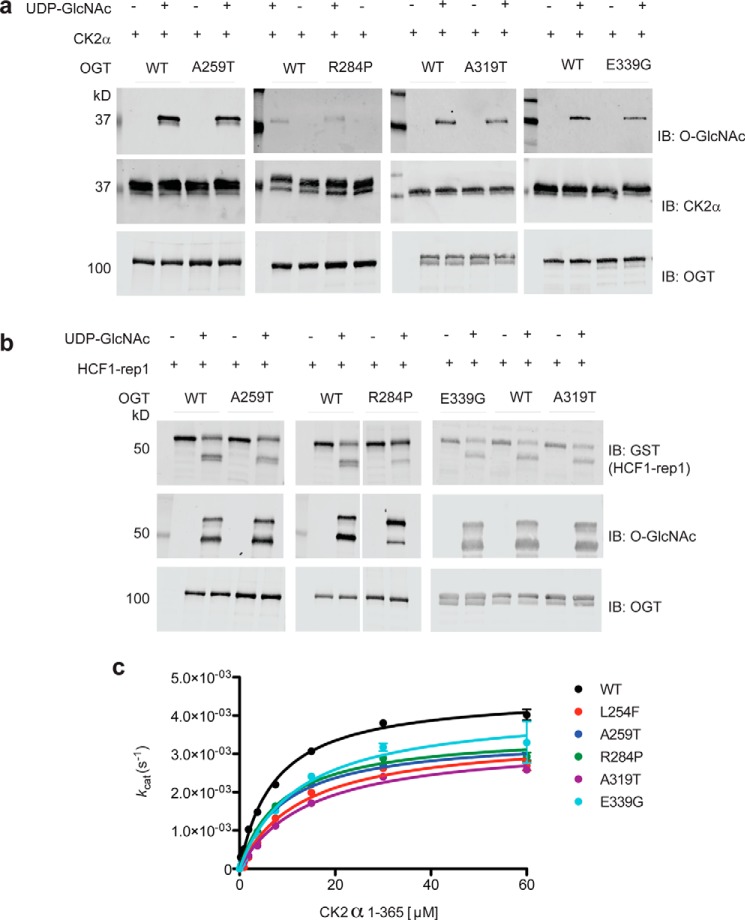
Having established that the XLID OGT-TPR mutants form functional dimers like OGT, but that their thermal stability is modestly reduced, we went on to assess the function of all of the enzyme variants, except for L254F, which we previously characterized (27), using recombinant full-length OGT variants. We performed an end-point assay wherein we incubated WT or mutant OGTs with the well-studied acceptor protein substrate CK2α (12) in the presence or absence of the donor substrate UDP-GlcNAc and then probed the samples for O-GlcNAc by Western blotting using the anti-O-GlcNAc antibody RL2 (Fig. 4a). We found that all of the XLID OGT-TPR variants glycosylated CK2α to an extent comparable with OGT-WT (Fig. 4a).
Figure 4.
XLID–TPR variants of OGT are active enzymes but exhibit impaired glycosyltransferase kinetics. a, O-GlcNAcylation of recombinant CK2α(1–365) protein substrate by WT or mutant OGT detected using the anti-O-GlcNAc antibody RL-2. Reactions were carried out at 37 °C for 1.5 h. Reactions containing no UDP-GlcNAc were included as a negative control. Samples were resolved in a 10% (A259T, R284P and E339G) or a 4–15% gradient (A319T) SDS-polyacrylamide gel, explaining the differences in the appearance of specific bands across independent experiments. Blots shown are representative of three independent replicates. b, proteolytic cleavage of a recombinant HCF1–derived construct (HCF1–rep1) by WT or mutant OGT. Uncleaved and cleaved HCF1–rep1 were detected using an anti-GST tag antibody, whereas the O-GlcNAcylation of the substrate/products were detected using the anti-O-GlcNAc antibody RL-2. Reactions were carried out at 37 °C for 6 h. Reactions containing no UDP-GlcNAc were included as a negative control. Samples were resolved in a 10% (A259T and R284P) or a 4–15% gradient (A319T and E339G) SDS-polyacrylamide gel, explaining the differences in the appearance of specific bands across independent experiments. Blots shown are representative of three independent replicates. c, Michaelis-Menten kinetics of WT or mutant OGT measured using varying amounts of CK2α(1–365) protein substrate and a fixed concentration of UDP-GlcNAc in excess of the Km. Reactions were carried out for 90 min at room temperature and read using the UDP-Glo assay system (Promega). Data points were fitted to the Michaelis-Menten equation using Prism (GraphPad). Experiments were performed in triplicate, and error bars represent mean ± S.E. IB, immunoblot.
OGT is not only a glycosyltransferase, but also carries out the proteolytic processing of the transcriptional cofactor HCF1 (35). Interestingly, mutations in HCFC1 have been implicated in causing XLID (36–38). We therefore evaluated the protease activity of OGT using the short GST-tagged substrate HCF1–rep1 (35, 39, 40) in an end-point assay similar to the one for CK2α O-GlcNAcylation (Fig. 4b). This assay revealed that all of the XLID OGT-TPR mutants are active HCF1 proteases, with activity comparable with OGT-WT, with the exception of OGT-R284P, which showed visibly reduced cleavage of the substrate compared with OGT-WT, although it still glycosylated the cleavage product efficiently (Fig. 4b). This is consistent with previously published data for OGT-R284P (29).
Having observed that the XLID OGT-TPR variants are competent enzymes, at least with respect to the conditions used in the end-point assays, we asked whether the enzyme kinetics of OGT would be affected by these mutations to explain the disease association. This time we titrated in increasing concentrations of the protein substrate CK2α into reaction mixtures containing a fixed concentration of UDP-GlcNAc in excess of the Km value and OGT-WT or one of the XLID variants and measured the Michaelis-Menten kinetics using a luminescence-based assay for the detection of UDP released (Fig. 4c). All of the XLID OGT-TPR variants showed a modest increase in Km accompanied by a reduction in turnover, resulting in an ∼2-fold decrease in catalytic efficiency (Fig. 4c and Table 2). The purity of the protein preparations used for these assays is demonstrated by the Coomassie G-250–stained SDS-polyacrylamide gel (Fig. 1b).
Table 2.
