Abstract
The bacterial species Streptomyces avermitilis is an important industrial producer of avermectins, which are widely utilized as effective anthelmintic and insecticidal drugs. We used gene deletion, complementation, and overexpression experiments to identify SAV4189, a MarR-family transcriptional regulator (MFR) in this species, as an activator of avermectin biosynthesis. SAV4189 indirectly stimulated avermectin production by altering expression of cluster-situated activator gene aveR, and directly repressed the transcription of its own gene (sav_4189) and adjacent cotranscribed gene sav_4190 (which encodes an unknown transmembrane efflux protein). A consensus 13-bp palindromic sequence, 5′-TTGCCYKHRSCAA-3′ (Y = T/C; K = T/G; H = A/C/T; R = A/G; S = C/G), was found within the SAV4189-binding sites of its own promoter region, and shown to be essential for binding. The SAV4189 regulon was thus predicted based on bioinformatic analysis. Night new identified SAV4189 targets are involved in transcriptional regulation, primary metabolism, secondary metabolism, and stress response, reflecting a pleiotropic role of SAV4189. sav_4190, the important target gene of SAV4189, exerted a negative effect on avermectin production. sav_4189 overexpression and sav_4190 deletion in S. avermitilis wild-type and industrial strains significantly increased avermectin production. SAV4189 homologs are widespread in other Streptomyces species. sav_4189 overexpression in the model species S. coelicolor also enhanced antibiotic production. The strategy of increasing yield of important antibiotics by engineering of SAV4189 homologs and target gene may potentially be extended to other industrial Streptomyces species. In addition, SAV4189 bound and responded to exogenous antibiotics hygromycin B and thiostrepton to modulate its DNA-binding activity and transcription of target genes. SAV4189 is the first reported exogenous antibiotic receptor among Streptomyces MFRs.
Keywords: Streptomyces avermitilis, avermectins, MarR-family regulator, SAV4189, sav_4190
Introduction
Streptomyces are filamentous soil bacteria characterized by complex life cycle and production of numerous antibiotics (van Wezel and McDowall, 2011). Antibiotic biosynthetic gene clusters (BGCs) are tightly controlled by cluster-situated regulators (CSRs) and higher-level pleiotropic regulators that respond to environmental and physiological signals (van Wezel and McDowall, 2011; Liu G. et al., 2013; Urem et al., 2016). However, the signals and their transduction systems that control expression of specific BGCs are poorly understood, because genome sequencing has shown that Streptomyces species contain numerous BGCs, some of them are not expressed under laboratory conditions and called cryptic BCGs. Activation of cryptic BGCs provides an important source for discovery of new compounds (Rutledge and Challis, 2015). Identification and manipulation of transcriptional regulators involved in antibiotic biosynthesis will facilitate discovery of new antibiotics and construction of antibiotic-overproducing strains for industrial applications.
At least 20 families of transcriptional regulators are found in Streptomyces, including TetR, LuxR, LysR, AraC, GntR, SARP, ROK, MerR, and MarR (multiple antibiotic resistance regulator). MarR-family transcriptional regulators (MFRs) comprise more than 19,000 members and are widespread in bacteria and archaea. MFRs control a variety of cellular processes, including multidrug resistance, pathogenicity, stress responses, metabolic pathways, and degradation of aromatic compounds (Perera and Grove, 2010; Romero-Rodríguez et al., 2015). They have characteristic winged helix-turn-helix (wHTH) DNA-binding domains and typically act as homodimeric transcriptional repressors (sometimes as activators, or both) by binding to palindromic sequences within target promoter regions. DNA-binding affinity of MFRs is reduced by conformational changes in response to small-molecule ligands, resulting in blocking of transcriptional regulation, but the natural ligands are often unknown (Perera and Grove, 2010; Grove, 2013). MFRs are the fourth most abundant family of transcriptional regulators in Streptomyces (average 50 per genome), but only a few members have been characterized in this genus, including Streptomyces coelicolor OhrR (Oh et al., 2007), TamR (Huang and Grove, 2013), PecS (Huang et al., 2013), and PcaV (Davis et al., 2013), S. exfoliatus PenR, S. treptomyces arenae PntR (Zhu et al., 2013), and S. roseosporus DptR3 (Zhang et al., 2015).
Avermectins produced by S. avermitilis are economically important, potent anthelmintic and insecticidal drugs widely utilized in human medicine, animal husbandry, and agriculture (Burg et al., 1979; Egerton et al., 1979). There are eight avermectin components (A1a, A1b, A2a, A2b, B1a, B1b, B2a, B2b), among which B1a has the highest insecticidal activity. The regulatory mechanisms of avermectin biosynthesis have been intensively studied. The 82-kb ave gene cluster contains only one CSR gene, aveR, which encodes a LAL family activator required for expression of ave biosynthetic genes (Kitani et al., 2009; Guo et al., 2010). Our recent studies have shown that PhoP (response regulator of two-component system PhoR/PhoP) (Yang et al., 2015), AvaR1 (avenolide autoregulator receptor) (Zhu et al., 2017), and AvaR2 (homolog of pseudo γ-butyrolactone receptor) (Zhu et al., 2016) are direct repressors of aveR, and that SA742 (AraC-family regulator) directly represses several ave structural genes, rather than aveR (Sun et al., 2016). Transcriptional regulators shown to indirectly control avermectin production include TetR-family regulators AveI (SAV4110) (Chen et al., 2008), SAV7471 (Liu Y. et al., 2013), SAV151 (He et al., 2014), SAV576 (Guo et al., 2013), SAV577 (Guo et al., 2014), and AveT (SAV3619) (Liu et al., 2015), ECF-σ25 (Luo et al., 2014), and alternative σ factor σ8 (Sun et al., 2017). Despite these findings, our knowledge of the complex regulatory network underlying avermectin biosynthesis is still limited and fragmentary, and presents an obstacle to rational design of avermectin high-producing strains through genetic manipulation.
Forty-two MFRs are encoded by the S. avermitilis genome, but none of them have been characterized. In a search for new regulators that affect avermectin biosynthesis, we previously applied a S. avermitilis whole-genome chip to compare transcriptomes of wild-type strain and avermectin high-producer 76–02-e (Guo et al., 2013). We observed that transcription level of sav_4189, which encodes a MFR, was greatly upregulated in 76–02-e, suggesting that SAV4189 is involved in control of avermectin biosynthesis. In the present study, we examined the regulatory role of SAV4189, and found that it functions as an indirect activator of avermectin production. An effective strategy was developed for enhancing avermectin yield in industrial strains through overexpression of SAV4189 and deletion of its target gene, sav_4190. Exogenous antibiotics hygromycin B (HygB) and thiostrepton (Thi), produced by other Streptomyces species, released SAV4189 from its target genes. Thus, SAV4189 appears to mediate interspecies communication in Streptomyces by responding to antibiotic signals.
Materials and Methods
Strains, Plasmids, and Growth Conditions
Plasmids and bacterial strains used/constructed in this work are summarized in Table 1, and primers are listed in Supplementary Table S1. Culture conditions for S. avermitilis and Escherichia coli were as described previously (Liu et al., 2015). YMS (Ikeda et al., 1988), SFM (Kieser et al., 2000), MM (Kieser et al., 2000), and RM14 (MacNeil and Klapko, 1987) agar were used for observation of S. avermitilis phenotype. S. coelicolor M145 was grown at 28°C and used as a heterogeneous host for overexpression of SAV4189. YBP agar (Ou et al., 2009) was used for observation of S. coelicolor phenotype. For routine avermectin production, insoluble fermentation medium FM-I was used (Chen et al., 2007). For growth analysis, soluble fermentation medium FM-II was used (Guo et al., 2010).
