Abstract
Protein folding in the cell is regulated by several quality-control mechanisms. Correct folding of glycoproteins in the endoplasmic reticulum (ER) is tightly monitored by the recognition of glycan signals by lectins in the ER-associated degradation (ERAD) pathway. In mammals, mannose trimming from N-glycans is crucial for disposal of misfolded glycoproteins. The mannosidases responsible for this process are ER mannosidase I and ER degradation–enhancing α-mannosidase–like proteins (EDEMs). However, the molecular mechanism of mannose removal by EDEMs remains unclear, partly owing to the difficulty of reconstituting mannosidase activity in vitro. Here, our analysis of EDEM3-mediated mannose-trimming activity on a misfolded glycoprotein revealed that ERp46, an ER-resident oxidoreductase, associates stably with EDEM3. This interaction, which depended on the redox activity of ERp46, involved formation of a disulfide bond between the cysteine residues of the ERp46 redox-active sites and the EDEM3 α-mannosidase domain. In a defined in vitro system consisting of recombinant proteins purified from HEK293 cells, the mannose-trimming activity of EDEM3 toward the model misfolded substrate, the glycoprotein T-cell receptor α locus (TCRα), was reconstituted only when ERp46 had established a covalent interaction with EDEM3. On the basis of these findings, we propose that disposal of misfolded glycoproteins through mannose trimming is tightly connected to redox-mediated regulation in the ER.
Keywords: endoplasmic reticulum (ER), endoplasmic-reticulum-associated protein degradation (ERAD), ER quality control, N-linked glycosylation, protein complex, alpha-mannosidase, EDEM3, ERp46
Introduction
Protein folding in all parts of the cell is regulated by quality control mechanisms. For proteins that function in the extracellular oxidative environment, folding reactions take place in the endoplasmic reticulum (ER)2 (1–3). Concomitant with polypeptide elongation and sequestration into the ER lumen, nearly 70% of proteins are modified with N-glycans. Trimming of sugar residues starts shortly after the transfer of N-glycans to the polypeptides and plays pivotal roles in both proper folding and degradation of glycoproteins that fail to obtain their native conformations (4–6). This mechanism is conserved among eukaryotes, and mannose trimming from N-glycans is crucial for the degradation of glycoproteins by ER-associated degradation (ERAD) (7–10).
Mannose is trimmed from N-glycans by α1,2-mannosidases (glycoside hydrolase family 47) in the ER. In the budding yeast Saccharomyces cerevisiae, the mechanism of creation of the glycan moiety critical for glycoprotein ERAD has been elucidated (11). First, the mannose is trimmed from the middle branch (branch B) of the Man9GlcNAc2 glycan (M9), forming Man8GlcNAc2 isomer B (M8B) (Fig. S1). Next, the terminal mannose on branch C is removed from the M8B glycan, yielding Man7GlcNAc2 isomer A (M7A), which serves as the glycan signal for degradation. Mns1p, a homologue of mammalian ER mannosidase I (ERManI), catalyzes the first step of creation of M8B, followed by trimming by Htm1p/Mnl1p, the homologue of mammalian EDEMs, to yield M7A. Sequential demannosylation is thought to ensure protein quality control by allowing enough time for nascent polypeptides to fold (12, 13). The mannose-trimming process is generally well-conserved in eukaryotes, except that, in vertebrates, in addition to M7A, smaller N-glycans containing five or six mannoses (Man5GlcNAc2 (M5) or Man6GlcNAc2 (M6)) are also generated and recognized as degradation signals. The three ER degradation–enhancing α-mannosidase–like proteins (EDEMs), EDEM1, -2, and -3, all contain an α-mannosidase (glycoside hydrolase family 47) domain homologous to that of ERManI (14–16). In vertebrates, EDEM2 (17) or ERManI (18) trims mannose to generate M8B, followed by mannose removal by EDEM1, EDEM3, and ERManI, yielding the M7, M6, and M5 glycans (6, 19). Demannosylated N-glycans that lack the terminal mannose of the C-branch are then recognized by lectins, such as OS-9 in mammals or Yos9p in yeast, and targeted for degradation by ERAD (20, 21).
The mechanism and specificity of trimming have been extensively studied in yeast, in which Htm1p/Mnl1p removes mannose from the N-glycan. Htm1p/Mnl1p forms a complex with Pdi1p, a homologue of mammalian protein-disulfide isomerase (PDI), which is required for the mannose-trimming activity of Htm1p/Mnl1p. PDI is a member of the oxidoreductases, which introduce and isomerize disulfide bonds during oxidative protein folding (22–24). The Htm1p-Pdi1p complex preferentially processes mannose from misfolded or nonnative glycoproteins but exhibits very low enzyme activity toward purified oligosaccharides (25–27). By contrast, little is known about the mechanism by which vertebrate EDEM proteins process mannose from N-glycans.
Among the three mammalian EDEMs, EDEM3 has a mannose-trimming activity that is readily detected when the protein is expressed in cultured cells (16). However, because the mannose-trimming activity of EDEM3 has not been successfully detected in vitro, it remains unclear how the reaction takes place in the ER by EDEM3.
In this study, we searched for proteins that interact with and regulate the mannosidase activity of EDEM3. We identified the oxidoreductase ERp46 (ER-resident protein 46) as a binding partner that accelerates the mannose-trimming activity of EDEM3 in vivo. By purifying recombinant proteins from HEK293 cells, we succeeded in analyzing the demannosylation activity of EDEM3 in vitro. We found that ERp46 triggers the mannose trimming of EDEM3 in vitro and that this effect requires a covalent interaction between the two proteins, most probably involving mixed disulfide bond formation. Our findings suggest that mammalian EDEM3 is the mannosidase that generates the glycan signal on glycoproteins that is crucial for ERAD and that interaction with ERp46 through its redox-active sites is required for this process.
Results
ERp46 associates with EDEM3 and promotes the mannose-trimming activity of EDEM3 in vivo
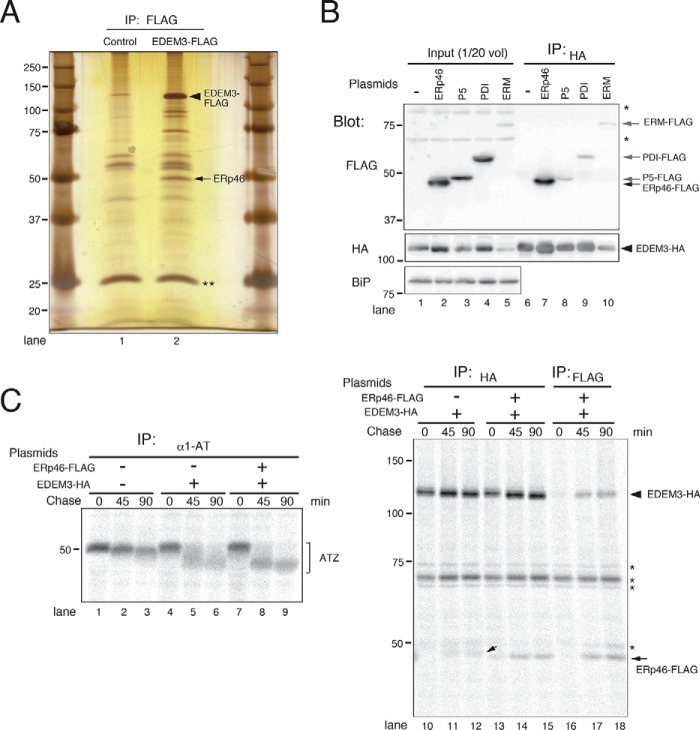
To identify proteins that regulate the mannose-trimming activity of EDEM3, we first searched for proteins that associate with EDEM3. For this purpose, we expressed FLAG-tagged EDEM3 in HEK293 cells, immunoprecipitated with anti-FLAG antibody, and then separated the co-immunoprecipitated proteins by SDS-PAGE, followed by silver staining (Fig. 1A). A ∼50-kDa protein that associated with EDEM3 (Fig. 1A, arrowhead) was identified by MS analysis as ERp46, a member of the PDI family (22, 28). Because this was reminiscent of the interaction between Htm1p/Mnl1p and Pdi1p (29, 30), we investigated whether EDEM3 also binds other PDI family members. When oxidoreductases ERp46, P5, and PDI were co-expressed with EDEM3, ERp46 was specifically associated with EDEM3, whereas P5 and PDI were not (Fig. 1B and Fig. S2). ERManI (ERM in Fig. 1) was included in the experiment because its co-expression with EDEM3 promotes mannose trimming from misfolded glycoproteins (31).