Michaelis-Menten kinetics of the XLID-associated TPR variants of OGT
| OGT variant | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | s−1 m−1 | |
| WT | 7 ± 0.6 | 4.5 × 10−3 ± 0.1 × 10−3 | 642.86 |
| L254F | 13 ± 3 | 3.5 × 10−3 ± 0.3 × 10−3 | 269.23 |
| A259T | 9 ± 0.6 | 3.5 × 10−3 ± 0.07 × 10−3 | 388.89 |
| R284P | 9 ± 1 | 3.5 × 10−3 ± 0.1 × 10−3 | 388.89 |
| A319T | 14.5 ± 1.5 | 3.3 × 10−3 ± 0.1 × 10−3 | 227.59 |
| E339G | 12 ± 1.5 | 4.0 × 10−3 ± 0.2 × 10−3 | 333.33 |
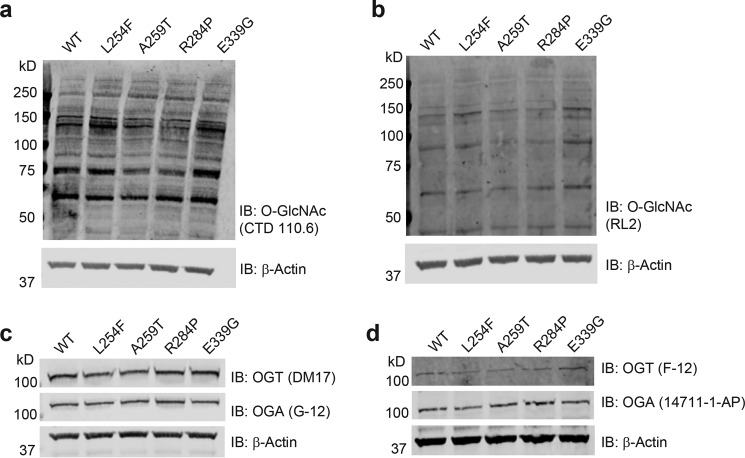
Human embryonic stem cells expressing the XLID OGT-TPR variants show no gross changes in basal O-GlcNAc, OGT, and OGA levels
After characterizing the properties of the XLID OGT-TPR variants using recombinantly isolated proteins in in vitro assays, we wanted to dissect the effects of the mutations in the context of a human isogenic ex vivo model in an attempt to identify and understand the molecular basis of the disease. To this end, we engineered the mutations into the RUES-1 male human embryonic stem cell line using CRISPR/Cas9 (Fig. 1). We chose stem cells so that we could investigate both differentiation pathways as well as multiple cell types. We were able to generate cell lines containing all of the mutations except for OGT-A319T, which presented as a technical challenge, likely because the gRNA design with the highest predicted efficiency did not straddle the site of the intended mutation, and no alternative gRNAs (with or without high off-target efficiency) were designable. All of the cell lines obtained were confirmed to contain the intended mutation by Sanger sequencing (Fig. 2), and no noticeable changes were observed in cell growth or maintenance of pluripotency (data not shown).
We previously showed that OGT steady-state levels in XLID-derived lymphoblasts containing the OGT-L254F mutation were lower than in unaffected controls from relatives; however, global O-GlcNAc levels assessed by Western blotting remained unchanged (27). This was demonstrated to be the consequence of a compensation mechanism whereby OGT, in complex with the mSin3A–HDAC1 complex, transcriptionally down-regulated OGA to maintain O-GlcNAc homeostasis (27). A similar mechanism was proposed to exist in patient-derived fibroblasts expressing OGT-R284P; however, as the authors noted, significant differences in the levels of OGT and OGA in the two separate control fibroblast lines expressing OGT-WT used in that study confounded the findings (29). In light of this, we set out to analyze and compare the levels of O-GlcNAc, OGT, and OGA in our RUES-1 cells expressing the different OGT variants by Western blotting using two antibodies against each of the targets (Fig. 5, a–d). It was unsurprising that we found no significant changes (CTD 110.6, p > 0.4; RL2, p > 0.7; Mann-Whitney U test, data not shown) in the levels of O-GlcNAc in the XLID OGT-TPR mutant-expressing lines compared with WT (Fig. 5, a and b). We also, however, saw no significant changes in the levels of OGT (DM17, p > 0.2; F-12, p > 0.1, Mann-Whitney U test, data not shown) or OGA (G-12, p > 0.7; 14711-1-AP, p > 0.4, Mann-Whitney U test, data not shown) (Fig. 5, c and d), an observation suggesting that there may be cell-type–specific differences in the regulation, and indeed function, of these enzymes.
Figure 5.
Human embryonic stem cells expressing the XLID OGT-TPR variants show no gross changes in steady-state O-GlcNAc, OGT, and OGA levels. The endogenous WT or variant OGT expressing RUES-1 hES cell lysates was probed for O-GlcNAc using the anti-O-GlcNAc antibody, CTD 110.6 (a), or a second anti-O-GlcNAc antibody, RL2 (b). Samples were also probed for OGT and OGA using the antibodies DM-17 and G-12 respectively (c) or antibodies F-12 and 14711-1-AP, respectively (d). All membranes were probed using an anti-β-actin antibody as a loading control. Blots shown are representative of three biological replicates. IB, immunoblot.
Differential transcriptomes of the hES cells expressing XLID OGT-TPR variants reveal changes in genes associated with neural development
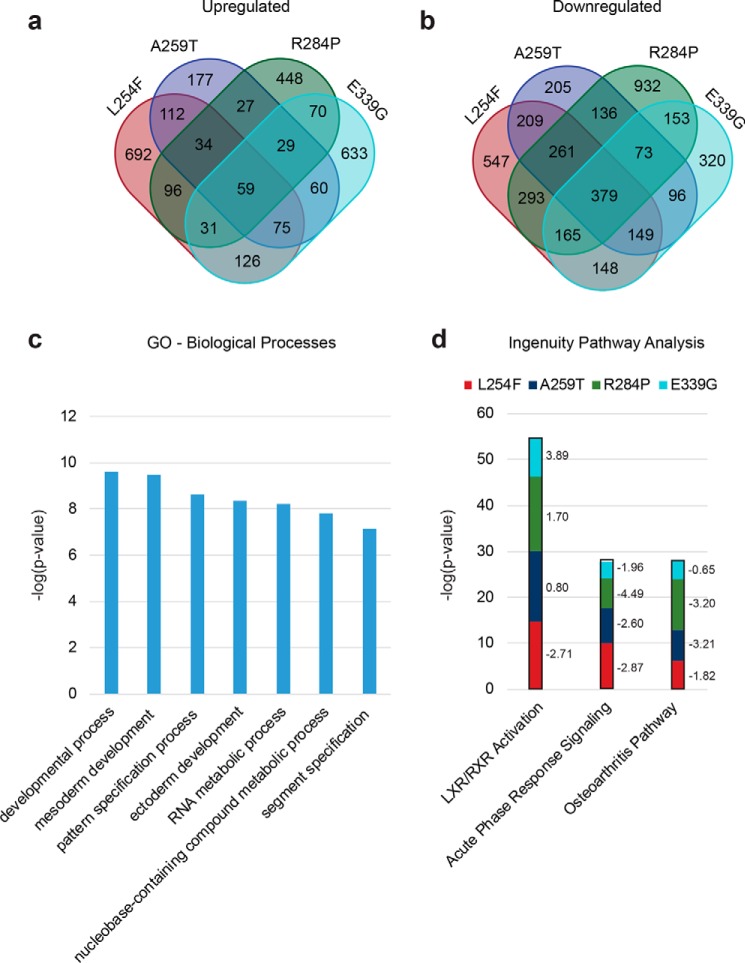
Given the developmental phenotypes of the patients and the fact that OGT, belonging to the polycomb group of developmental genes (24), regulates the transcription of a number of these genes (22), we went on to investigate changes in gene expression between OGT-WT and XLID OGT-TPR mutant–expressing stem cells by RNA-seq. A total of 20,930 genes were identified across all five cell lines combined (supporting Dataset 1). We found that a larger number of genes were 2-fold down-regulated than 2-fold up-regulated in all of the mutant lines compared with WT (Fig. 6, a and b, and supporting Datasets 2–5). This is in contrast to our previous findings from patient lymphoblasts expressing OGT-L254F, where a larger number of genes were up-regulated than down-regulated (27), once again pointing at cell-type–specific differences in the regulation and function of OGT.