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Description | Source |
|---|---|---|
| S. avermitilis | ||
| ATCC31267 | Wild-type (WT) strain | Laboratory stock |
| A-144 | Industrial strain | Qilu Pharmaceutical |
| D4189 | sav_4189 deletion mutant | This study |
| C4189 | sav_4189 complemented strain | This study |
| O4189 | sav_4189 overexpression strain | This study |
| O4189/A-144 | sav_4189 overexpression strain based on A-144 | This study |
| D4190 | sav_4190 deletion mutant | This study |
| D4190/A-144 | sav_4190 deletion mutant based on A-144 | This study |
| S. coelicolor | ||
| M145 | WT strain | Laboratory stock |
| O4189/M145 | sav_4189 overexpression strain based on M145 | This study |
| E. coli | ||
| JM109 | Routine cloning host | Laboratory stock |
| ET12567 | Methylation-deficient strain | MacNeil and Klapko, 1987 |
| BL21(DE) | Host for His6-SAV4189 overexpression | Novagen |
| Plasmids | ||
| pKC1139 | Multiple-copy, temperature-sensitive E. coli–Streptomyces shuttle plasmid | Bierman et al., 1992 |
| pSET152 | Integrative E. coli–Streptomyces shuttle plasmid | Bierman et al., 1992 |
| pET-28a (+) | Plasmid for His6-tagged protein overexpression in E. coli | Novagen |
| pMD18-T | TA cloning vector | TaKaRa |
| pJL117 | Plasmid carrying Streptomyces strong constitutive promoter ermE∗p | Li et al., 2010 |
| pD4189 | pKC1139 derivative for sav_4189 deletion | This study |
| pKC1139-4189 | pKC1139 derivative for sav_4189 overexpression | This study |
| pSET152-4189 | pSET152 derivative for sav_4189 complementation | This study |
| pD4190 | pKC1139 derivative for sav_4190 deletion | This study |
| pET28-4189 | pET-28a (+) derivative for His6-SAV4189 overexpression | This study |
Gene Disruption, Complementation, and Overexpression
To construct a sav_4189 deletion mutant, two flanking fragments of sav_4189 were prepared by PCR using genomic DNA of wild-type (WT) strain ATCC31267 as template. A 511-bp 5′ flanking region (from positions -314 to +197 relative to the sav_4189 start codon) was amplified with primers ZX141 and ZX142, and a 522-bp 3′ flanking region (from positions +502 to +1023) was amplified with primers ZX143 and ZX144. The two fragments were fused by PCR using primers ZX141 and ZX144, and ligated into pKC1139 to generate sav_4189 deletion vector pD4189, which was transformed into WT protoplasts. sav_4189-deleted mutant D4189 was selected by the method described previously (Zhao et al., 2007), confirmed by PCR analysis using primers ZX235, ZX236, ZX237, and ZX238 (Supplementary Figure S1A), and subjected to DNA sequencing. When using primers ZX235/ZX236 (flanking the exchange regions), a 1.4-kb band was observed in D4189, whereas a 1.7-kb band was produced from WT genomic DNA. When using primers ZX237/ZX238 (located within the deletion region), only WT produced a 0.4-kb PCR fragment as predicted (data not shown).
For complementation of D4189, a 607-bp DNA fragment carrying the sav_4189 ORF was obtained by PCR with primers ZX149 and ZX150, and ligated simultaneously with a 264-bp fragment containing promoter ermE∗p from pJL117 into pSET152 to generate sav_4189-complemented vector pSET152-4189, which was then introduced into D4189 to obtain complemented strain C4189. The two fragments containing sav_4189 ORF and ermE∗p were ligated simultaneously into pKC1139 to produce sav_4189 overexpression vector pKC1139-4189, which was then introduced into S. avermitilis WT and industrial strain A-144 to obtain sav_4189 overexpression strains O4189 and O4189/A-144, respectively. pKC1139-4189 was transformed into S. coelicolor M145 to obtain O4189/M145.
To construct a sav_4190 deletion mutant, a 585-bp 5′ flanking region (from positions -481 to +104 relative to the sav_4190 start codon), and a 575-bp 3′ flanking region (from positions +2446 to +3020) were amplified with primer pairs ZX177/ZX178 and ZX179/ZX180, respectively. The two fragments were ligated into pKC1139 to obtain sav_4190 deletion vector pD4190, which was transformed into WT protoplasts. The resulting sav_4190-deleted mutant D4190 was confirmed by PCR using ZX239, ZX240, ZX241, and ZX236 as primers (Supplementary Figure S1B). When using primers ZX239/ZX240 (flanking the exchange regions), a 1.5-kb band appeared in D4190, whereas a 3.8-kb band was produced from WT. When using primers ZX241/ZX236 (located within the deletion region), only WT produced a 0.5-kb PCR fragment as predicted (data not shown). To delete sav_4190 in A-144, the vector pD4190 was transformed into A-144 protoplasts. The expected mutant, termed D4190/A-144, was isolated by selection of the D4190 mutant and confirmed by PCR using the same primer pairs.
Analysis of Avermectin Production
Avermectins were extracted by methanol from S. avermitilis fermentation cultures, and analyzed by HPLC as described previously (Chen et al., 2007).
Scanning Electron Microscopy (SEM)
Spores and mycelia of S. avermitilis WT and D4189 strains grown on SFM agar for 4 days were observed by SEM. Specimens were prepared and examined as described previously (Sun et al., 2016).
Quantitative Real-Time RT-PCR (qRT-PCR) Analysis
Total RNAs were isolated at various times from S. avermitilis cultures grown in FM-I, and qRT-PCR was conducted as described previously (Luo et al., 2014) to determine transcription levels of tested genes using primers listed in Supplementary Table S1. hrdB gene (sav_2444, encoding the principal sigma factor) was used as internal control for normalization of transcription levels. Gene expression was determined in triplicate and repeated for three independent samples.
Overexpression and Purification of His6- SAV4189
The 516-bp sav_4189 coding region (171 amino acids) was amplified by PCR with primers ZX205 and ZX206. The PCR fragment was digested with NdeI/EcoRI and inserted into expression vector pET-28a (+) to generate pET28-4189, which was verified by DNA sequencing. pET28-4189 was transformed into E. coli BL21 (DE3), and recombinant His6-SAV4189 protein overexpression was induced by 0.2 mM IPTG for 8 h at 16°C. Cells were collected, washed, and disrupted in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) by sonication on ice. The preparation was centrifuged, and the supernatant containing soluble His6-SAV4189 was loaded onto a Ni2+-NTA column (Qiagen). After extensive washing with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 50 mM imidazole, pH 8.0), the His6-SAV4189 protein was specifically eluted from the resin with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 200 mM imidazole, pH 8.0). Concentrations of purified proteins were determined by Bradford assay, and proteins were stored at -80°C.
Electrophoretic Mobility Shift Assays (EMSAs)
Electrophoretic mobility shift assays were performed as described previously (Luo et al., 2014). DNA probes harboring promoter regions of tested genes were obtained by PCR using corresponding primers (Supplementary Table S1), labeled with non-radioactive digoxigenin (DIG) at the 3′ end, and incubated individually with various amounts of His6-SAV4189 in binding reaction. To verify specificity of SAV4189-probe 4189p interaction, a ∼100-fold excess of unlabeled specific probe 4189p or non-specific probe hrdB was added to the reaction mixture.
For EMSAs with antibiotics, avermectin B1 (AveB1), oligomycin (Oli), and thiostrepton (Thi) were dissolved in DMSO, while hygromycin B (HygB), apramycin (Apr), and kanamycin (Kan) were dissolved in deionized water. Dissolved antibiotics were added at various final concentrations, and solvent control DMSO was added at 8 μl in the reaction mixture.
Determination of Transcriptional Start Site (TSS)
5′ rapid amplification of cDNA ends (5′ RACE) analysis was performed to identify the sav_4189 TSS using a 5′/3′ RACE kit (Roche). Total RNA (2 μg) prepared from 84-h culture of WT grown on YMS was used for reverse transcription with 20 pmol of gene-specific primer 4189SP1. An oligo(dA) tail was added to the 3′ end of purified cDNA using terminal deoxynucleotidyltransferase, followed by PCR amplification of the tailed cDNA using specific nested primer 4189SP2 and oligo(dT) anchor primer. The resulting PCR product (diluted 1000-fold) was amplified in an additional round of PCR with nested primer 4189SP3 and an anchor primer, to yield a single specific band. The purified final PCR product was inserted into pMD18-T vector (TaKaRa) for sequencing. The first nucleotide following oligo(dA) sequence was determined as the complementary base of sav_4189 TSS.
DNase I Footprinting Assay
A non-radiochemical capillary electrophoresis technique (Zianni et al., 2006) was used for DNase I footprinting with some modifications (Guo et al., 2013). To investigate binding sites of SAV4189 on its own promoter region, a 511-bp 5′ FAM fluorescence-labeled DNA probe covering the entire sav_4188-sav_4189 intergenic region was synthesized by PCR using primers FAM-89footS/89footAS, and purified. The probe (400 ng) and various concentrations of His6-SAV4189 were mixed in a 25-μl volume, and incubated for 30 min at 25°C. Following DNase I digestion, DNA samples were purified and subjected to capillary electrophoresis with 3730XL DNA analyzer (Applied Biosystems). Results were processed with GeneMarker v2.2.