Figure 1.
ERp46 associates with EDEM3 and promotes mannose-trimming activity in vivo. A, silver staining of proteins associated with EDEM3. FLAG-tagged EDEM3 expressed in HEK293 cells was purified using FLAG-agarose beads, and co-immunoprecipitated proteins were separated by SDS-PAGE. Closed triangle, EDEM3-FLAG; arrow, co-immunoprecipitated ERp46. **, immunoglobulin light chains eluted from the FLAG-agarose beads. B, specific interaction of ERp46 with EDEM3. HEK293 cells were transfected with EDEM3-HA and FLAG-tagged oxidoreductases or ERManI (ERM) and immunoprecipitated using anti-HA antibody. Co-immunoprecipitation of FLAG-tagged proteins was analyzed by Western blotting with anti-FLAG antibody. *, signals nonspecifically detected by anti-HA antibody. C, co-expression of ERp46 promotes mannose-trimming activity of EDEM3 in vivo. HEK293 cells transfected with ATZ were pulse-labeled for 15 min and chased for the indicated periods. Proteins were immunoprecipitated using specific antibodies and separated by SDS-PAGE. Small arrow, ATZ co-immunoprecipitated with EDEM3. *, bands nonspecifically detected by anti-HA antibody.
To examine the effect of ERp46 on the mannose-trimming activity of EDEM3, we transfected HEK293 cells with ATZ, the Z variant of α1-antitrypsin, which misfolds and is retained in the ER. ATZ has three N-glycans, and thus, we evaluated their demannosylation by the mobility shift on SDS-PAGE. Consistent with our previous report (16), mannose trimming from ATZ was strongly promoted by co-transfection of EDEM3 (Fig. 1C, compare lanes 4–6 with lanes 1–3). Co-expression of ERp46 with EDEM3 further enhanced mannose processing from ATZ (Fig. 1C, lanes 7–9). ATZ co-immunoprecipitated with both EDEM3 and ERp46 (Fig. 1C (small arrow) and Fig. S3). Collectively, these data suggest that ERp46 interacts specifically with EDEM3 and promotes the mannose-trimming activity of EDEM3 in vivo.
ERp46 alters the redox state of EDEM3
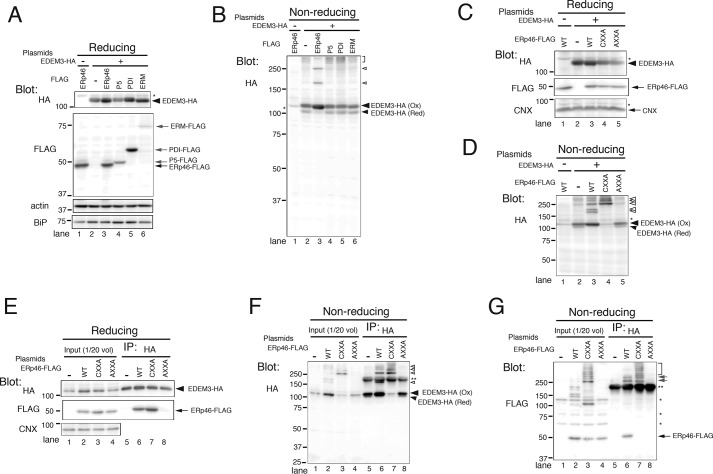
PDI family proteins, which have multiple thioredoxin-like domains containing CXXC redox-active sequences, promote the formation and exchange of disulfide bonds in nascent polypeptides synthesized in the ER (22–24). Mature EDEM3 contains nine Cys residues (Fig. S4). To examine the redox state of EDEM3, we resolved cell lysates under nonreducing conditions. EDEM3 migrated as two discrete bands (Fig. 2B, lane 2), and we noticed that the slower migrating band disappeared under reducing conditions. Hence, we assigned the corresponding signal as the oxidized (Ox) and the faster migrating band as the reduced (Red) form, respectively (Fig. S5; see Fig. 3 as discussed below). Interestingly, when EDEM3 was co-expressed with ERp46, the level of reduced EDEM3 decreased, and EDEM3-containing complexes appeared (Fig. 2B, lane 3, open arrowheads), but this effect was not observed when EDEM was co-expressed with PDI or P5. Co-expression of P5 and PDI did not alter the Ox/Red ratio of EDEM3 (Fig. 2B, compare lanes 4 and 5 with lane 2). Furthermore, in cells expressing P5 and PDI, the levels of high-molecular weight EDEM3-containing complexes were elevated, probably due to up-regulation of ER oxidation capacity by overexpression of P5 and PDI (Fig. 2B, bracket). However, in contrast to ERp46-transfected cells, no distinct EDEM3-containing complexes were detected.
Figure 2.
Covalent interaction with ERp46 alters the redox states of EDEM3. A and B, redox states of EDEM3. HEK293 cells transfected with EDEM3-HA and ER oxidoreductases or ERManI were analyzed by Western blotting under reducing (A) and nonreducing (B) conditions. Open arrowheads, complexes covalently associated with EDEM3; bracket, high-molecular weight complexes containing EDEM3. *, signals nonspecifically detected by the anti-HA antibody used for blotting. C and D, covalent interaction of EDEM3 with ERp46. EDEM3 co-expressed with ERp46 or ERp46 active-site mutants (CXXA and AXXA) was analyzed by Western blotting under reducing (C) and nonreducing (D) conditions. Open arrowheads, same species as in B. E–G, association of EDEM3 with ERp46 or its active-site mutants. Cells were transfected with EDEM3-HA and WT or redox-active-site mutants of ERp46 and immunoprecipitated with anti-HA antibody. Samples were separated by SDS-PAGE under reducing (E) and nonreducing (F and G) conditions. Open arrowheads, complexes covalently associated with EDEM3; gray arrows and bracket, EDEM3-containing complexes covalently associated with WT or CXXA mutant of ERp46. *, signals nonspecifically detected by anti-HA and anti-FLAG antibodies; **, native Igs used for the precipitation of EDEM3-HA.
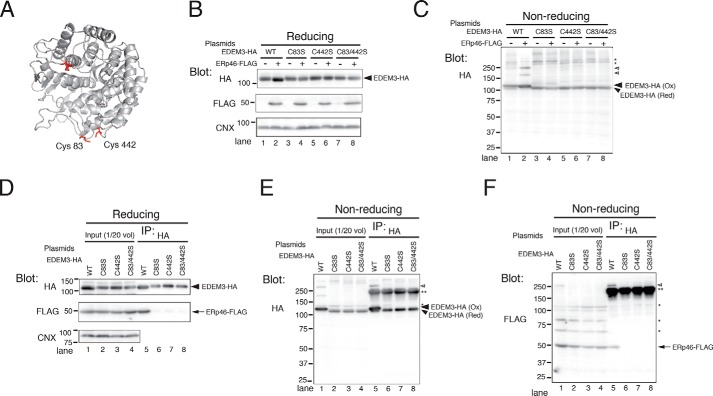
Figure 3.