Figure 6.
Differential transcriptomes of the hES cells expressing XLID OGT-TPR variants reveal changes in genes associated with neural development. a, number of genes that are >2-fold up-regulated in each of the TPR mutants analyzed compared with WT. b, number of genes that are >2-fold down-regulated in each of the TPR mutants analyzed compared with WT. c, GO analysis (PANTHER Classification System) showing significantly represented (over- or under-represented, p < 1 × 10−7, Bonferroni correction for multiple testing applied) biological processes in genes that are differentially expressed (up- or down-regulated) in at least three mutants compared with WT. d, pathway analysis (ingenuity pathway analysis, Qiagen) showing significantly represented (over- or under-represented, p < 0.001) pathways in genes found in each of the mutants compared with WT. The z-scores for the pathways in each of the mutants is shown to the right of the bars. A z-score of < −2 denotes the pathway as a whole is down-regulated, and a score of > +2 denotes it is up-regulated. Only those significantly represented pathways with a z-score of ±2 in at least two mutants are shown.
59 of the total number of genes that were over 2-fold up-regulated were common to all mutants, whereas for down-regulated genes, this number was 379 (Fig. 6, a and b, and supporting Datasets 2–5). Interestingly, Gene Ontology (GO) analysis (PANTHER Classification System) (Fig. 6c, supporting Datasets 6–9) (41, 42) also showed that genes involved in DNA-dependent transcription and the regulation of transcription by RNA polymerase II were significantly over-represented (p < 0.005, Bonferroni correction for multiple testing applied, supporting Dataset 9) within the subset of down-regulated genes in the mutant cell lines, perhaps explaining why more genes are down-regulated than up-regulated in the mutants. It is to be noted, however, that in general the over- or under-representation of down-regulated genes within a Biological Process-GO term does not directly imply that the process itself is down-regulated. Significantly, genes involved in mesoderm and ectoderm development were also differentially expressed (down-regulated) in all of the mutants (Fig. 6c), indicating that cellular differentiation of the mutant cell lines along these lineages may proceed at a rate that is altered (faster or slower than WT cells), which could then result in defects that cause intellectual disability (as a consequence of effects on differentiation of neural stem cells derived from ectoderm) and the dysmorphic facial features and clinodactyly seen in many of the patients (as a consequence of effects on differentiation of neural crest cells and mesenchymal stem cells derived from the ectoderm and mesoderm, respectively). Other significantly represented (p < 1 × 10−7, Bonferroni correction for multiple testing applied) biological processes include development and pattern/segment specification, as would be expected of a developmental disease such as XLID and RNA and nucleobase metabolic processes, which would influence cellular transcription (Fig. 6c).
Ingenuity pathway analysis (Fig. 6d and supporting Datasets 10–13) revealed components of the LXR/RXR activation pathway to be the most significantly represented in all of the mutants compared with WT (p < 0.001) and predicted the pathway itself to be down-regulated (z-score, L254F, −2.71; A259T, −0.80; R284P, −1.7; E339G, −3.89). This is interesting given that LXR has been shown to have a role in the generation of midbrain dopaminergic neurons, which have roles in memory and learning, from human embryonic stem cells (43). Other significantly represented (p < 0.001) pathways predicted to be down-regulated (z-score ≤ −2) in at least two of the four mutants were the acute-phase response pathway and the osteoarthritis pathway (Fig. 6d and supporting Datasets 6–9). RT-qPCR analysis was performed on GPR18, which is only slightly up-regulated (3–7-fold by RNA-seq), from biological triplicates of the WT and mutant cell lines, to orthogonally validate the directionality and fold change of the expression data (Fig. 4 and Fig. S3).
Discussion
Advances in genome sequencing and the reduction in associated costs have made it possible in the past decade or so to investigate the etiology of rare (genetic) disorders (2). Etiological studies on cohorts of patients with ID, however, show that 29% of these individuals still have an unknown (genetic or otherwise) cause of ID, suggesting further need for the use of massive parallel sequencing approaches in the clinic for the investigation of disease causality (2). This facilitates not only genetic counseling for families but may, in some cases, inform strategies for therapeutic intervention for patients. From a biochemical standpoint, large-scale sequencing studies reveal the involvement of novel genes/proteins in disease, as in the case of OGT and XLID, allowing for a detailed examination and thereby understanding of the roles of such genes/proteins in specific cellular processes/pathways in both disease and normal physiology.
Three missense mutations were previously shown to segregate in patients with XLID (26–29), and in this study we describe two more novel mutations (Figs. 1 and 2). OGT-dependent XLID appears to be syndromic; however, not all phenotypes found in patients with one mutation are present in patients with another mutation (Table 1). This is conceivably because OGT has a plethora of substrates/interactors, and although all of the mutations may affect a common pool of these substrates, they may also each affect a unique subset, as all are present in slightly different regions of the protein–protein interaction domain, TPRs, of OGT. Our previous study on the family with the OGT-L254F mutation (27) and the family with the OGT-E339G mutation reported here show that related patients with the same mutations also exhibit a range of severity of phenotypes as well as unique phenotypes, indicating there is variable expressivity and incomplete penetrance. Nonetheless, a phenotype does emerge across all patients consisting of intellectual disability, developmental delay, dysmorphic features, and eye abnormalities. Many of the clinical findings are not uncommon in and of themselves in the XLID population, but a male presenting with this constellation of clinical findings should perhaps be considered for OGT testing.