Results
SAV4189 Is an Activator of Avermectin Production
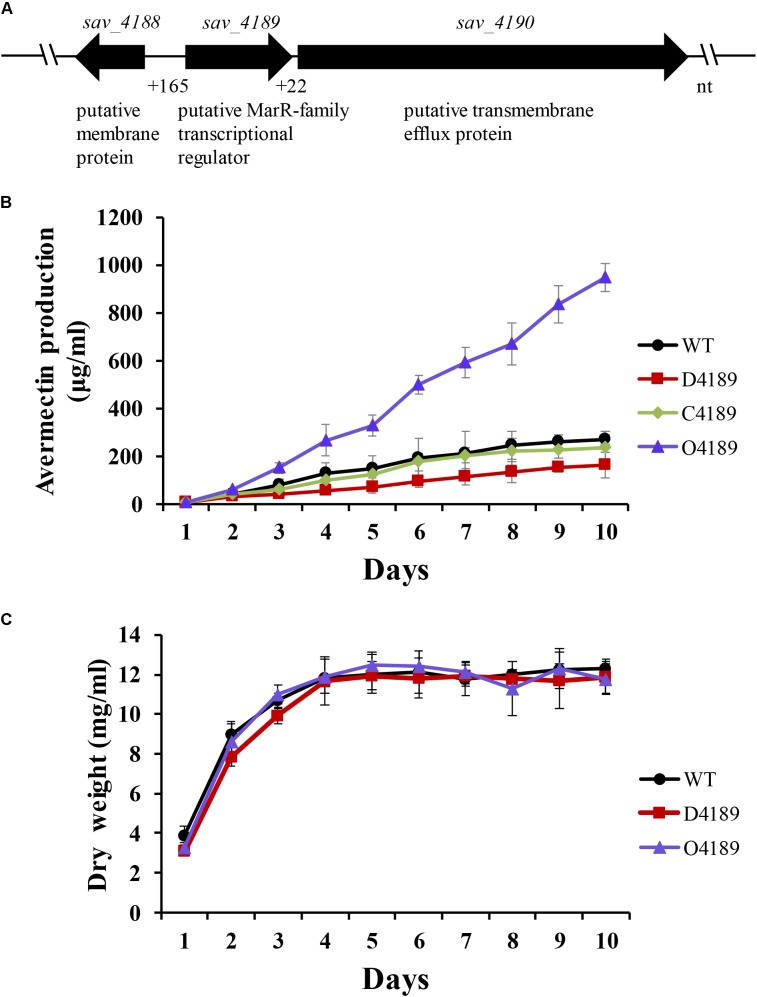
The sav_4189 gene contains 516 nucleotides (nt) and encodes a MFR of 171 amino acids (including a conserved wHTH DNA-binding motif homologous to MarR) whose function is unknown. Divergently transcribed gene sav_4188, located 165 nt upstream of sav_4189, encodes an unknown membrane protein. Convergently transcribed gene sav_4190, located downstream of sav_4189, encodes a putative transmembrane efflux protein (Figure 1A). The sav_4189-sav_4190 intergenic region in only 22 nt long, suggesting that the two genes might be cotranscribed, and this was confirmed by RT-PCR (Supplementary Figure S2). BLAST analysis revealed that the genetic organization of sav_4188, sav_4189 and sav_4190 homologous genes is conserved among different Streptomyces species and SAV4189 homologs have high amino acid sequence identities (72–80%) (Supplementary Figure S3), reflecting the important biological function of this MFR in the genus.
FIGURE 1.
Effects of SAV4189 on avermectin production and growth in Streptomyces avermitilis. (A) Organization of sav_4189 and its adjacent genes. (B) Avermectin yield in WT, sav_4189 deletion mutant (D4189), complemented strain (C4189), and overexpression strain (O4189) cultured in FM-I. Avermectin yield contains total yield of all eight components A1a, A1b, A2a, A2b, B1a, B1b, B2a, and B2b. NS, not significant; ∗∗P< 0.01 (comparison with WT by Student’s t-test). (C) Growth curves of WT, D4189, and O4189 cultured in FM-II. In this and subsequent figures, error bars represent SD of three replicates.
To clarify the role of sav_4189 in avermectin biosynthesis, we constructed sav_4189 deletion mutant D4189, complemented strain C4189, and overexpression strain O4189, and compared their total avermectin yields (from 1 to 10 days culture in FM-I) with that of WT. HPLC analysis showed that avermectin yield in WT strain was not observable until day 2, then increased gradually from day 2 onward (Figure 1B). Relative to WT level, yields were lower for D4189, higher for O4189, and not significantly different for C4189. On the end of fermentation day 10, overexpression of sav_4189 (strain O4189) increased avermectin yield by ∼2.5-fold, and deletion of sav_4189 (strain D4189) led to a clear reduction (∼40%) in avermectin yield (Figure 1B). These findings indicate that SAV4189 plays an activator role in avermectin production.
To evaluate the effect of SAV4189 on cell growth, we measured biomass (dry cell weight) of WT, D4189, and O4189 cultured in soluble FM-II. Cell weights of the three strains were similar (Figure 1C), indicating that SAV4189 is not involved in regulation of cell growth, and that the changed avermectin yields of D4189 and O4189 did not result from alteration of cell growth. D4189 and O4189 grew normally on YMS, SFM, RM14, and MM plates (Supplementary Figure S4A). In SEM observations, degree of separation of aerial hyphae, spore size, and spore shape for D4189 were nearly identical as for WT (Supplementary Figure S4B), indicating that SAV4189 has no effect on morphological development.
SAV4189 Activates aveR but Represses Its Own Gene and Adjacent Gene sav_4190
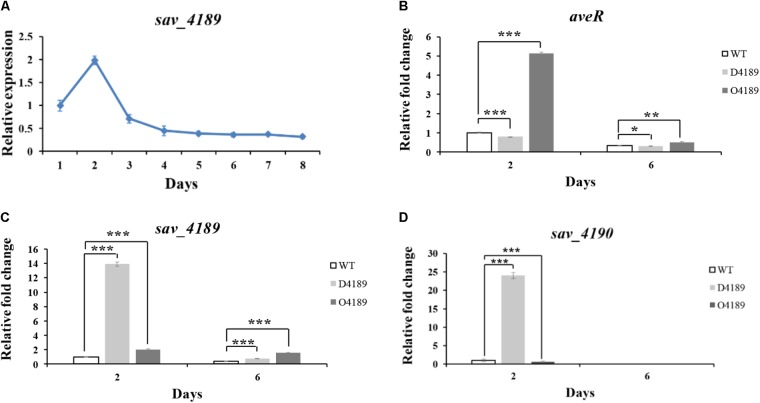
To elucidate the relationship between SAV4189 and avermectin production, we examined the sav_4189 transcription profile of WT grown in FM-I by qRT-PCR. sav_4189 transcription level varied during the avermectin production process; it reached a maximum on day 2 when avermectin production was initiated, then declined sharply, and remained low from day 4 onward (Figure 2A). Thus, SAV4189 evidently exerts its regulatory function mainly during early fermentation stage.
FIGURE 2.
Transcriptional analysis of aveR, sav_4189, and sav_4190 by qRT-PCR. (A) Transcriptional profile of sav_4189 during the avermectin fermentation process in WT grown in FM-I. Value of sav_4189 on day 1 was defined as 1. (B) aveR, (C) sav_4189, and (D) sav_4190 transcription levels in WT, D4189, and O4189 grown in FM-I. Value of each gene was calculated relative to WT value on day 2, defined as 1. sav_4189: 91-bp transcript amplified from remainder sav_4189 ORF in D4189 with primers ZX9 and ZX10. ∗P< 0.05, ∗∗P< 0.01, ∗∗∗P< 0.001.
To determine whether SAV4189 regulates avermectin production through CSR AveR, we performed qRT-PCR to examine aveR transcription levels of WT, D4189, and O4189 cultured in FM-I for 2 or 6 days. On both days, level relative to WT was reduced in D4189 and increased in O4189 (Figure 2B), consistently with avermectin yields. Notably, aveR transcription level was particularly high for O4189 on day 2. These findings indicate that SAV4189 regulates avermectin production by stimulating cluster-situated activator gene aveR, primarily during early fermentation stage.
A common regulatory mechanism of MFRs involves MarR protein regulating transcription of its own gene and a divergently oriented gene by binding to their intergenic region, which contains TSSs for both genes (Perera and Grove, 2010; Grove, 2013). sav_4189 and sav_4190 are cotranscribed. We therefore predicted that SAV4189 regulates expression of sav_4189-sav_4190 operon and adjacent divergently transcribed gene sav_4188. We measured transcription levels of these three genes using the same RNA samples as for examining aveR expression. No sav_4188 transcription was detectable in any of the three strains WT, D4189, and O4189, suggesting that this gene transcription was at very low level, or was not expressed under our fermentation conditions. We therefore did not investigate sav_4188 in subsequent experiments. In O4189, sav_4189 transcription level was higher than in WT, confirming sav_4189 overexpression (Figure 2C). sav_4190 transcription was slightly reduced on day 2, and undetectable on day 6 (Figure 2D). In D4189, transcription of both sav_4189 and sav_4190 was strongly upregulated on day 2. Transcription of sav_4189 was slightly increased, and that of sav_4190 was undetectable on day 6 (Figures 2C,D). These findings indicate that SAV4189 functions as a repressor of its own operon sav_4189-sav_4190, mainly during early fermentation stage.