ERp46 covalently associates with the mannosidase domain of EDEM3. A, ribbon model of EDEM3 mannosidase domain. The 3D structure of EDEM3 mannosidase domain (Mus musculus) was modeled based on the crystallographic structure of human ERM (Protein Data Bank code 1FMI) using SWISS-MODEL. Four Cys residues (Cys83, Cys226, Cys229, and Cys442) in the mannosidase domain are highlighted in red. B and C, redox states of EDEM3 and its Cys mutants. Cell lysates were analyzed by Western blotting under reducing (B) and nonreducing (C) conditions. Open arrowheads, complexes covalently associated with EDEM3; *, signals nonspecifically detected by the anti-HA antibody. D–F, Cys residues in the EDEM3 mannosidase domain are required for the covalent association with ERp46. EDEM3 and its Cys mutants were immunoprecipitated and electrophoresed under reducing (D) and nonreducing (E and F) conditions. Open arrowhead, complexes covalently associated with EDEM3; **, native Igs used to pull down EDEM3-HA.
ERp46 has three thioredoxin-like domains, all of which contain CGHC (Cys-Gly-His-Cys) redox-active sequences. To determine whether the oxidoreductase activity of ERp46 is required to alter the redox state of EDEM3, we co-expressed ERp46 mutants with EDEM3 and analyzed them by Western blotting (Fig. 2, C and D). The CXXA mutant contains three CGHA motifs in place of the WT CGHC motifs. We assumed that this mutant lacked the isomerase activity (i.e. the ability to exchange disulfides) (32) so that the intermolecular disulfide bond cannot be resolved, thereby trapping the substrates in disulfide-bonded complexes (33). Remarkably, the ERp46 CXXA mutant trapped all of the EDEM3 protein into high-molecular weight complexes, resulting in the disappearance of EDEM3 monomer (Fig. 2D, lane 4). On the other hand, the ERp46 AXXA mutant, in which all three redox-active sites were replaced by AGHA, did not affect the redox state of EDEM3, similar to what was observed in cells expressing only EDEM3 (Fig. 2D, compare lane 5 with lane 2). Collectively, these results suggest that ERp46 causes a specific change in the redox state of EDEM3 via its oxidoreductase activity.
Oxidoreductase activity of ERp46 is required for association with EDEM3
To determine whether the oxidoreductase activity of ERp46 is required for its interaction with EDEM3, we co-expressed EDEM3 and WT ERp46 or its redox-active site mutants. WT ERp46 and CXXA mutant co-immunoprecipitated with EDEM3, whereas the ERp46 AXXA mutant did not (Fig. 2E, lanes 5–8). In cells co-expressing the CXXA mutant, all of the EDEM3 formed disulfide-bonded complexes, which were detected by SDS-PAGE under nonreducing conditions (Fig. 2F, lane 7, open arrowheads). In WT ERp46-expressing cells, similar disulfide-bonded complexes were detected in addition to the EDEM3 monomer (Fig. 2F, lane 6), indicating that the interaction of ERp46 with EDEM3 depends on the redox-active sites of ERp46. ERp46 was present in these covalently associated complexes (Fig. 2G, gray arrows). These results suggest that ERp46 associates with EDEM3 by forming disulfide bonds through the redox-active sites.
ERp46 covalently associates with the EDEM3 α-mannosidase domain
Mammalian EDEM3 has an α-mannosidase domain at the N terminus that contains four Cys residues (Fig. S4). A structural model of the EDEM3 α-mannosidase domain based on the crystallographic structure of human ERManI suggests that two Cys residues, Cys83 and Cys442, are close to each other (Fig. 3A). Because recombinant EDEM3 α-mannosidase domain purified from bacteria can adopt either the oxidized or reduced conformation, depending on the redox conditions,3 we predicted that Cys83 and Cys442 form a disulfide bond in the ER. As expected, all three EDEM3 mutants in which Ser replaced Cys83 and/or Cys442 (C83S, C442S, and C83S/C442S) adopted only the reduced conformation (Fig. 3, B and C), even in cells co-expressing ERp46. The high-molecular weight disulfide-bonded complexes of EDEM3 were present only in cells expressing WT EDEM3 (Fig. 3C, open arrowheads). The stability of EDEM3 C83S/C442S mutant was similar to that of WT EDEM3 (Fig. 4A (IP: HA) and Fig. S6), suggesting that these mutations did not cause gross misfolding.
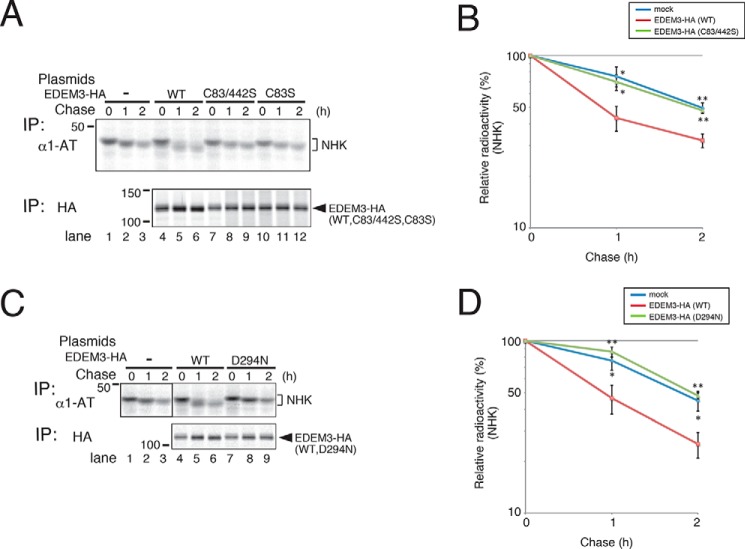
Figure 4.
EDEM3 mutants lacking mannose-trimming activity inhibit ERAD of misfolded NHK. NHK degradation was inhibited by the co-expression of EDEM3 Cys mutants (A, quantified in B) and the D294N mutant (C, quantified in D). HEK293 cells transfected with NHK and EDEM3 mutants were pulse-labeled for 15 min and chased for the indicated periods. Error bars, S.D. of three independent experiments. *, p < 0.05; **, p < 0.01 (two-tailed Student's t test, compared with WT EDEM3-transfected cells).
We next analyzed the association of ERp46 with the Cys mutants in the EDEM3 α-mannosidase domain. ERp46 co-immunoprecipitated only with WT EDEM3 (Fig. 3D, lane 5), indicating that both Cys83 and Cys442 are required for the interaction. As in Fig. 3C, the covalent complex was detected only in lysates from WT EDEM3-expressing cells resolved under nonreducing conditions (Fig. 3, E and F, open arrowheads). These data suggest that ERp46 forms a disulfide bond between Cys83 and Cys442 in the α-mannosidase domain of EDEM3.