In an attempt to study the direct effects of the XLID mutations on OGT stability and function, we purified recombinant OGT variants and subjected these to size-exclusion chromatography, the Thermofluor assay, and both end-point and kinetic activity assays. We observed no adverse effects on the ability of the OGT variants to dimerize (Fig. 3a), and we found modest decreases in their thermodynamic stability compared with OGT-WT (Fig. 3b). The end-point activity assays showed that all of the OGT variants investigated in this study were able to glycosylate the substrate CK2α. They were also able to glycosylate and proteolytically cleave HCF1 (Fig. 4, a and b), although the assay conditions utilized may not detect subtle changes in the reaction rates. Given mutations in the gene encoding HCF1 have previously been implicated in XLID pathogenesis, it is possible that minor undetected changes in proteolysis of HCF1 by OGT could be responsible for some of the observed pathology (36–38). Analysis of the kinetics of CK2α glycosylation revealed that although catalytically competent, the OGT variants demonstrated reduced turnover (Fig. 4c and Table 2). Overall, these findings, while important to establish, are perhaps not surprising given that the mutations all cluster to the TPR domains and not the catalytic domains of the enzyme.
To delineate the molecular mechanisms underpinning this developmental disease, we introduced all of the patient mutations into a male human embryonic stem cell line. We chose this over deriving induced pluripotent stem cells from patient fibroblasts to eliminate effects arising from the different genetic backgrounds, which could confound comparative analyses. Examination of the steady-state global O-GlcNAc, OGT, and OGA levels revealed no significant changes in these isogenic cells expressing the OGT variants compared with OGT-WT (Fig. 5, a–d). This, combined with the contrasting observation that there are changes in the rate of catalysis by the OGT variants (Fig. 4c), suggests the need to investigate temporal dynamics of an in-depth O-GlcNAcome beyond what can be observed by a steady-state 1D Western blotting. We are currently in the process of developing such an approach. One hypothesis to be tested is that alterations in the kinetics of O-GlcNAc cycling on certain substrates, rather than the complete loss of O-GlcNAc on these substrates, is disability-causing.
OGT interacts with a number of proteins in the cell through its N-terminal TPRs (44). Although the TPRs are also implicated in the recognition of certain substrates for O-GlcNAcylation (15), it has been shown using hypomorphic OGT mutants of the fly that post-pupal development proceeds in the near-absence of the catalytic activity of OGT (45), suggesting that the scaffolding function of OGT in forming functional protein complexes is also critical. Given that the XLID mutations lie in the TPR domain of OGT, we also hypothesize that an altered OGT-interactome in these patients may be responsible for disease and are in the process of defining the differential OGT-interactome using multiple parallel approaches, including looking at cell-type–specific interactomes derived from the human embryonic stem cells harboring the disorder-causing mutations.
Because OGT and protein O-GlcNAcylation have roles in transcription (4, 7, 16), we subjected mRNA samples from the OGT variant and OGT-WT–expressing stem cells to a differential transcriptomics analysis using RNA-seq (Fig. 6, a–d, and supporting Datasets 1–11). Our data show, as predicted from the differences in patient phenotypes, that a common pool of genes is regulated by all of the mutants but that each mutant also regulates the expression of a unique subset of genes (Fig. 6, a and b). Although all of the cell lines expressed comparable levels of the pluripotency markers POU5F1 (Oct4), SOX2, and NANOG (supporting Dataset 1), GO analysis on genes that were found to be differentially regulated in all of the mutants revealed them to be involved in ectoderm and mesoderm development (Fig. 6c). Pathway analysis of all of the differentially expressed genes in each of the mutants led to the identification of common pathways affected in OGT-dependent XLID, although the members of the pathway affected or the extent to which members found in all mutants were affected was different between each of the mutants (Fig. 6d). Modulation of the LXR/RXR transcriptional network is of particular interest given the established role of this pathway in cell fate specification and neurogenesis (46, 47).
In conclusion, we have identified a list of genes that are differentially expressed in the XLID OGT-TPR variant-expressing cells that clearly correlate with patient phenotypes. We are now taking both biased and unbiased approaches using our isogenic stem cells and derived neuronal lineages to reveal the impact of these variants on the cell-type–specific dynamic O-GlcNAcome and OGT interactome to provide mechanistic insight into these XLIDs.
Experimental procedures
Study samples
Individuals with XLID and control relatives with normal cognitive function were recruited from the Children's Hospital of Michigan (Detroit) and the Genetic Health Services, Northern Hub, New Zealand (Auckland, New Zealand). Human subject research protocols for these studies were approved by the respective institutional review boards and abide by the Declaration of Helsinki principles. Informed consent was obtained from each study member and/or their parents or legal guardians. All individuals were found to have a normal karyotype and a negative molecular test for Fragile X syndrome.
Full-exome sequencing and validation
Whole-exome sequencing of genomic DNA from Patient 1, his parents, and an unaffected brother was performed through Ambry's (Ambry Genetics) clinical diagnostic exome assay. DNA was isolated from the whole blood of the subjects, and short tandem repeat markers were used to confirm the relationship of proband and parents. Samples were prepared using the SeqCap EZ VCRome 2.0 (NimbleGen, Roche Applied Science). Each DNA sample was sheared, adaptor ligated, PCR-amplified, and incubated with the exome baits. Captured DNA was eluted and PCR-amplified. Final quantified libraries were seeded onto an Illumina flow cell and sequenced using paired-end, 100 cycle chemistry on the Illumina HiSeq 2000 or HiSeq 2500.
Initial data processing, base calling, alignments, and variant calls were generated using various bioinformatics tools. Approximately 90% of the bases had a quality score of Q20 or higher, which translated to an expected base-calling rate of 99% and an error rate of 1:100. Exons and at least two bases into the 5′ and 3′ ends of all introns were analyzed. 90% of exome was covered at 10 times or higher depth, sufficient for high-quality heterozygous or homozygous variant calling for germline variants. Exons plus at least two bases into the 5′ and 3′ ends of all the introns were analyzed and reported. Sequencing of the mitochondrial genome and screening for characterized mutations were performed. The mean depth of coverage for targeted mitochondrial bases was greater than 1000×.