SAV4189 Binds Specifically to Its Own Gene Promoter Region
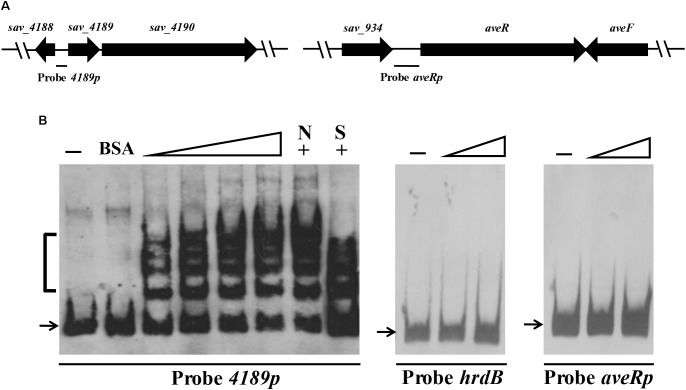
To determine whether SAV4189 directly regulates aveR and sav_4189-sav_4190 operon, we performed EMSAs using soluble His6-SAV4189 protein expressed in E. coli. The aveR and sav_4189 promoter regions were labeled as probes aveRp and 4189p, respectively (Figure 3A). Probe hrdB and BSA were used as negative probe and protein controls. His6-SAV4189 did not bind to probe aveRp or hrdB, but bound specifically to probe 4189p and generated several clearly retarded bands (Figure 3B), suggesting that SAV4189 binds more than two sites on its own promoter region. Binding specificity was confirmed by competition experiments using ∼100-fold excesses of unlabeled specific probe 4189p (lane S), which reduced binding of SAV4189 to labeled probe 4189p, and of non-specific probe hrdB (lane N), which did not reduce the retarded signals. BSA did not retard probe 4189p, even at high protein concentration (750 nM). These findings indicate that the positive regulatory effect of SAV4189 on aveR expression is indirect, and that SAV4189 directly regulates sav_4189-sav_4190 operon through binding to the sav_4189 promoter region.
FIGURE 3.
Electrophoretic mobility shift assays (EMSAs) of SAV4189 binding to its own promoter region. (A) Schematic diagram of probes used for EMSAs. Probe 4189p: 148-bp DNA fragment, positions –145 to +3 relative to sav_4189 translational start codon. Probe aveRp: 501-bp DNA fragment, positions –476 to +25 relative to aveR translational start codon. (B) Interaction of His6-SAV4189 with probes 4189p and aveRp. Negative probe: hrdB, 118-bp DNA fragment within hrdB ORF. Negative protein control: 750 nM BSA. 0.15 nM labeled probe was added in each reaction. Concentrations of His6-SAV4189 for probes: for 4189p, 12.5, 25, 50, and 62.5 nM; for aveRp and hrdB, 125 and 250 nM. 62.5 nM His6-SAV4189 was used for competition experiments (lanes +). Lanes –: EMSAs without His6-SAV4189. Lanes N and S: competition assays with ∼100-fold excess of unlabeled non-specific competitor DNA hrdB (N) and specific probe 4189p (S). Arrows: free probes. Bracket: SAV4189-DNA complex.
Determination of Precise SAV4189-Binding Sites
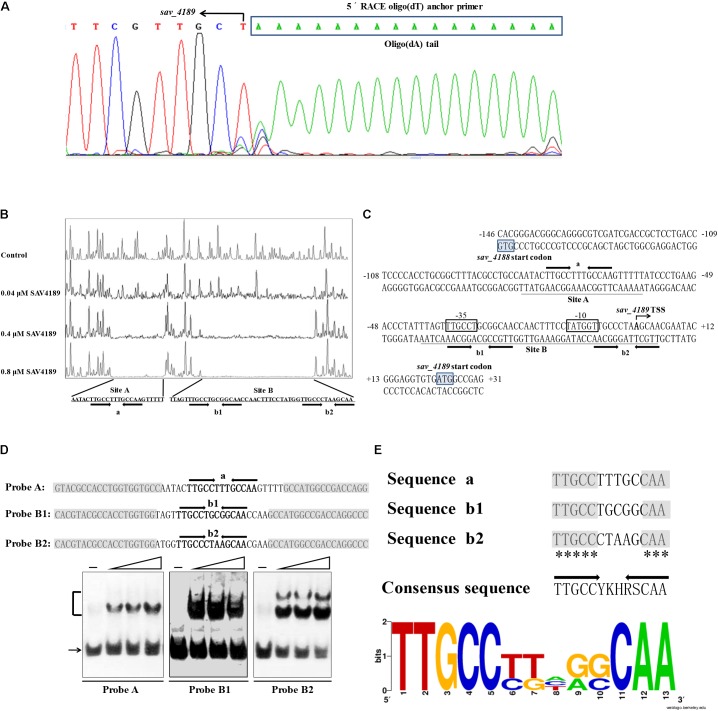
To identify precise binding sites and elucidate the regulatory mechanism of SAV4189 on its target promoter, we performed 5′ RACE and DNase I footprinting assays. The sav_4189 TSS was localized by 5′ RACE to an A residue at position 22 nt upstream of the sav_4189 translational start codon (Figures 4A,C), leading to the probable -10 and -35 promoter regions shown by boxes in Figure 4C.
FIGURE 4.
sav_4189 promoter structure and SAV4189-binding sites. (A) Determination of sav_4189 TSS by 5′-RACE PCR. Box: complementary sequence of oligo(dT) anchor primer. Arrow: complementary base of TSS. (B) DNase I footprinting assay of SAV4189 on its own promoter region. Top fluorogram: reaction with 10 μM BSA (control). Protection patterns were acquired with increasing His6-SAV4189 concentrations. (C) Nucleotide sequences of sav_4189 promoter region and SAV4189-binding sites. Numbers: distance (nt) from sav_4189 TSS. Shading: translational start codons. Bent arrow: sav_4189 TSS and transcription orientation. Boxes: potential –10 and –35 regions. Underlining: SAV4189-binding sites. Straight arrows: inverted repeats. (D) EMSAs of His6-SAV4189 with probes A, B1, and B2. Each lane contained 0.3 nM labeled probe. Each probe was 59 bp. Non-specific DNA sequences from hrdB ORF (shaded) were fused with intact 13-bp palindromic sequences a, b1, and b2 (boldfaced) to generate probes A, B1, and B2, respectively. Lanes –: EMSAs without His6-SAV4189. Lanes 2, 3, and 4: 62.5, 125, and 250 nM His6-4189. (E) Consensus sequence analysis of SAV4189-binding sites by WebLogo program. Asterisks: consensus bases. Arrows: inverted repeats. Height of each letter is proportional to appearance frequency of corresponding base.
DNase I footprinting assay with a 511-bp FAM-labeled probe covering the sav_4188-sav_4189 intergenic region revealed that SAV4189 protected two sites (termed A and B) on the sav_4189 coding strand (Figure 4B). The two sites are 17 nt apart. Site A extends from positions -82 to -59, and site B extends from positions -41 to +5, relative to the sav_4189 TSS (Figure 4C). Site A is close to, and site B overlaps the putative -35 and -10 regions of sav_4189 promoter, indicating that SAV4189 represses transcription of sav_4189-sav_4190 operon by hindering recruitment of RNA polymerase.
MarR-family transcriptional regulators generally form homodimers and bind palindromic sequences within target promoter regions (Perera and Grove, 2010; Grove, 2013). DNAMAN analysis of the SAV4189-binding sites revealed that site A contained one palindromic sequence (termed “a”), and site B contained two such sequences (termed “b1” and “b2”) (Figures 4B,C). The three palindromic sequences were all 13 bp (5-nt inverted repeats separated by 3 nt) and highly similar. To assess contributions of the three 13-bp sequences to SAV4189 binding, we performed EMSAs using 59-bp probes containing intact 13-bp sequences; i.e., both sides of sequences a, b1 and b2 were fused with non-specific DNA fragments from hrdB ORF by PCR to create probes A, B1 and B2, respectively (Figure 4D). SAV4189 bound to each of these probes, indicating that all three 13-bp sequences are important for SAV4189 binding, and that SAV4189 binds at the three palindromic sequences on its own promoter region. These findings are consistent with that of several retarded bands in EMSAs for interaction of probe 4189p with purified His6-SAV4189, as described in the preceding section.