EDEM3 Cys mutants lack mannose-trimming activity and inhibit ERAD of misfolded glycoproteins
To examine the effect of Cys mutation on the mannose-trimming and ERAD-enhancing activities of EDEM3, we transfected HEK293 cells with terminally misfolded glycoprotein NHK, a variant of α1-antitrypsin. Mobility shift of NHK on SDS-PAGE revealed that demannosylation activity was strongly promoted by co-expression of WT EDEM3 (Fig. 4A, compare lanes 4–6 with lanes 1–3) (16). However, co-transfection of the EDEM3 Cys mutants described above did not alter the electrophoretic mobility of NHK as the WT did (Fig. 4A, compare lanes 7–12 with lanes 1–3). Measurements of the rate of disappearance of NHK from cells (Fig. 4A, quantified in B) revealed that the EDEM3 Cys mutations abolished the ERAD-promoting activity of EDEM3. In addition, the D294N mutant of EDEM3, which corresponds to a mutation in proton donor Asp-463 of human ERManI, exhibited no mannose-processing or ERAD-promoting activity (Fig. 4, C and D). Collectively, these findings show that Cys83 and Cys442 in the EDEM3 α-mannosidase domain are necessary for the mannose-trimming and subsequent ERAD-promoting activity of EDEM3.
ERp46 triggers the mannose-trimming activity of EDEM3 in vitro
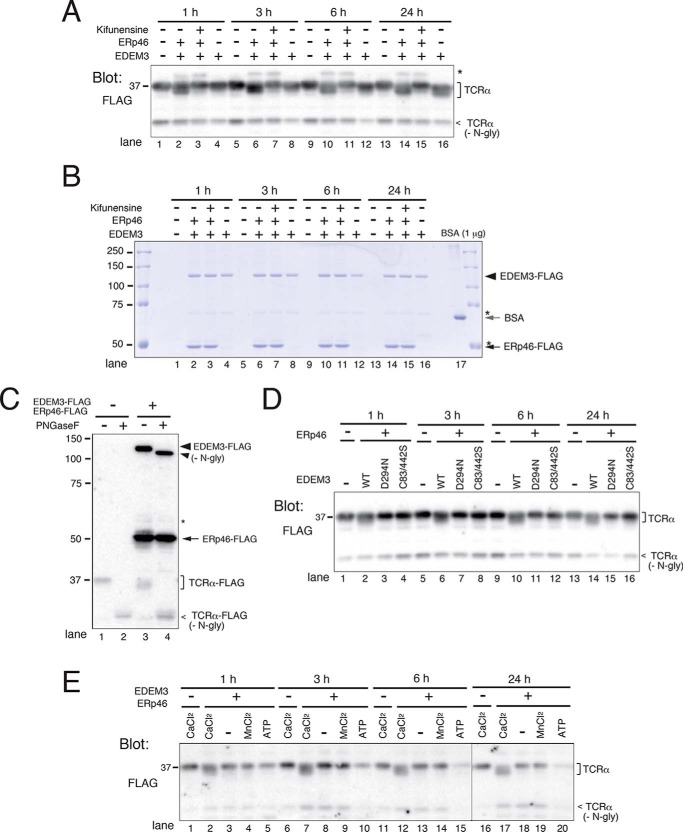
To clarify the molecular mechanism by which ERp46 promotes the mannose-trimming activity of EDEM3, we monitored mannose removal from misfolded glycoprotein TCRα in vitro. TCRα is a glycoprotein with four N-glycans that is degraded rapidly by ERAD when expressed in cells. FLAG-tagged TCRα was purified from HEK293 cells using FLAG-agarose beads followed by FLAG-peptide elution. To purify recombinant EDEM3 and ERp46, EDEM3-FLAG was expressed in HEK293 cells with or without ERp46-FLAG. After incubating the purified proteins at 37 °C, mannose trimming of TCRα was monitored by Western blot analysis (Fig. 5A). When EDEM3 was co-purified with ERp46, mannose trimming from TCRα was clearly detected even after 1 h of incubation (Fig. 5A, lane 2), whereas in samples containing only EDEM3, demannosylation was only faintly detected even after 24 h (Fig. 5A, lane 13). The addition of kifunensine, an inhibitor of processing α1,2-mannosidases, completely inhibited the mannose removal from TCRα (Fig. 5A, Kifunensine +). CBB staining of the recombinant proteins revealed that the amounts of EDEM3 and ERp46 purified were ∼2.5 and ∼3.5 μg, respectively, corresponding to final concentrations of ∼15 and ∼20 μg/ml, respectively (Fig. 5B). TCRα migrated at the same position after removal of the N-glycans by PNGase F treatment (Fig. 5C, compare lane 2 with lane 4), indicating that the electrophoretic mobility shift of TCRα was the result of N-glycan trimming.
Figure 5.
ERp46 triggers mannose-trimming activity of EDEM3 in vitro. A, mannose-trimming activity of EDEM3 on TCRα in vitro. FLAG-tagged proteins were expressed in HEK293 cells, and recombinant proteins were purified from cell lysates using FLAG-agarose beads. TCRα was incubated with EDEM3 or EDEM3 and ERp46 (EDEM3 +, ERp46 +) for the indicated periods and analyzed by Western blotting. Kifunensine (5 μg/ml) was added as indicated. *, signal nonspecifically detected by the anti-FLAG antibody used for blotting. B, CBB staining of the recombinant EDEM3 and ERp46 proteins analyzed in A. The top part of the gel used for the Western blotting in A was stained with CBB. C, PNGase F treatment of TCRα. TCRα was incubated in vitro in the presence or absence of EDEM3 and ERp46 for 24 h and then digested with PNGase F. Samples were separated by SDS-PAGE and analyzed by Western blotting using anti-FLAG antibody. D, EDEM3 D294N and C83S/C442S mutants lack mannose-trimming activity in vitro. HEK293 cells were co-transfected with ERp46 and WT or mutant EDEM3. Mannose trimming from TCRα was analyzed by Western blotting. E, Ca2+ is required for the EDEM3 mannose-trimming activity in vitro. TCRα was incubated with EDEM3 and ERp46 in vitro in the presence or absence of CaCl2 (5 mm), MnCl2 (0.1 mm), and ATP (10 mm).
Next, we analyzed the mannose-trimming activity of EDEM3 D294N and C83S/C442S mutants in vitro. This analysis confirmed that the mutants lacked enzyme activity even in the presence of ERp46 (Fig. 5D). We then tested the requirement for divalent cations and ATP in vitro (Fig. 5E). Ca2+ was necessary for the mannosidase activity of EDEM3, whereas Mn2+ was not. ATP did not affect the mannose-trimming activity of EDEM3. It should be noted that decrease of TCRα signal in Fig. 5E was not reproduced in other experiments, suggesting that the observed loss was caused by a contamination of nonspecific ATP-dependent or regulatable proteases. The amounts of recombinant proteins in all reactions were monitored by CBB staining (Fig. S7).
Covalent association of ERp46 with EDEM3 is required for the mannose-trimming activity of EDEM3
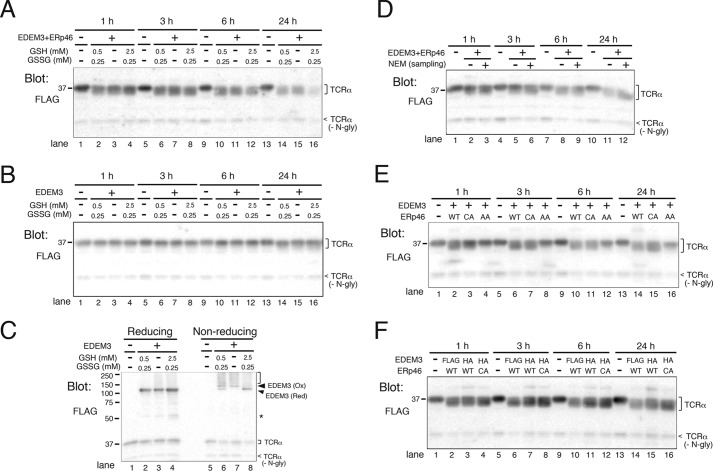
We next investigated how redox conditions influence EDEM3 mannosidase activity in vitro. For this purpose, we co-purified EDEM3 and ERp46 and incubated them in three redox buffers containing various concentrations of GSH and GSSG, a physiological condition that mimicked the normal redox environment in the ER ([GSH]2/[GSSG] = 1.0), a reducing condition ([GSH]2/[GSSG] = 25), and an oxidizing milieu (no GSH or GSSG) (34). Unexpectedly, mannose trimming proceeded effectively under all redox conditions examined (Fig. 6A).