Variants were analyzed for population frequency, predicted functional impact, nucleotide and amino acid conservation, and biochemical nature of amino acid substitution. Human Gene Mutation Database, the dbSNP, 1000 genomes, HapMap data, and on-line search engine (e.g. Pubmed) sites were used to search for previously described gene mutations and polymorphisms. Variants were then filtered based on possible inheritance models and family cosegregation.
Alteration(s) detected via exome sequencing with Q-score and read depth above established confidence thresholds did not systematically undergo confirmation by Sanger sequencing. But Sanger sequencing was used for validation of candidate variant and family cosegregation studies. Candidate mutations were evaluated from among the genes in Ambry's (Ambry Genetics) internal, dynamic gene database, which classifies genes as characterized or novel. Characterized genes include those currently understood to be associated with a clinical phenotype based on data from the medical literature, human gene mutation databases, and novel gene findings identified through clinical exome. Novel genes are those not currently known to underlie a genetic condition. Characterized genes were analyzed first. As no positive findings were identified in characterized genes, reflex analysis of novel genes occurred. Each candidate mutation was analyzed by a board-certified molecular geneticist/laboratory director to identify the most likely causative mutation(s). The evidence to support the likelihood that a novel gene alteration was involved in the patient's phenotype included knowledge of the gene function based on in vitro or in vivo functional studies, knowledge of the gene family or gene pathway, and familial cosegregation analysis.
X-chromosome exome sequencing and validation
DNA from Patient 2 was extracted from blood using the QIAamp DNA blood maxi kit (Qiagen). From this, X-exome sequencing, followed by data analysis, was performed as described before (48). Confirmation of variants and segregation analysis in further family members, including Patient 3, was performed by Sanger sequencing using the following oligonucleotides: g1F, AGC ATT ACC AGC CAT TAG GC; g1R, TCT CTT GAA CTG TGG GTT AAT GT.
Plasmids and site-directed mutagenesis
Constructs used in this study have been described before (27). The plasmid encoding the TPR domain of OGT was obtained from Elena Conti (Max Planck Institute for Biochemistry). Site-directed mutagenesis was performed as described before (49).
Protein expression and purification
All proteins were expressed in Escherichia coli and purified as described before (14, 27). Briefly, all proteins except for ncOGT were purified to homogeneity using affinity chromatography followed by size-exclusion chromatography. ncOGT was purified just using affinity chromatography. Protein preparations were quantified using a NanoDropTM spectrophotometer (ThermoFisher Scientific). For ncOGT, relative quantification using BSA standards was performed, whereby Coomassie G-250–stained SDS-polyacrylamide gels were scanned on an Odyssey® CLx Imaging System (LI-COR Biosciences), and bands of interest were quantified using the Image StudioTM software (LI-COR Biosciences).
Thermal stability assay
The thermal stability of the proteins was measured using differential scanning fluorimetry (Thermofluor assay). Briefly, 45 μl of 1 mg/ml solutions of protein in a buffer composed of 25 mm Tris-HCl, pH 7.5, and 150 mm NaCl were combined with 5 μl of 200× SYPRO Orange diluted from a 5000× concentrate (ThermoFisher Scientific) in the aforementioned buffer. The mixture was allowed to incubate at room temperature for 15 min. The sealed PCR plate containing the mixtures was exposed to a temperature gradient program (25–95 °C, 0.5 °C intervals) with fluorescence intensity measured after a 30-s dwell time at each particular temperature in a MyIQTM Single Color Real-Time PCR Detection System (485/20 nm for excitation and 530/30 nm for emission) equipped with an iCycler (Bio-Rad). The fluorescence intensities plotted against their corresponding temperatures were exported to Microsoft Excel and then to Prism (GraphPad). Wells containing no protein, but only buffer and SYPRO Orange, were used for background subtraction. Fluorescence values obtained between 40 and 65 °C were fitted to the Boltzmann Sigmoidal equation to obtain Tm values. All experiments were performed in technical triplicate.
CK2α glycosylation and HCF1 proteolysis
The Western blotting assays for CK2α glycosylation and HCF1 proteolysis were carried out using full-length OGT as described before (27). Reactions were run either on a 10 or 4–15% gradient Mini-PROTEAN® TGX Stain-FreeTM precast gel (Bio-Rad) before being blotted onto nitrocellulose membranes, explaining the differences in the appearance of specific bands across independent experiments. Blots were probed with the antibodies described before (27). All experiments were performed in biological triplicate (with three separate preparations of ncOGT).
Enzyme kinetics
The UDP-GloTM glycosyltransferase assay kit (Promega) was used to measure the steady-state kinetics. Recombinantly purified full-length OGT was used in these experiments. Reaction mixtures contained 50 nm of the OGT variants (20 nm OGT-WT), 500 μm ultrapure donor substrate UDP-GlcNAc (Promega), and varying concentrations (0.234 to 60 μm of the acceptor substrate), human recombinant CK2α(1–365)), in a final volume of 18 μl. The reaction buffer was made up of 20 mm Tris-HCl, pH 8.0, 20 mm MgCl2, 150 mm NaCl, and 1 mm DTT. Reactions were incubated for 90 min at room temperature and stopped by the addition of 18 μl of the UDP detection reagent prepared as recommended by the manufacturer. 10 μl of the samples were transferred to a white 384-well plate, and the luminescence was measured using the GloMax® multidetection system (Promega). Relative luminescence units were converted to the amount of UDP released using a UDP standard curve. All assays were performed in technical triplicate, and measurements were corrected for background emission from reactions containing no acceptor substrate. For all assays performed, UDP-GlcNAc turnover was under 10%, and reactions were linear past the 90-min incubation time used. Nonlinear regression curves were fitted with Prism (GraphPad).
Cell lines and culture maintenance
RUES-1 male human ES cells were obtained from the Laboratory of Molecular Embryology at the Rockefeller University. XLID OGT-TPR point mutations in these cells were introduced using CRISPR/Cas9 as a pay-for-service by Applied StemCell. Cells were maintained in TeSRTM E8TM media (STEMCELL Technologies) in 5% CO2. The media were changed every day, and at appropriate confluency, cells were split 1:3 or 1:5 using an enzyme-free dissociation buffer (1× DPBS - Ca2+ and Mg2+-free (ThermoFisher Scientific) supplemented with sterile-filtered 0.5 mm EDTA and 150 mm NaCl).