WebLogo program1 analysis of these three palindromic sequences generated a 13-bp consensus SAV4189-binding sequence 5′-TTGCCYKHRSCAA-3′ (Y = T/C; K = T/G; H = A/C/T; R = A/G; S = C/G) (Figure 4E).
Affinity of SAV4189 Binding to Different Target Sequences
To compare affinity of SAV4189 for its three target sequences, we performed competitive EMSAs using labeled probes A, B1 and B2 that contained sequences a, b1 and b2, respectively, and ∼50-fold and 150-fold excesses of unlabeled specific competitor DNAs. In experiments using 50-fold excesses, unlabeled probe B1 dissociated less SAV4189 from labeled probe A than the other two unlabeled probes (Figure 5A). Further analysis using labeled probe B1 (Figure 5B) confirmed that affinity of SAV4189 to probe B1 was the lowest among the three probes. In experiments using 150-fold excesses, the dissociation of SAV4189 from labeled probe B2 caused by unlabeled probe B2 was more than that of the other two unlabeled probes (Figure 5C), indicating the highest affinity of SAV4189 for probe B2, which was confirmed by comparison of intensities of SAV4189-probe B2 complex corresponding to 50-fold excess of unlabeled probes (Figure 5C). The competitive ability of 150-fold excess of unlabeled probe A was higher than that of the same excess of unlabeled probe B1 (Figures 5B,C). Therefore, we determined the affinity of SAV4189 to its target sequences in the order b2 > a > b1; however, the differences are slight.
FIGURE 5.
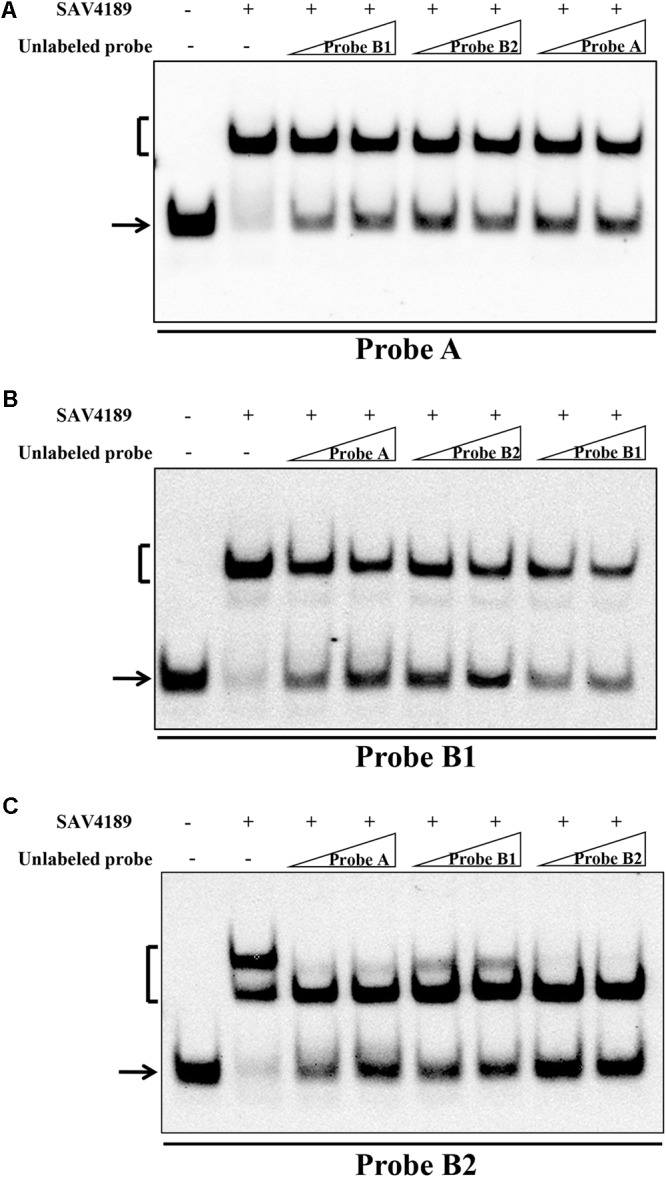
Comparison of the relative affinities of SAV4189 for different target sequences. (A) EMSA of His6-SAV4189 with labeled probe A and unlabeled probes (A, B1, B2). (B) EMSA of His6-SAV4189 with labeled probe B1 and unlabeled probes. (C) EMSA of His6-SAV4189 with labeled probe B2 and unlabeled probes. 0.15 nM labeled probe and ∼50- and 150-fold amounts of unlabeled specific competitor probe were used in competition assays. SAV4189 concentration: 125 nM. Arrows: free labeled probes. Brackets: SAV4189-DNA complexes.
Prediction of SAV4189 Regulon and Verification of New SAV4189 Target Genes
To decipher SAV4189 regulon in S. avermitilis, we used the 13 bp SAV4189 consensus binding sequence to scan the genome with the PREDetector software program2. We identified 250 putative SAV4189 target genes using cut-off score ≥ 7. Of these genes, 115 are unknown or unclassified (Supplementary Table S2), and the remaining 135 were assigned to 17 functional groups based on the KEGG pathway database for S. avermitilis3. 49 of these 135 genes are associated with regulatory functions. These findings reflect the degree of biological significance of SAV4189 in S. avermitilis.
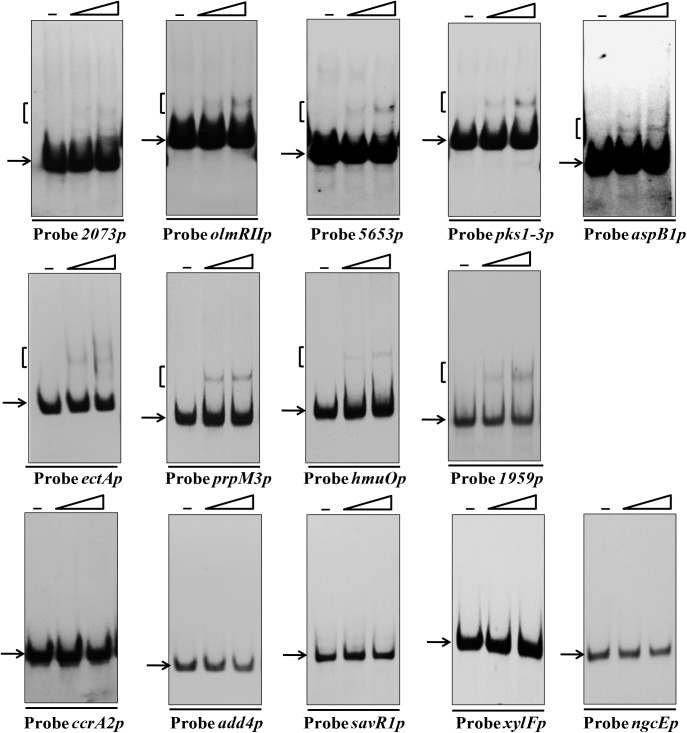
To test the accuracy of bioinformatic analysis, 14 putative target genes from various groups that have relatively high score or are well annotated were selected and confirmed experimentally by EMSAs. Among these, 9 targets were confirmed to bind directly to SAV4189 (Figure 6). The newly identified SAV4189 target genes were sav_2073 (encoding a putative GntR-family transcriptional regulator), olmRII (sav_2901, encoding a LuxR-family transcriptional regulator), sav_5653 (encoding a putative ROK-family transcriptional regulator), pks1-3 (sav_7362, encoding a putative modular polyketide synthase), aspB1 (sav_2008, encoding a putative aspartate aminotransferase), ectA (sav_6398, encoding a L-2,4-diaminobutyrate acetyltransferase), prpM3 (sav_3185, encoding a putative magnesium or manganese-dependent protein phosphatase), hmuO (sav_5930, encoding a putative heme oxygenase), and sav_1959 (encoding a putative secreted acyl esterase). SAV4189 did not interact with promoter regions of ccrA2 (sav_1911, encoding a putative crotonyl-CoA reductase), add4 (sav_4906, encoding a putative adenosine deaminase), savR1 (sav_3309, encoding a putative Mrr restriction system protein), xylF (sav_2247, encoding a putative simple sugar ABC transporter substrate-binding protein), and ngcE (sav_2251, encoding a putative ABC transporter substrate-binding protein).
FIGURE 6.