Figure 6.
EDEM3-ERp46 complex trims mannose from TCRα in vitro. A and B, GSH buffers had no effects on the mannose-trimming activity of EDEM3 co-expressed with ERp46 (A) or of EDEM3 in the absence of co-expressed ERp46 (B). GSH and GSSG were added at the indicated concentrations and incubated for the indicated periods. C, redox states of EDEM3 incubated under different redox conditions for 24 h (same samples as in B, lanes 13–16). D, alkylation of EDEM3 and ERp46 does not affect the mannose-trimming activity of EDEM3. HEK293 cells co-expressing EDEM3-FLAG and ERp46-FLAG were lysed in a buffer supplemented with (+) or without (−) the alkylating agent NEM (2 mm). Recombinant proteins were incubated with TCRα in vitro for the indicated periods and analyzed by Western blotting. E, covalent interaction with ERp46 is required for the mannose-trimming activity of EDEM3. HEK293 cells were transfected with EDEM3-FLAG and WT or redox-active site mutants of ERp46, and both EDEM3 and ERp46 were purified from lysates using FLAG-agarose beads. GSH/GSSG was not added to the reaction in vitro. Mannose trimming from TCRα was analyzed by Western blotting. F, EDEM3-ERp46 complexes enhance mannose trimming from TCRα in vitro. WT and CXXA active-site mutant of ERp46 were co-transfected with EDEM3-FLAG or EDEM3-HA, and recombinant proteins were purified using FLAG-agarose beads.
Because ERp46 alters the redox state of EDEM3 (Fig. 2B), we investigated whether EDEM3 purified without co-expressed ERp46 can process mannose by simply changing the redox environment in vitro. For this purpose, EDEM3-FLAG was purified and incubated in the redox buffers described above. However, mannose trimming proceeded minimally under all three conditions (Fig. 6B), although the Ox/Red ratio of EDEM3 changed dependent on the redox buffer used (Fig. 6C, Non-reducing). These results strongly imply that the mannose-trimming activity of EDEM3 requires the oxidoreductase activity of ERp46.
ERp46 is thought to function on nascent polypeptides (22, 35). Hence, we investigated whether ERp46 acts on the mature EDEM3 in vitro, thereby turning on its mannosidase activity of EDEM3. To this end, we co-purified EDEM3 with ERp46 from cell lysates in the presence or absence of the alkylating agent NEM (Fig. 6D). Unexpectedly, NEM did not affect the demannosylation activity of EDEM3, suggesting that post-translational disulfide bond exchange between ERp46 and EDEM3 and/or within EDEM3 is not required for the mannose-trimming activity of EDEM3.
To analyze the role of redox-active sites of ERp46 on the demannosylation activity of EDEM3 in vitro, we co-purified EDEM3 with WT or mutant ERp46. As expected, ERp46 AXXA mutant did not promote mannose removal from TCRα (Fig. 6E). However, to our surprise, the ERp46 CXXA mutant stimulated the mannose-trimming activity of EDEM3 as effectively as WT ERp46 (Fig. 6E). The result of CXXA mutant, which formed stable disulfide-bonded complexes with EDEM3, suggested the requirement of covalent interaction of EDEM3 with ERp46 for the expression of enzyme activity. Next, to isolate the EDEM3-ERp46 complex, HA-tagged EDEM3 was co-transfected with WT or the CXXA mutant of ERp46-FLAG, and ERp46-FLAG–associated EDEM3-HA was pulled down with anti-FLAG beads. Mannose was efficiently trimmed from TCRα by EDEM3 complexes containing WT ERp46 or the CXXA mutant (Fig. 6F). The amounts of recombinant proteins in the reactions were confirmed by CBB staining (Fig. S8). Taken together, these results strongly suggest that the covalent interaction of ERp46 with EDEM3 triggers mannose trimming of EDEM3.
Discussion
In this study, we showed that ERp46 covalently associates with EDEM3 through its redox-active sites and that this interaction triggers the mannose-trimming activity of EDEM3. Among the PDI family of proteins, relatively little is known about the function of ERp46. Besides its abilities to interact with and reduce peroxiredoxin 4 (36) and to oxidize regulatory disulfides of Ero1α, resulting in its inactivation (37), ERp46 can introduce disulfide bonds into newly synthesized proteins at an early stage; these bonds are subsequently edited by PDI (35, 38). We observed that the intermolecular disulfide bonds between EDEM3 and ERp46 were stable under our experimental conditions and that EDEM3 did not interact with other members of the PDI family (Figs. 1A and 2B). Importantly, the mannose-trimming activity of EDEM3 was detected only when the covalent dimer was formed (Fig. 6). Analyses of a set of Cys mutants revealed that the disulfide bond was formed between the redox-active sites of ERp46 and the Cys residues in the EDEM3 α-mannosidase domain (Fig. 3).
To elucidate the mechanism by which EDEM3 processes mannose, we purified recombinant proteins expressed in HEK293 cells and monitored their mannose-trimming activity in vitro. As the substrate, we used TCRα purified from HEK293 cells. TCRα contains four N-glycans and is rapidly degraded by ERAD when expressed without its other subunits (39). Mannose trimming from TCRα in vitro is readily detected by the mobility shift on 10% SDS-PAGE (Fig. 5A). Because its N-glycans were removed by the cytosolic PNGase of HEK293 cells during purification in the absence of the alkylating agent NEM (40) (Fig. S9), TCRα was probably purified in a denatured conformation such that the Asn-attachment portion of the core N-glycans was exposed. The shift was not observed when N-glycans were removed by PNGase F (Fig. 5C). Further, the mobility shift disappeared in the presence of kifunensine, a specific inhibitor of α1,2-mannosidase (Fig. 5A). In addition, loss of enzyme activity upon mutation of the predicted proton donor Asp-294 (Fig. 5D), along with the requirement of Ca2+ ion for activity (Fig. 5E), indicated that the assay properly reflects the in vivo reaction. Overall, the in vitro demannosylation activity was insensitive to the redox conditions, which are buffered by GSH/GSSG (Fig. 6, A and B). Indeed, the disulfide bond exchange reaction in vitro was probably dispensable for trimming (Fig. 6D). Instead, the results strongly suggest that EDEM3 must form a mixed disulfide bond with ERp46 (Figs. 6 (E and F) and 7 (2)). We consider two underlying mechanisms for this unique regulation of sugar processing: 1) covalent interaction with ERp46 changes the conformation of the EDEM3 α-mannosidase domain, and/or 2) ERp46 is required for recognition of the substrate by EDEM3. In either case, the EDEM3-ERp46 redox cycle is likely to be regulated by another disulfide acceptor protein(s) (Fig. 7, yellow), which may adjust mannose trimming of cargo by EDEM3.