Cell lysis and Western blotting
Harvested stem cells were lysed using 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% IGEPAL® CA-630, and 1 mm DTT supplemented with 10 μm PUGNAc (hexosaminidase inhibitor), 1× EDTA-free Protease Inhibitor Mixture Set V (Millipore), and 1× Phosphatase Inhibitor Mixture Set II (Millipore). Lysates were resolved on BoltTM 4–12% BisTris Plus Gels (ThermoFisher Scientific) and blotted onto nitrocellulose membranes. Membranes were probed with the following antibodies: O-GlcNAc CTD 110.6 (1:500, ascites generated in the lab of Gerald Hart at Johns Hopkins University); O-GlcNAc RL2 (1:1000, ThermoFisher Scientific, MA1-072); OGT DM17 (1:1000, Sigma, O6264); OGT F-12 (1:1000, Santa Cruz Biotechnology, sc-74546); OGA/MGEA5 (1:1000, Proteintech, 14711-1-AP); OGA/NCOAT G-12 (1:1000, Santa Cruz Biotechnology, sc-376429); and β-actin 13E5 (1:1000, Cell Signaling, 4970). Secondary antibodies were purchased from LI-COR and used at a dilution of 1:10,000. Experiments were conducted in biological triplicate.
RNA-seq and bioinformatics
Total RNA was isolated from cells harvested from confluent 60-mm dishes using the RNeasy Plus mini kit (Qiagen). The concentration and integrity of the extracted total RNA were estimated using the Qubit® 2.0 Fluorometer (Invitrogen) and Agilent 2100 Bioanalyzer (Applied Biosystems), respectively. Five hundred nanograms of total RNA was required for downstream RNA-seq applications. Polyadenylated RNAs were isolated using NEBNext Magnetic Oligo(dT)25 beads. Next, first-strand synthesis was performed using NEBNext RNA first-strand synthesis module (New England Biolabs). Immediately, directional second-strand synthesis was performed using NEBNExt Ultra Directional second-strand synthesis kit. The NEBNext® DNA Library Prep Master Mix Set for Illumina® was then used to prepare individually bar-coded next-generation sequencing expression libraries as per the manufacturer's recommended protocol. Library quality was assessed using the Qubit 2.0 Fluorometer, and the library concentration was estimated by utilizing a DNA 1000 Chip on an Agilent 2100 Bioanalyzer. Accurate quantification for sequencing applications was determined using the qPCR-based KAPA Biosystems Library Quantification Kit (Kapa Biosystems). Paired-end sequencing (50 bp) was performed on an Illumina HiSeq2500 sequencer to obtain ∼50 million reads per sample. Raw reads were de-multiplexed using bcl2fastq Conversion Software (Illumina) with default settings.
Post-processing of the sequencing reads from RNA-seq experiments for each sample was performed using HudsonAlpha's unique in-house RNA-seq data analysis pipeline. Briefly, quality control checks on raw sequence data for each sample were performed using FastQC (Babraham Bioinformatics, Cambridge, UK). Raw reads were mapped to the reference hg19 using TopHat version 2.0. The alignment metrics of the mapped reads were estimated using SAMtools. Aligned reads were imported to the commercial data analysis platform AvadisNGS (Strand Scientifics). After quality inspection, the aligned reads were filtered on the basis of read quality metrics; reads with a base quality score of less than 30, alignment score of less than 95, and mapping quality of less than 40 were removed. Remaining reads were then filtered on the basis of their read statistics; missing mates, translocated, unaligned, and flipped reads were removed. The reads list was then filtered to remove duplicates. Samples were grouped, and transcript abundance was quantified for this final read list using Trimmed Means of M-values as the normalization method. Output data utilized for all subsequent comparisons were summarized as normalized signal values generated by AvadisNGS. Differential expression of genes was calculated on the basis of fold changes (using the default cutoff ≥ ±2.0) observed in comparisons between defined conditions.
RT-qPCR analysis
Total RNA was isolated from cells harvested from confluent 60-mm dishes using the RNeasy Plus mini kit (Qiagen). The concentration of the extracted total RNA was estimated using a NanoDropTM spectrophotometer (ThermoFisher Scientific). iScript cDNA synthesis kit (Bio-Rad) was used for cDNA synthesis and iTaqTM Universal SYBR® Green Supermix (Bio-Rad) or iQTM SYBR® Green Supermix (Bio-Rad) was used for qPCR, performed as described before (50, 51). Briefly, the ΔΔCt method was used to quantify differences in GPR18 gene expression and ACTB (β-actin) was used as the housekeeping gene. Experiments were performed in biological triplicate, and data analysis was performed as described before (50, 51) Primer sequences are provided in Table S1.
Author contributions
N. S. and L. W. conceptualization; N. S., F. J. S., M. S., N. P., C. E. S., and L. W. data curation; N. S., S. G., F. J. S., M. S., L. H., V. M. K., N. P., C. E. S., and L. W. formal analysis; N. S., S. G., F. J. S., M. S., and L. W. validation; N. S., S. G., F. J. S., M. S., L. H., V. M. K., J. T., S. A., C. E. S., A. M. H., J. G., and L. W. investigation; N. S., S. G., F. J. S., J. T., S. A., A. M. H., and L. W. visualization; N. S., S. G., F. J. S., M. S., L. H., V. M. K., and L. W. methodology; N. S. writing-original draft; N. S., M. S., C. E. S., A. M. H., J. G., and L. W. writing-review and editing; F. J. S., M. S., and N. P. software; S. E. L., C. E. S., A. M. H., J. G., and L. W. supervision; S. E. L., C. E. S., J. G., and L. W. funding acquisition; S. E. L., J. G., and L. W. project administration; L. W. resources.
Supplementary Material
Acknowledgments
We thank Dr. Gerald Hart (The Johns Hopkins School of Medicine), Dr. Suzanne Walker (Harvard University), Dr. Alison Nairn (University of Georgia), and Dr. Winship Herr (University of Lausanne) for providing plasmids and/or technical advice.
This work was supported in part by a grant from the W. M. Keck Foundation (to L. W.), National Institutes of Health Grant P41GM103490 from NIGMS (to L. W.), a grant from the South Carolina Department of Disabilities and Special Needs (SCDDSN) (to C. E. S.), National Institutes of Health Grant RO1N5073854 from NINDS (to C. E. S.), and National Health and Medical Research Council of Australia Grants APP1091593 and APP1041920 (to J. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3, Table S1, and supporting Datasets S1–S13.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, accession no. GSE110616.