Confirmation of new SAV4189 target genes. EMSAs of His6-SAV4189 with promoter regions of 14 predicted target genes. Each lane contained 0.15 nM labeled probe. Lanes –, EMSAs without His6-SAV4189. Lanes 2–3 contained 200 and 400 nM His6-SAV4189, respectively.
sav_4189 Overexpression and sav_4190 Deletion Enhance Avermectin Yield in Industrial Strain A-144
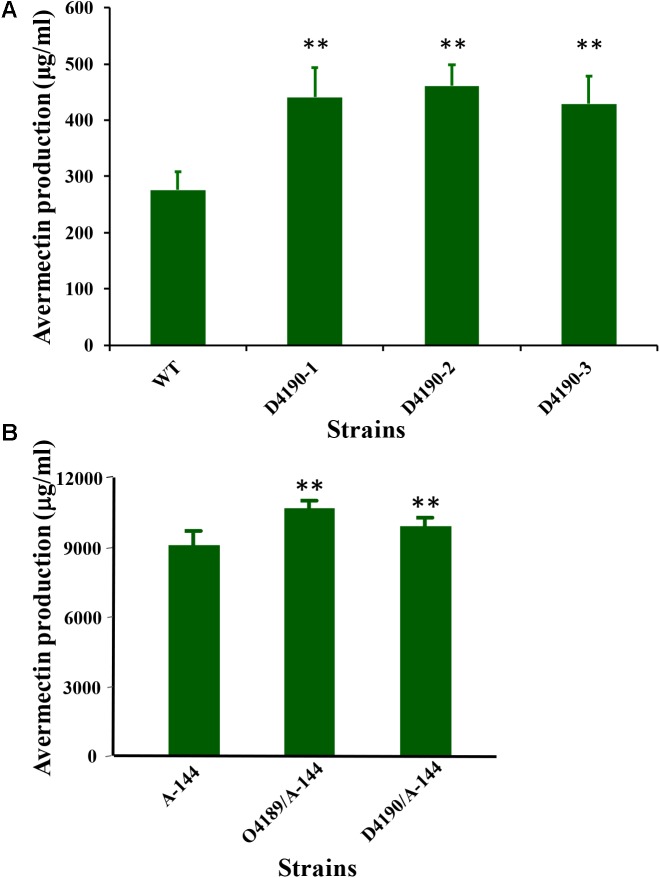
The findings that SAV4189 positively regulates avermectin production, and that sav_4190 is a target gene of SAV4189, suggest that sav_4190 has a functional role in avermectin production. To test this concept, we constructed sav_4190 deletion mutant D4190 (Supplementary Figure S1B). Avermectin yields were ∼56–67% higher in strains D4190-1, -2, and -3 than in WT (Figure 7A), indicating that sav_4190 plays a negative role in avermectin production. sav_4190 expression level on day 2 for D4189 grown in FM-I was ∼24-fold higher than that of WT (Figure 2B), indicating that altered sav_4190 expression contributes to the reduced avermectin yield of D4189.
FIGURE 7.
Avermectin yield in various S. avermitilis strains grown in FM-I for 10 days. (A) WT and sav_4190 deletion mutant strains D4190-1, -2, and -3. (B) Industrial strain A-144 and its derivatives (O4189/A-144: sav_4189 overexpression strain; D4190/A-144: sav_4190 deletion strain). ∗∗P< 0.01.
Because avermectin yield was strikingly increased by sav_4189 overexpression and by sav_4190 deletion, we experimentally engineered these two genes in industrial strain A-144 and evaluated effects on avermectin yield. sav_4189 overexpression vector pKC1139-4189 and sav_4190 deletion vector pD4189 were introduced into A-144 to construct mutants O4189/A-144 and D4190/A-144, respectively. Avermectin yield in shake-flask fermentation was ∼21% higher for O4189/A-144 and ∼15% higher for D4190/A-144 relative to parental A-144 (Figure 7B). sav_4189 overexpression and sav_4190 deletion thus appear to be effective strategies for further enhancement of avermectin production in industrial strains.
sav_4189 Overexpression in S. coelicolor Promotes Antibiotic Production
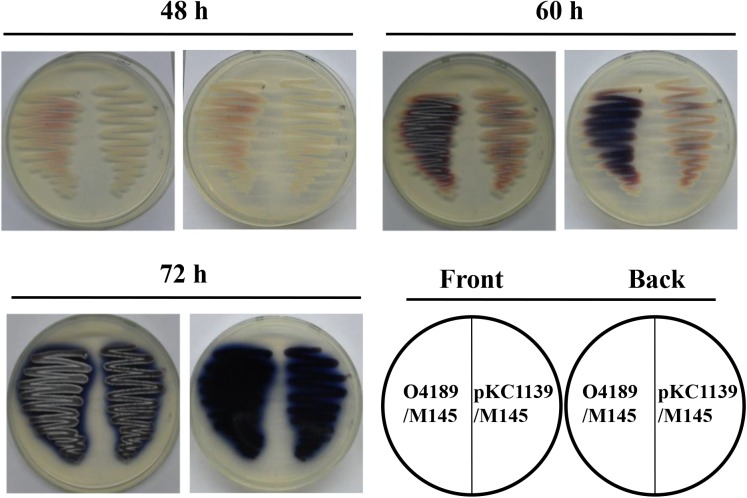
SAV4189 homologs are distributed widely in Streptomyces. To investigate possible regulation of antibiotic production by SAV4189 in other Streptomyces species, we transformed sav_4189 overexpression vector pKC1139-4189 into model strain S. coelicolor M145. Resulting transformant O4189/M145 was grown on YBP agar, and earlier, higher antibiotic production was visually observed (Figure 8). This finding suggests that SAV4189 (or its homolog) acts as an activator of antibiotic production in Streptomyces species other than S. avermitilis. Thus, a strategy for strain improvement based on engineering of SAV4189 and its target gene may be extended to other industrial Streptomyces strains having SAV4189 homologs, to promote antibiotic production.
FIGURE 8.
Effect of sav_4189 overexpression on antibiotic production in S. coelicolor M145. S. coelicolor strains were grown on YBP agar at 28°C. O4189/M145: sav_4189 overexpression strain of M145. pKC1139/M145: M145 carrying control plasmid pKC1139.
HygB and Thi Act as Ligands of SAV4189
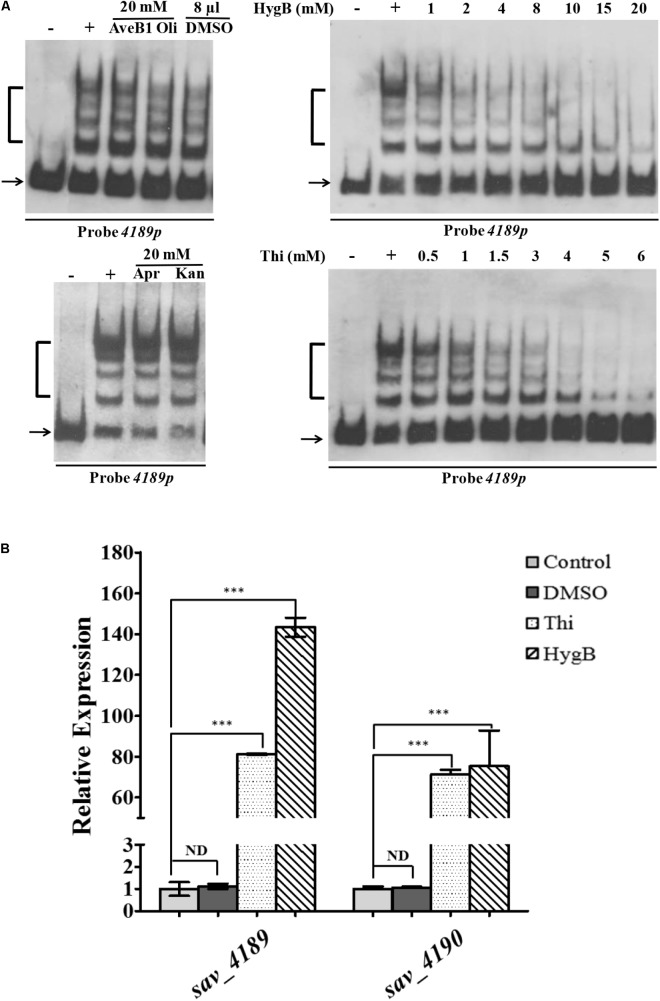
A defining characteristic of MFRs is their response to specific ligands, such as antibiotics and phenolic compounds, resulting in attenuated DNA-binding activity and derepressed gene expression (Perera and Grove, 2010; Grove, 2013). In view of the finding that SAV4189 regulates antibiotic production, we performed EMSAs to evaluate effects of various antibiotics on SAV4189 affinity for probe 4189p. SAV4189 DNA-binding activity was not inhibited by endogenous antibiotics Oli and AveB1, nor by exogenous antibiotics Kan and Apr, even at high concentration (20 mM) (Figure 9A). In contrast, SAV4189-4189p complex was disrupted in dose-dependent manner by exogenous HygB and Thi (Figure 9A), suggesting that SAV4189 may sense and respond to these antibiotics to reduce repression of its target genes. This concept was tested by measuring expression of SAV4189 targets sav_4189 and sav_4190 in WT in response to exogenous HygB and Thi. Cells were cultured in FM-II to stationary phase (96 h), added with these antibiotics at subinhibitory concentration, and total RNAs were isolated from fermentation culture after 3 h and analyzed by qRT-PCR. sav_4189 and sav_4190 transcription levels were notably increased by addition of HygB or Thi, but not of DMSO (Figure 9B). HygB caused ∼143-fold increase of sav_4189 and ∼75-fold increase of sav_4190 expression. Thi caused ∼81-fold increase of sav_4189 and ∼71-fold increase of sav_4190 expression. These findings, taken together, indicate that SAV4189 recognizes various exogenous antibiotics such as HygB and Thi as ligands to modulate its DNA-binding activity and transcription of target genes.