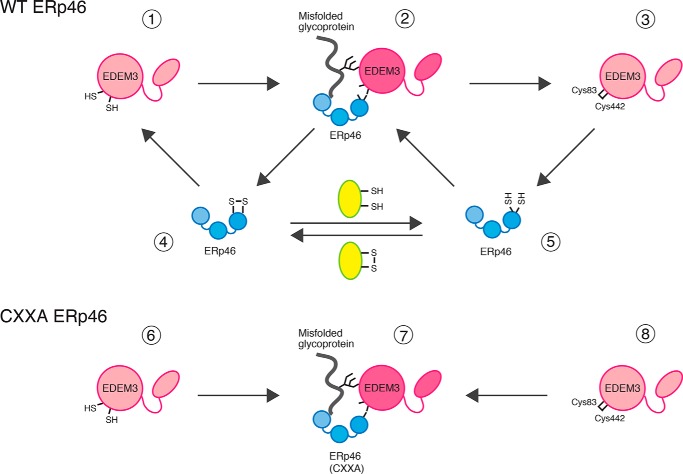
Figure 7.
Model: Redox cycle of the EDEM3-ERp46 complex, which regulates the mannose-trimming activity of EDEM3. ERp46 (4 or 5) binds to EDEM3 monomer (1 or 3) to form an intermolecular disulfide bridge between its redox-active sites and Cys83/Cys442 of EDEM3 (2) by transferring a disulfide bond to the reduced active site Cys. Covalent interaction of ERp46 with EDEM3 induces structural changes in the EDEM3 mannosidase domain, which triggers mannose-trimming activity and/or provides the scaffold for the substrate recognition. The cycle could be facilitated by another acceptor protein (yellow oval), regulating the demannosylation activity of EDEM3. In cells overexpressing CXXA mutant of ERp46 (bottom), disulfide bond of the complex (7) cannot be transferred owing to the lack of isomerase activity, leading to the depletion of EDEM3 monomer (6 or 8). ERp46 has three redox-active sites, but only one Cys pair is shown in the cartoon.
The observed interaction between EDEM3 and ERp46 appeared to be stable and long-lived unlike the transient interactions of oxidoreductases with nascent proteins. Particularly, dissociation was negligible in ERp46 CXXA mutant. On the other hand, WT ERp46 allowed dissociation to some extent so that about half of EDEM3 existed as monomer in the cells (Fig. 2D). It should be noted that the intermolecular disulfide bond between EDEM3 and ERp46 (Fig. 7, 2) needs to be resolved in vivo at an appropriate time by the action of another electron donor, such that the cargo proteins can be further processed by other machinery. Thus, the covalent complex could be reduced by another electron donor (Fig. 7, yellow), resulting in the formation of EDEM3 monomer (1 or 3) and ERp46 (4 or 5), terminating the mannosidase reaction. It is reasonable to assume that, under physiological conditions, the mannosidase activity of EDEM3 is strictly regulated by the cellular redox condition and is therefore elicited only when ERp46 renders an electron to the Cys83–Cys442 disulfide of EDEM3. This feature may protect against excessive disposal of glycoprotein cargo.
The mannosidase activity of yeast Htm1p/Mnl1p has been successfully measured in vitro (25–27). Htm1p/Mnl1p is purified in a complex with Pdi1p, which is required for mannose-trimming activity. This interaction is reminiscent of that between mammalian EDEM3 and ERp46. Htm1p/Mnl1p and Pdi1p associate in part by forming a disulfide bridge (30), also similar to the association between EDEM3 and ERp46. In yeast, however, Pdi1p forms a mixed disulfide with Cys in the C-terminal region of Htm1p/Mnl1p, followed by disulfide bond formation in the mannosidase domain, which is reported to be essential for ERAD activity (30). Although the corresponding Cys residues are mostly conserved (Fig. S4), our results suggest that ERp46 forms a covalent bridge directly with the Cys residues in the mannosidase domain of EDEM3. Consistent with this, oxidation of the mannosidase domain per se did not induce EDEM3 enzyme activity (Fig. 6, B and C).
Collectively, our results reveal that proper regulation of EDEM3 mannosidase activity requires association with, and redox activity of, ERp46. Because the enzyme activity of EDEM3 plays an important role in glycoprotein ERAD (6, 16, 21), we predict that other redox regulators, such as peroxiredoxin 4, Ero1α, and other oxidoreductases in the ER, might affect glycoprotein quality control through interactions with ERp46 as well as external redox alteration.
Experimental procedures
Cell culture and transfection
HEK293 cells were cultured in DMEM supplemented with 10% fetal bovine serum and antibiotics (penicillin, 100 units/ml; streptomycin, 100 μg/ml). Plasmids were purified using the Maxi-prep plasmid purification kit (Qiagen) and transfected into cells using polyethyleneimine (branched; Sigma-Aldrich) or Lipofectamine 2000 (Invitrogen).
Construction of plasmids
EDEM3-FLAG was constructed by PCR-amplifying the coding region of mouse EDEM3 and then subcloning the amplicon into the BamHI-EcoRI site of the pCMV-Tag4A vector (Stratagene). EDEM3 mutants (C83S, C442S, C83S/C442S, and D294N) were constructed by in vitro site-directed mutagenesis using Pfu Turbo DNA polymerase (Agilent Technology). All resultant plasmids were confirmed by sequencing. Construction of EDEM3-HA and α1-antitrypsin mutant NHK was as described previously (14, 16). FLAG-tagged oxidoreductase (ERp46, PDI, and P5) and its redox-active site mutants were kindly provided by Dr. K. Araki (National Institute of Advanced Industrial Science and Technology, Tokyo, Japan) (41). TCRα-FLAG was a generous gift from Dr. R. Kopito (Stanford University, Stanford, CA) (39), and the Z variant of α1-antitrypsin was provided by Dr. K. Kokame (National Cerebral and Cardiovascular Center, Suita, Japan).
Antibodies
Antibodies were purchased from the following sources: mouse monoclonal anti-HA (HA-7) (Sigma-Aldrich), rabbit polyclonal anti-HA (Recenttec), mouse anti-FLAG (M2) (Sigma-Aldrich), rabbit anti-FLAG (Sigma-Aldrich), rabbit anti-α1-antitrypsin (DAKO), goat anti-ERp46 (Santa Cruz Biotechnology), rabbit anti-EDEM3 (Sigma-Aldrich), rabbit anti-calnexin (Enzo Life Sciences), mouse monoclonal anti-BiP (BD Transduction Laboratories). Horseradish peroxidase (HRP)–conjugated secondary antibodies used for Western blotting were anti-rabbit IgG (BTI), anti-mouse IgG (Zymed Laboratories Inc.), and anti-goat IgG (Jackson Immunoresearch).
Silver staining and MS analysis
HEK293 cells were plated on 10-cm dishes and transfected with EDEM3-FLAG. Cells were lysed in a buffer containing 1% Nonidet P-40, 150 mm NaCl, and 50 mm Tris-HCl (pH 7.6), supplemented with protease inhibitors (2 mm N-ethylmaleimide, 0.2 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml pepstatin). After centrifugation at 13,000 × g for 20 min at 4 °C, FLAG-agarose was added, and the sample was rotated overnight at 4 °C. After the agarose beads were washed twice with a buffer containing 1% Nonidet P-40, 400 mm NaCl, and 50 mm Tris-HCl and once with a buffer used for cell extraction, immunoprecipitated proteins were eluted with FLAG peptide. The eluates were separated by 10% SDS-PAGE, followed by staining with the PlusOne silver staining kit (GE Healthcare). After the gels were washed several times in H2O, the bands of interest were cut out. The proteins were digested and recovered using the In-gel tryptic digestion kit (Thermo Fisher Scientific), separated on a Nano-LC-Ultra 2D-plus system equipped with cHiPLC Nanoflex (Eksigent) in trap-and-elute mode, and then analyzed on a TripleTOF5600+ system (SCIEX) in information-dependent acquisition method. Proteins were identified using ProteinPilot software version 4.5beta (SCIEX) with the UniProtKB/Swiss-Prot database (Homo sapiens, June 2014) appended with the known contaminant database (SCIEX). Protein identifications were evaluated based on Unused ProtScores and the number of identified peptides with confidence ≥95%, as determined by the ProteinPilot software.