- ID
- intellectual disability
- XLID
- X-linked intellectual disability
- OGT
- O-GlcNAc transferase
- TPR
- tetratricopeptide
- LXR
- liver X receptor
- RXR
- retinoid X receptor
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- OGA
- O-GlcNAc hydrolase
- ADHD
- attention deficit hyperactivity disorder
- SNP
- single nucleotide polymorphism
- dbSNP
- database of SNP
- qPCR
- quantitative PCR
- PDB
- Protein Data Bank
- RNA-seq
- RNA-sequencing
- GO
- Gene Ontology
- hES
- human embryonic stem cell.
References
- 1. Vissers L. E., Gilissen C., and Veltman J. A. (2016) Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 17, 9–18 10.1038/nrg3999 [DOI] [PubMed] [Google Scholar]
- 2. Kvarnung M., and Nordgren A. (2017) Intellectual disability and rare disorders: a diagnostic challenge. Adv. Exp. Med. Biol. 1031, 39–54 10.1007/978-3-319-67144-4_3 [DOI] [PubMed] [Google Scholar]
- 3. Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., and Marth J. D. (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 10.1073/pnas.100471497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 10.1146/annurev-biochem-060608-102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu Y., Liu T. W., Cecioni S., Eskandari R., Zandberg W. F., and Vocadlo D. J. (2015) O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol. 11, 319–325 10.1038/nchembio.1774 [DOI] [PubMed] [Google Scholar]
- 6. Ranuncolo S. M., Ghosh S., Hanover J. A., Hart G. W., and Lewis B. A. (2012) Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem. 287, 23549–23561 10.1074/jbc.M111.330910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., et al. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 10.1038/emboj.2012.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang F., Su K., Yang X., Bowe D. B., Paterson A. J., and Kudlow J. E. (2003) O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–725 10.1016/S0092-8674(03)00974-7 [DOI] [PubMed] [Google Scholar]
- 9. Guo B., Liang Q., Li L., Hu Z., Wu F., Zhang P., Ma Y., Zhao B., Kovács A. L., Zhang Z., Feng D., Chen S., and Zhang H. (2014) O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 16, 1215–1226 10.1038/ncb3066 [DOI] [PubMed] [Google Scholar]
- 10. Yang Y. R., Song M., Lee H., Jeon Y., Choi E. J., Jang H. J., Moon H. Y., Byun H. Y., Kim E. K., Kim D. H., Lee M. N., Koh A., Ghim J., Choi J. H., Lee-Kwon W., et al. (2012) O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11, 439–448 10.1111/j.1474-9726.2012.00801.x [DOI] [PubMed] [Google Scholar]
- 11. Keembiyehetty C., Love D. C., Harwood K. R., Gavrilova O., Comly M. E., and Hanover J. A. (2015) Conditional knock-out reveals a requirement for O-linked N-acetylglucosaminase (O-GlcNAcase) in metabolic homeostasis. J. Biol. Chem. 290, 7097–7113 10.1074/jbc.M114.617779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lazarus M. B., Nam Y., Jiang J., Sliz P., and Walker S. (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 10.1038/nature09638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanover J. A., Yu S., Lubas W. B., Shin S. H., Ragano-Caracciola M., Kochran J., and Love D. C. (2003) Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409, 287–297 10.1016/S0003-9861(02)00578-7 [DOI] [PubMed] [Google Scholar]
- 14. Jínek M., Rehwinkel J., Lazarus B. D., Izaurralde E., Hanover J. A., and Conti E. (2004) The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α. Nat. Struct. Mol. Biol. 11, 1001–1007 10.1038/nsmb833 [DOI] [PubMed] [Google Scholar]
- 15. Lazarus B. D., Love D. C., and Hanover J. A. (2006) Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 16, 415–421 10.1093/glycob/cwj078 [DOI] [PubMed] [Google Scholar]
- 16. Yang X., Zhang F., and Kudlow J. E. (2002) Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69–80 10.1016/S0092-8674(02)00810-3 [DOI] [PubMed] [Google Scholar]
- 17. Okuyama R., and Marshall S. (2003) UDP-N-acetylglucosaminyl transferase (OGT) in brain tissue: temperature sensitivity and subcellular distribution of cytosolic and nuclear enzyme. J. Neurochem. 86, 1271–1280 10.1046/j.1471-4159.2003.01939.x [DOI] [PubMed] [Google Scholar]
- 18. Wang Z., Udeshi N. D., O'Malley M., Shabanowitz J., Hunt D. F., and Hart G. W. (2010) Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 9, 153–160 10.1074/mcp.M900268-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., and Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 10.1074/mcp.O112.018366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Webster D. M., Teo C. F., Sun Y., Wloga D., Gay S., Klonowski K. D., Wells L., and Dougan S. T. (2009) O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev. Biol. 9, 28 10.1186/1471-213X-9-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kenwrick S., Amaya E., and Papalopulu N. (2004) Pilot morpholino screen in Xenopus tropicalis identifies a novel gene involved in head development. Dev. Dyn. 229, 289–299 10.1002/dvdy.10440 [DOI] [PubMed] [Google Scholar]
- 22. Selvan N., Williamson R., Mariappa D., Campbell D. G., Gourlay R., Ferenbach A. T., Aristotelous T., Hopkins-Navratilova I., Trost M., and van Aalten D. M. (2017) A mutant O-GlcNAcase enriches Drosophila developmental regulators. Nat. Chem. Biol. 13, 882–887 10.1038/nchembio.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ingham P. W. (1984) A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell 37, 815–823 10.1016/0092-8674(84)90416-1 [DOI] [PubMed] [Google Scholar]
- 24. Gambetta M. C., Oktaba K., and Müller J. (2009) Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96 10.1126/science.1169727 [DOI] [PubMed] [Google Scholar]