FIGURE 9.
Effects of ligands on SAV4189 DNA-binding activity. (A) EMSAs of His6-SAV4189 with various antibiotics. 125 nM His6-SAV4189 was used for Apr and Kan; 62.5 nM His6-SAV4189 was used for other antibiotics. DMSO was used as solvent control. (B) Effects of HygB (10 μg/ml) and Thi (diluted in DMSO; final concentration 5 μg/ml) on transcription of SAV4189 target genes in vivo. Control: no antibiotic or DMSO added. DMSO: DMSO (but no antibiotic) added. Value of each gene was shown as fold change relative to control value, defined as 1. NS, not significant; ∗∗∗P< 0.001.
Discussion
In this study, we characterized a previously unknown MFR, SAV4189, as a strong activator of avermectin biosynthesis, and identified the SAV4189 regulon in S. avermitilis. Of the 42 MFRs of S. avermitilis, SAV4189 is the first to be characterized. The present findings enhance our understanding of control of secondary metabolism by MFRs in Streptomyces. Results of qRT-PCR and EMSAs indicate that SAV4189 plays an indirect role in stimulation of avermectin production by altering transcription of the CSR gene aveR, whose product is essential for activating all ave structural genes (Guo et al., 2010). The observation that SAV4189 indirectly activates aveR transcription suggests a possibility that it controls aveR through a yet-unknown cascade mechanism. However, the candidate SAV4189 targets listed in Supplementary Table S2 do not include reported aveR upstream regulator genes phoP (Yang et al., 2015), avaR1 (Zhu et al., 2017), or avaR2 (Zhu et al., 2016). Future studies will lead to identification of direct regulator(s) of aveR that mediate SAV4189 activation, and clarify the molecular mechanism underlying SAV4189 function in avermectin production.
We identified two SAV4189 target genes: sav_4189 and its adjacent cotranscribed gene sav_4190. Overexpression of sav_4189 in both WT and industrial strains strongly promoted avermectin production, indicating that endogenous SAV4189 is not at saturated level. Enhancement of SAV4189 expression is therefore an effective approach for increasing avermectin yield. sav_4189 is negatively autoregulated, indicating that SAV4189 utilizes this mechanism to strictly regulate its expression level and consequent avermectin production. We proposed that SAV4189 mainly activates avermectin production during early growth of S. avermitilis when sav_4189 transcription reaches a maximum level. As the level of SAV4189 expression increases, the transcription of its own gene is repressed, resulting in low expression level during late stage to avoid overproduction of avermectins. SAV4189 homologs are widespread in Streptomyces, and sav_4189 overxpression in S. coelicolor enhances antibiotic production. Therefore, the strategy of increasing yield by engineering of SAV4189 or its homologs (e.g., SCO4032 in S. coelicolor) may be extended to other industrially important antibiotics produced by Streptomyces.
sav_4190, which encodes a transmembrane efflux protein belonging to the major facilitator superfamily (MFS), is repressed directly by SAV4189. sav_4190 expression level is low in WT strain; however, deletion of this gene results in increased avermectin yield in both WT and industrial strains, suggesting that sav_4190 plays an important negative role in avermectin production. In O4189, though sav_4190 transcription was undetectable on day 6, there is still a very low sav_4190 transcription level on day 2. Thus, deletion of sav_4190 in sav_4189 overexpression strain may further improve avermectin yield.
The MFS transport system is widespread in bacteria, archaea, and eukarya, and has great diversity of function. According to Pao et al. (1998), MFS members can be classified into 17 distinct families, each of which transports a single class of structurally related compounds. Getsin et al. (2013) categorized transport proteins in S. coelicolor on the basis of their substrates. The putative substrates transported by SCO4031, a SAV4190 homolog in S. coelicolor, are multiple drugs. However, the substrates of SAV4190 could not be avermectins. If SAV4190 pumps out avermectins directly, deletion of sav_4190 will reduce avermectin yield. The fact is that SAV4190 has a negative effect on avermectin production. Thus, SAV4190 may be involved in expulsion from cells of intermediates of the avermectin biosynthetic pathway or some other precursors needed for avermectin biosynthesis when they accumulate to a certain level, thereby avoiding excessive avermectin production. We demonstrated recently that another MFS transporter in S. avermitilis, SAV7490 (AveM), also has a strong inhibitory effect on avermectin production, and that sav_7490 deletion significantly enhances avermectin yields in WT and industrial strains (Liu et al., 2015). Engineering of this type of transport proteins is therefore a promising approach for increasing antibiotic production, and studies are required to elucidate the functions of these proteins in Streptomyces.
MarR-family transcriptional regulators are completely conserved in the wHTH DNA-binding domain, but display high variability in the ligand-binding domain, which interacts with a wide variety of ligands (Perera and Grove, 2010). The effective component AveB1 is not the ligand of SAV4189, perhaps because of indirect control of avermectin production by SAV4189. It is not possible at this point to rule out the possibility that avermectin precursors act as SAV4189 ligands. MFR ligands are often related to the regulated genes and the important SAV4189 target sav_4190 encodes a transmembrane efflux protein which may expel avermectin precursors. It is therefore possible that SAV4190 pumps out SAV4189 ligands during fermentation.
SAV4189 did not bind to endogenous antibiotics AveB1 and Oli, but it did sense exogenous antibiotics HygB and Thi, which are produced by other Streptomyces species, to release repression of its target genes. There is increasing evidence for functioning of antibiotics as interspecies signals (Romero et al., 2011; Wang et al., 2014; Zhu et al., 2016). However, only a few receptors for exogenous antibiotic signals have been identified to date. In S. coelicolor, pseudo γ-butyrolactone receptor ScbR2 was reported to be the receptor for exogenous antibiotic jadomycin B (JadB), which modulates morphological differentiation and endogenous undecylprodigiosin (Red) production (Wang et al., 2014). We found recently that AvaR2, a ScbR2 homolog in S. avermitilis, acts as receptor for exogenous antibiotic signals JadB, HygB, and Apr (Zhu et al., 2016). AvaR2 and ScbR2 are members of the TetR-family transcriptional regulators (TFRs). TcaR from Staphylococcus epidermidis is an MFR involved in control of antibiotic resistance. Crystal structures have been determined for complexes of TcaR with the antibiotics ampicillin (Amp), Kan, streptomycin (Strep), methicillin (Meth), penicillin G (PnG), and chloramphenicol (Chl), indicating the remarkable plasticity of the multidrug-binding pocket of this protein (Chang et al., 2010, 2013). The above findings for TcaR, and our observation that SAV4189 responds to various structurally distinct antibiotics, indicate that some MFR proteins also function as receptors for exogenous antibiotic signals. This concept is consistent with the well-known variability of antibiotics produced by different Streptomyces species. These species may require differing types of receptors (such as TFRs and MFRs) to sense antibiotic signals produced by neighboring organisms, to elicit appropriate cellular responses. To our knowledge, SAV4189 is the first example of an exogenous antibiotic receptor among Streptomyces MFRs, and presumably plays an important role in interspecies communication. Our findings inform further investigations of ecologically relevant interactions between Streptomyces MFRs and exogenous antibiotic signals.