Western blotting and immunoprecipitation followed by Western blotting
Western blotting was performed as described previously (42). To separate proteins under nonreducing conditions, cell lysates were prepared in Laemmli buffer without the addition of the reducing reagent DTT. To block disulfide bond exchange during cell lysis, cells were incubated in PBS containing 10 mm iodoacetamide for 5 min before cell lysis. Western blotting signals were detected using Pierce Western blotting substrate (Thermo Scientific) and visualized on a LAS-4000 digital imaging system (GE Healthcare). Signal intensity was quantified using the ImageQuant software (GE Healthcare). For immunoprecipitation followed by Western blotting, cell lysates were mixed with specific antibodies and incubated at 4 °C for several hours to overnight. After the addition of Protein A- or Protein G-Sepharose, cell lysates were rotated at 4 °C for several additional hours, for a total incubation period of 12–15 h. After the Sepharose beads were washed as described for FLAG-agarose, the immunoprecipitates were eluted with Laemmli buffer. The eluates were separated by SDS-PAGE and blotted onto a polyvinylidine difluoride membrane (Millipore). For signal detection by Western blotting, Clean-BlotTM IP detection reagent (HRP) was used as the secondary antibody (Thermo Fisher Scientific).
Metabolic labeling and immunoprecipitation
Metabolic labeling pulse-chase experiments were performed as described previously (42). Immunoprecipitation was performed as described above. Gels were dried and exposed to imaging plates (Fuji Film/GE Healthcare), and signals were visualized on a Typhoon PhosphorImager (GE Healthcare). Incorporation of radioactive materials was quantified using ImageQuant software (GE Healthcare).
Expression and purification of recombinant proteins from HEK293 cells
HEK293 cells plated on 10-cm dishes were transfected with FLAG-tagged plasmids using polyethyleneimine. Transfection medium was removed after 6 h, and cells were incubated for an additional 24–28 h in fresh growth medium. Cells were lysed in buffer containing 1% Nonidet P-40, 150 mm NaCl, and 20 mm MES (pH 7.0) (43), supplemented with protease inhibitors as described above. For preparation of EDEM3 in the presence or absence of co-transfected ERp46, NEM was excluded unless otherwise specified. Cell extracts were centrifuged at 13,000 × g for 20 min at 4 °C, FLAG-agarose (Sigma-Aldrich) was added, and the sample was rotated for 12 h at 4 °C. After the beads were washed twice with MES buffer containing 400 mm NaCl and twice again with the same buffer used for cell extraction, FLAG-tagged proteins were eluted with FLAG peptide.
In vitro mannose-trimming assay
Recombinant proteins prepared as described above were mixed and supplemented with GSH/GSSG to a final concentration of 0.5 mm/0.25 mm, along with 5 mm CaCl2 unless stated otherwise. Aliquots ( volume) were collected after rotation at 37 °C for the indicated times and separated by 10% SDS-PAGE. Gels were cut below the 50 kDa marker, and the lower part was blotted onto a polyvinylidine difluoride membrane, followed by detection of TCRα-FLAG using rabbit anti-FLAG antibody and Clean-BlotTM IP detection reagent (HRP). The other part of the gel was stained with CBB to estimate the amount of recombinant EDEM3 and ERp46 in the reaction.
Author contributions
S. Y., S. I., and N. H. data curation; S. Y., I. W., and N. H. formal analysis; S. Y., S. I., and N. H. investigation; S. Y. and N. H. writing-original draft; S. I. and N. H. methodology; I. W. and N. H. conceptualization; I. W. and N. H. supervision; I. W. and N. H. validation; I. W. and N. H. writing-review and editing; N. H. funding acquisition; N. H. project administration.
Supplementary Material
Acknowledgments
We thank Dr. K. Araki (National Institute of Advanced Industrial Science and Technology, Tokyo, Japan) for the generous gift of the plasmid encoding ERp46-FLAG and its redox-active site mutants (CXXA and AXXA), PDI-FLAG, and P5-FLAG. We thank Dr. M. Yamasaki (Ryukoku University, Otsu, Japan) for the modeling of the EDEM3 mannosidase domain.
This work was supported by a Joint Research grant from the Institute for Molecular Science (IMS); Scientific Research C of KAKENHI from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and the Mizutani Foundation for Glycosciences (to N. H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S9.
I. Wada and N. Hosokawa, unpublished observations.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- ERManI
- ER mannosidase I
- EDEM
- ER degradation–enhancing α-mannosidase–like protein
- M9
- Man9GlcNAc2
- M8B
- Man8GlcNAc2 isomer B
- Man7A
- Man7GlcNAc2 isomer A
- M5
- Man5GlcNAc2
- M6
- Man6GlcNAc2
- PDI
- protein-disulfide isomerase
- ERp46
- ER-resident protein 46
- Ox
- oxidized
- Red
- reduced
- CBB
- Coomassie Brilliant Blue
- IP
- immunoprecipitation
- HRP
- horseradish peroxidase
- NEM
- N-ethylmaleimide
- PNGase F
- peptide:N-glycosidase F.