- 25. Sinclair D. A., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., Vocadlo D. J., Brock H. W., and Honda B. M. (2009) Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432 10.1073/pnas.0904638106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Niranjan T. S., Skinner C., May M., Turner T., Rose R., Stevenson R., Schwartz C. E., and Wang T. (2015) Affected kindred analysis of human X chromosome exomes to identify novel X-linked intellectual disability genes. PLoS ONE 10, e0116454 10.1371/journal.pone.0116454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vaidyanathan K., Niranjan T., Selvan N., Teo C. F., May M., Patel S., Weatherly B., Skinner C., Opitz J., Carey J., Viskochil D., Gecz J., Shaw M., Peng Y., Alexov E., et al. (2017) Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J. Biol. Chem. 292, 8948–8963 10.1074/jbc.M116.771030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bouazzi H., Lesca G., Trujillo C., Alwasiyah M. K., and Munnich A. (2015) Nonsyndromic X-linked intellectual deficiency in three brothers with a novel MED12 missense mutation [c.5922G>T (p.Glu1974His)]. Clin. Case Rep. 3, 604–609 10.1002/ccr3.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Willems A. P., Gundogdu M., Kempers M. J. E., Giltay J. C., Pfundt R., Elferink M., Loza B. F., Fuijkschot J., Ferenbach A. T., van Gassen K. L. I., van Aalten D. M. F., and Lefeber D. J. (2017) Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J. Biol. Chem. 292, 12621–12631 10.1074/jbc.M117.790097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., O'Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B., Tukiainen T., Birnbaum D. P., Kosmicki J. A., Duncan L. E., Estrada K., et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siva N. (2008) 1000 Genomes project. Nat. Biotechnol. 26, 256 10.1038/nbt0308-256b [DOI] [PubMed] [Google Scholar]
- 32. Sherry S. T., Ward M., and Sirotkin K. (1999) dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 9, 677–679 [PubMed] [Google Scholar]
- 33. Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., Kondrashov A. S., and Sunyaev S. R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ng P. C., and Henikoff S. (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Capotosti F., Guernier S., Lammers F., Waridel P., Cai Y., Jin J., Conaway J. W., Conaway R. C., and Herr W. (2011) O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144, 376–388 10.1016/j.cell.2010.12.030 [DOI] [PubMed] [Google Scholar]
- 36. Huang L., Jolly L. A., Willis-Owen S., Gardner A., Kumar R., Douglas E., Shoubridge C., Wieczorek D., Tzschach A., Cohen M., Hackett A., Field M., Froyen G., Hu H., Haas S. A., et al. (2012) A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am. J. Hum. Genet. 91, 694–702 10.1016/j.ajhg.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jolly L. A., Nguyen L. S., Domingo D., Sun Y., Barry S., Hancarova M., Plevova P., Vlckova M., Havlovicova M., Kalscheuer V. M., Graziano C., Pippucci T., Bonora E., Sedlacek Z., and Gecz J. (2015) HCFC1 loss-of-function mutations disrupt neuronal and neural progenitor cells of the developing brain. Hum. Mol. Genet. 24, 3335–3347 10.1093/hmg/ddv083 [DOI] [PubMed] [Google Scholar]
- 38. Koufaris C., Alexandrou A., Tanteles G. A., Anastasiadou V., and Sismani C. (2016) A novel HCFC1 variant in male siblings with intellectual disability and microcephaly in the absence of cobalamin disorder. Biomed. Rep. 4, 215–218 10.3892/br.2015.559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bhuiyan T., Waridel P., Kapuria V., Zoete V., and Herr W. (2015) Distinct OGT-binding sites promote HCF-1 cleavage. PLoS ONE 10, e0136636 10.1371/journal.pone.0136636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kapuria V., Röhrig U. F., Bhuiyan T., Borodkin V. S., van Aalten D. M., Zoete V., and Herr W. (2016) Proteolysis of HCF-1 by Ser/Thr glycosylation-incompetent O-GlcNAc transferase:UDP-GlcNAc complexes. Genes Dev. 30, 960–972 10.1101/gad.275925.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mi H., and Thomas P. (2009) PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 563, 123–140 10.1007/978-1-60761-175-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mi H., Muruganujan A., Casagrande J. T., and Thomas P. D. (2013) Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun G., and Shi Y. (2010) Nuclear receptors in stem cells and their therapeutic potential. Adv. Drug Deliv. Rev. 62, 1299–1306 10.1016/j.addr.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Deng R. P., He X., Guo S. J., Liu W. F., Tao Y., and Tao S. C. (2014) Global identification of O-GlcNAc transferase (OGT) interactors by a human proteome microarray and the construction of an OGT interactome. Proteomics 14, 1020–1030 10.1002/pmic.201300144 [DOI] [PubMed] [Google Scholar]
- 45. Mariappa D., Zheng X., Schimpl M., Raimi O., Ferenbach A. T., Müller H. A., and van Aalten D. M. (2015) Dual functionality of O-GlcNAc transferase is required for Drosophila development. Open Biol. 5, 150234 10.1098/rsob.150234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sacchetti P., Sousa K. M., Hall A. C., Liste I., Steffensen K. R., Theofilopoulos S., Parish C. L., Hazenberg C., Richter L. A., Hovatta O., Gustafsson J. A., and Arenas E. (2009) Liver X receptors and oxysterols promote ventral midbrain neurogenesis in vivo and in human embryonic stem cells. Cell Stem Cell 5, 409–419 10.1016/j.stem.2009.08.019 [DOI] [PubMed] [Google Scholar]
- 47. Simandi Z., Horvath A., Cuaranta-Monroy I., Sauer S., Deleuze J. F., and Nagy L. (2017) RXR heterodimers orchestrate transcriptional control of neurogenesis and cell fate specification. Mol. Cell. Endocrinol. 2017, S0303–7207 10.1016/j.mce.2017.07.033 [DOI] [PubMed] [Google Scholar]
- 48. Hu H., Haas S. A., Chelly J., Van Esch H., Raynaud M., de Brouwer A. P., Weinert S., Froyen G., Frints S. G., Laumonnier F., Zemojtel T., Love M. I., Richard H., Emde A. K., Bienek M., et al. (2016) X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol. Psychiatry 21, 133–148 10.1038/mp.2014.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Selvan N., Mariappa D., van den Toorn H. W., Heck A. J., Ferenbach A. T., and van Aalten D. M. (2015) The early metazoan Trichoplax adhaerens possesses a functional O-GlcNAc system. J. Biol. Chem. 290, 11969–11982 10.1074/jbc.M114.628750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nairn A. V., dela Rosa M., and Moremen K. W. (2010) Transcript analysis of stem cells. Methods Enzymol. 479, 73–91 10.1016/S0076-6879(10)79004-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guo H., Zhang B., Nairn A. V., Nagy T., Moremen K. W., Buckhaults P., and Pierce M. (2017) O-Linked N-acetylglucosamine (O-GlcNAc) expression levels epigenetically regulate colon cancer tumorigenesis by affecting the cancer stem cell compartment via modulating expression of transcriptional factor MYBL1. J. Biol. Chem. 292, 4123–4137 10.1074/jbc.M116.763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schimpl M., Zheng X., Borodkin V. S., Blair D. E., Ferenbach A. T., Schüttelkopf A. W., Navratilova I., Aristotelous T., Albarbarawi O., Robinson D. A., Macnaughtan M. A., and van Aalten D. M. (2012) O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis. Nat. Chem. Biol. 8, 969–974 10.1038/nchembio.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.