References
- 1. Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., and Hartl F. U. (2013) Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- 2. Braakman I., and Hebert D. N. (2013) Protein folding in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5, a013201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hegde R. S., and Ploegh H. L. (2010) Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 22, 437–446 10.1016/j.ceb.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aebi M., Bernasconi R., Clerc S., and Molinari M. (2010) N-Glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 35, 74–82 10.1016/j.tibs.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 5. Helenius A., and Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 10.1146/annurev.biochem.73.011303.073752 [DOI] [PubMed] [Google Scholar]
- 6. Caramelo J. J., and Parodi A. J. (2015) A sweet code for glycoprotein folding. FEBS Lett. 589, 3379–3387 10.1016/j.febslet.2015.07.021 [DOI] [PubMed] [Google Scholar]
- 7. Preston G. M., and Brodsky J. L. (2017) The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem. J. 474, 445–469 10.1042/BCJ20160582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith M. H., Ploegh H. L., and Weissman J. S. (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334, 1086–1090 10.1126/science.1209235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hampton R. Y., and Sommer T. (2012) Finding the will and the way of ERAD substrate retrotranslocation. Curr. Opin. Cell Biol. 24, 460–466 10.1016/j.ceb.2012.05.010 [DOI] [PubMed] [Google Scholar]
- 10. Berner N., Reutter K. R., and Wolf D. H. (2018) Protein quality control of the endoplasmic reticulum and ubiquitin-proteasome-triggered degradation of aberrant proteins: yeast pioneers the path. Annu. Rev. Biochem. 10.1146/annurev-biochem-062917-012749 [DOI] [PubMed] [Google Scholar]
- 11. Xie W., and Ng D. T. (2010) ERAD substrate recognition in budding yeast. Semin. Cell Dev. Biol. 21, 533–539 10.1016/j.semcdb.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 12. Helenius A. (1994) How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol. Biol. Cell 5, 253–265 10.1091/mbc.5.3.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hebert D. N., and Molinari M. (2012) Flagging and docking: dual roles for N-glycans in protein quality control and cellular proteostasis. Trends Biochem. Sci. 37, 404–410 10.1016/j.tibs.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., and Nagata K. (2001) A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2, 415–422 10.1093/embo-reports/kve084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mast S. W., Diekman K., Karaveg K., Davis A., Sifers R. N., and Moremen K. W. (2005) Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology 15, 421–436 10.1093/glycob/cwi014 [DOI] [PubMed] [Google Scholar]
- 16. Hirao K., Natsuka Y., Tamura T., Wada I., Morito D., Natsuka S., Romero P., Sleno B., Tremblay L. O., Herscovics A., Nagata K., and Hosokawa N. (2006) EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 281, 9650–9658 10.1074/jbc.M512191200 [DOI] [PubMed] [Google Scholar]
- 17. Ninagawa S., Okada T., Sumitomo Y., Kamiya Y., Kato K., Horimoto S., Ishikawa T., Takeda S., Sakuma T., Yamamoto T., and Mori K. (2014) EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J. Cell Biol. 206, 347–356 10.1083/jcb.201404075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benyair R., Ogen-Shtern N., Mazkereth N., Shai B., Ehrlich M., and Lederkremer G. Z. (2015) Mammalian ER mannosidase I resides in quality control vesicles, where it encounters its glycoprotein substrates. Mol. Biol. Cell 26, 172–184 10.1091/mbc.e14-06-1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Benyair R., Ogen-Shtern N., and Lederkremer G. Z. (2015) Glycan regulation of ER-associated degradation through compartmentalization. Semin. Cell Dev. Biol. 41, 99–109 10.1016/j.semcdb.2014.11.006 [DOI] [PubMed] [Google Scholar]
- 20. Kanehara K., Kawaguchi S., and Ng D. T. (2007) The EDEM and Yos9p families of lectin-like ERAD factors. Semin. Cell Dev. Biol. 18, 743–750 10.1016/j.semcdb.2007.09.007 [DOI] [PubMed] [Google Scholar]
- 21. Hosokawa N., Kamiya Y., and Kato K. (2010) The role of MRH domain-containing lectins in ERAD. Glycobiology 20, 651–660 10.1093/glycob/cwq013 [DOI] [PubMed] [Google Scholar]
- 22. Appenzeller-Herzog C., and Ellgaard L. (2008) The human PDI family: versatility packed into a single fold. Biochim. Biophys. Acta 1783, 535–548 10.1016/j.bbamcr.2007.11.010 [DOI] [PubMed] [Google Scholar]
- 23. Hatahet F., and Ruddock L. W. (2009) Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 11, 2807–2850 10.1089/ars.2009.2466 [DOI] [PubMed] [Google Scholar]
- 24. Benham A. M. (2012) The protein disulfide isomerase family: key players in health and disease. Antioxid. Redox Signal. 16, 781–789 10.1089/ars.2011.4439 [DOI] [PubMed] [Google Scholar]
- 25. Gauss R., Kanehara K., Carvalho P., Ng D. T., and Aebi M. (2011) A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol. Cell 42, 782–793 10.1016/j.molcel.2011.04.027 [DOI] [PubMed] [Google Scholar]
- 26. Liu Y. C., Fujimori D. G., and Weissman J. S. (2016) Htm1p-Pdi1p is a folding-sensitive mannosidase that marks N-glycoproteins for ER-associated protein degradation. Proc. Natl. Acad. Sci. U.S.A. 113, E4015–E4024 10.1073/pnas.1608795113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pfeiffer A., Stephanowitz H., Krause E., Volkwein C., Hirsch C., Jarosch E., and Sommer T. (2016) A complex of Htm1 and the oxidoreductase Pdi1 accelerates degradation of misfolded glycoproteins. J. Biol. Chem. 291, 12195–12207 10.1074/jbc.M115.703256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kozlov G., Määttänen P., Thomas D. Y., and Gehring K. (2010) A structural overview of the PDI family of proteins. FEBS J. 277, 3924–3936 10.1111/j.1742-4658.2010.07793.x [DOI] [PubMed] [Google Scholar]
- 29. Clerc S., Hirsch C., Oggier D. M., Deprez P., Jakob C., Sommer T., and Aebi M. (2009) Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 184, 159–172 10.1083/jcb.200809198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sakoh-Nakatogawa M., Nishikawa S., and Endo T. (2009) Roles of protein-disulfide isomerase-mediated disulfide bond formation of yeast Mnl1p in endoplasmic reticulum-associated degradation. J. Biol. Chem. 284, 11815–11825 10.1074/jbc.M900813200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fujimori T., Kamiya Y., Nagata K., Kato K., and Hosokawa N. (2013) Endoplasmic reticulum lectin XTP3-B inhibits endoplasmic reticulum-associated degradation of a misfolded α1-antitrypsin variant. FEBS J. 280, 1563–1575 10.1111/febs.12157 [DOI] [PubMed] [Google Scholar]
- 32. Schwaller M., Wilkinson B., and Gilbert H. F. (2003) Reduction-reoxidation cycles contribute to catalysis of disulfide isomerization by protein-disulfide isomerase. J. Biol. Chem. 278, 7154–7159 10.1074/jbc.M211036200 [DOI] [PubMed] [Google Scholar]
- 33. Zito E., Melo E. P., Yang Y., Wahlander Å., Neubert T. A., and Ron D. (2010) Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol. Cell 40, 787–797 10.1016/j.molcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hwang C., Sinskey A. J., and Lodish H. F. (1992) Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496–1502 10.1126/science.1523409 [DOI] [PubMed] [Google Scholar]
- 35. Kojima R., Okumura M., Masui S., Kanemura S., Inoue M., Saiki M., Yamaguchi H., Hikima T., Suzuki M., Akiyama S., and Inaba K. (2014) Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family. Structure 22, 431–443 10.1016/j.str.2013.12.013 [DOI] [PubMed] [Google Scholar]
- 36. Tavender T. J., Springate J. J., and Bulleid N. J. (2010) Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 29, 4185–4197 10.1038/emboj.2010.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shepherd C., Oka O. B., and Bulleid N. J. (2014) Inactivation of mammalian Ero1α is catalysed by specific protein disulfide-isomerases. Biochem. J. 461, 107–113 10.1042/BJ20140234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sato Y., Kojima R., Okumura M., Hagiwara M., Masui S., Maegawa K., Saiki M., Horibe T., Suzuki M., and Inaba K. (2013) Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci. Rep. 3, 2456 10.1038/srep02456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu H., Kaung G., Kobayashi S., and Kopito R. R. (1997) Cytosolic degradation of T-cell receptor α chains by the proteasome. J. Biol. Chem. 272, 20800–20804 10.1074/jbc.272.33.20800 [DOI] [PubMed] [Google Scholar]
- 40. Hirsch C., Blom D., and Ploegh H. L. (2003) A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22, 1036–1046 10.1093/emboj/cdg107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Araki K., Iemura S., Kamiya Y., Ron D., Kato K., Natsume T., and Nagata K. (2013) Ero1-α and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J. Cell Biol. 202, 861–874 10.1083/jcb.201303027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fujimori T., Suno R., Iemura S. I., Natsume T., Wada I., and Hosokawa N. (2017) Endoplasmic reticulum proteins SDF2 and SDF2L1 act as components of the BiP chaperone cycle to prevent protein aggregation. Genes Cells 22, 684–698 10.1111/gtc.12506 [DOI] [PubMed] [Google Scholar]
- 43. Aikawa J., Matsuo I., and Ito Y. (2012) In vitro mannose trimming property of human ER α-1,2 mannosidase I. Glycoconj. J. 29, 35–45 10.1007/s10719-011-9362-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.