The predicted SAV4189 regulon contains 250 genes, which have been extensively cataloged and are involved in numerous physiological processes. EMSA analysis led to identification of 9 new SAV4189 targets. Among these, sav_2073, olmRII, and sav_5653 are involved in transcriptional regulation, reflecting the complex cascades of SAV4189 regulatory network. olmRII encodes an oligomycin pathway-specific activator (Yu et al., 2012). pks1-3 encodes a putative modular polyketide synthase that belongs to pks1 cluster for biosynthesis of an unknown secondary metabolite. These findings indicate a pleiotropic role of SAV4189 in regulation of secondary metabolism. We selected three putative SAV4189 target genes (aspB1, ccrA2, add4) associated with primary metabolism for evaluation and found that aspB1 involved in aspartate metabolism is SAV4189 target. ect genes encode enzymes for biosynthesis of compatible solute ectoine, which plays an important osmoprotective role (Bursy et al., 2008). The finding that SAV4189 binds to probe ectAp for ectA-ectB-ectC-ectD operon suggests that SAV4189 mediates protective responses to stress signals, such as osmotic stress. The other three identified SAV4189 targets are prpM3 involved in protein modification, hmuO involved in porphyrin metabolism, and sav_1959 with unknown function. Taken together, the present findings clearly demonstrate the pleiotropic effects of SAV4189 on cell physiological processes in S. avermitilis. Further studies in this important species will experimentally confirm the SAV4189 targets, clarify the broader regulatory roles of this protein, and eventually provide a comprehensive picture of cellular responses triggered by antibiotic and stress signals and mediated by SAV4189.
Author Contributions
YW and XZ designed the research. JG, XZ, XL, and WL performed the experiments. ZC, LD, and JL contributed study materials. YW and JG wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. S. Anderson for English editing of the manuscript.
Funding. This study was supported by the National Key Research and Development Program of China (Grant No. 2017YFD0201207) and the Project for Extramural Scientists of the State Key Laboratory of Agrobiotechnology (Grant No. 2017SKLAB7-8).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01358/full#supplementary-material
References
- Bierman M., Logan R., O’Brien K., Seno E. T., Rao R. N., Schoner B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116 43–49. 10.1016/0378-1119(92)90627-2 [DOI] [PubMed] [Google Scholar]
- Burg R. W., Miller B. M., Baker E. E., Birnbaum J., Currie S. A., Hartman R., et al. (1979). Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15 361–367. 10.1128/AAC.15.3.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursy J., Kuhlmann A. U., Pittelkow M., Hartmann H., Jebbar M., Pierik A. J., et al. (2008). Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl. Environ. Microbiol. 74 286–7296. 10.1128/AEM.00768-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. M., Chen C. K., Ko T. P., Chang-Chien M. W., Wang A. H. (2013). Structural analysis of the antibiotic-recognition mechanism of MarR proteins. Acta Crystallogr. D Biol. Crystallogr. 69 1138–1149. 10.1107/S0907444913007117 [DOI] [PubMed] [Google Scholar]
- Chang Y. M., Jeng W. Y., Ko T. P., Yeh Y. J., Chen C. K., Wang A. H. (2010). Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis. Proc. Natl. Acad. Sci. U.S.A. 107 8617–8622. 10.1073/pnas.0913302107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Lu Y., Chen J., Zhang W., Shu D., Qin Z., et al. (2008). Characterization of a negative regulator AveI for avermectin biosynthesis in Streptomyces avermitilis NRRL8165. Appl. Microbiol. Biotechnol. 80 277–286. 10.1007/s00253-008-1545-8 [DOI] [PubMed] [Google Scholar]
- Chen Z., Wen J., Song Y., Wen Y., Li J. (2007). Enhancement and selective production of avermectin B by recombinants of Streptomyces avermitilis via intraspecific protoplast fusion. Chin. Sci. Bull. 52 616–622. 10.1007/s11434-007-0119-y [DOI] [Google Scholar]
- Davis J. R., Brown B. L., Page R., Sello J. K. (2013). Study of PcaV from Streptomyces coelicolor yields new insights into ligand-responsive MarR family transcription factors. Nucleic Acids Res. 41 3888–3900. 10.1093/nar/gkt009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerton J. R., Ostlind D. A., Blair L. S., Eary C. H., Suhayda D., Cifelli S., et al. (1979). Avermectins, new family of potent anthelmintic agents: efficacy of the B1a component. Antimicrob. Agents Chemother. 15 372–378. 10.1128/AAC.15.3.372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getsin I., Nalbandian G. H., Yee D. C., Vastermark A., Paparoditis P. C., Reddy V. S., et al. (2013). Comparative genomics of transport proteins in developmental bacteria: Myxococcus xanthus and Streptomyces coelicolor. BMC Microbiol. 13:279. 10.1186/1471-2180-13-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove A. (2013). MarR family transcription factors. Curr. Biol. 23 R142–R143. 10.1016/j.cub.2013.01.013 [DOI] [PubMed] [Google Scholar]
- Guo J., Zhang X., Chen Z., Wen Y., Li J. (2014). Two adjacent and similar TetR family transcriptional regulator genes, SAV577 and SAV576, co-regulate avermectin production in Streptomyces avermitilis. PLoS One 9:e99224. 10.1371/journal.pone.0099224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Zhang X., Luo S., He F., Chen Z., Wen Y., et al. (2013). A novel TetR family transcriptional regulator, SAV576, negatively controls avermectin biosynthesis in Streptomyces avermitilis. PLoS One 8:e71330. 10.1371/journal.pone.0071330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Zhao J., Li L., Chen Z., Wen Y., Li J. (2010). The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol. Genet. Genomics 283 123–133. 10.1007/s00438-009-0502-2 [DOI] [PubMed] [Google Scholar]
- He F., Liu W., Sun D., Luo S., Chen Z., Wen Y., et al. (2014). Engineering of the TetR family transcriptional regulator SAV151 and its target genes increases avermectin production in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 98 399–409. 10.1007/s00253-013-5348-1 [DOI] [PubMed] [Google Scholar]
- Huang H., Grove A. (2013). The transcriptional regulator TamR from Streptomyces coelicolor controls a key step in central metabolism during oxidative stress. Mol. Microbiol. 87 1151–1166. 10.1111/mmi.12156 [DOI] [PubMed] [Google Scholar]
- Huang H., Mackel B. J., Grove A. (2013). Streptomyces coelicolor encodes a urate-responsive transcriptional regulator with homology to PecS from plant pathogens. J. Bacteriol. 195 4954–4965. 10.1128/JB.00854-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Kotaki H., Tanaka H., Omura S. (1988). Involvement of glucose catabolism in avermectin production by Streptomyces avermitilis. Antimicrob. Agents Chemother. 32 282–284. 10.1128/AAC.32.2.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000). Practical Streptomyces Genetics: A Laboratory Manual. Norwich: John Innes Foundation. [Google Scholar]
- Kitani S., Ikeda H., Sakamoto T., Noguchi S., Nihira T. (2009). Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 82 1089–1096. 10.1007/s00253-008-1850-2 [DOI] [PubMed] [Google Scholar]
- Li L., Guo J., Wen Y., Chen Z., Song Y., Li J. (2010). Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J. Ind. Microbiol. Biotechnol. 37 673–679. 10.1007/s10295-010-0710-0 [DOI] [PubMed] [Google Scholar]
- Liu G., Chater K. F., Chandra G., Niu G., Tan H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77 112–143. 10.1128/MMBR.00054-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Zhang Q., Guo J., Chen Z., Li J., Wen Y. (2015). Increasing avermectin production in Streptomyces avermitilis by manipulating the expression of a novel TetR-family regulator and its target gene product. Appl. Environ. Microbiol. 81 5157–5173. 10.1128/AEM.00868-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yan T., Jiang L., Wen Y., Song Y., Chen Z., et al. (2013). Characterization of SAV7471, a TetR-family transcriptional regulator involved in the regulation of coenzyme A metabolism in Streptomyces avermitilis. J. Bacteriol. 1954365–4372. 10.1128/JB.00716-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Sun D., Zhu J., Chen Z., Wen Y., Li J. (2014). An extracytoplasmic function sigma factor, σ25, differentially regulates avermectin and oligomycin biosynthesis in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 987097–7112. 10.1007/s00253-014-5759-7 [DOI] [PubMed] [Google Scholar]
- MacNeil D. J., Klapko L. M. (1987). Transformation of Streptomyces avermitilis by plasmid DNA. J. Ind. Microbiol 2 209–218. 10.1007/BF01569542 [DOI] [Google Scholar]
- Oh S. Y., Shin J. H., Roe J. H. (2007). Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J. Bacteriol. 189 6284–6292. 10.1128/JB.00632-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Zhang B., Zhang L., Zhao G., Ding X. (2009). Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor. Appl. Environ. Microbiol. 75 2158–2165. 10.1128/AEM.02209-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao S. S., Paulsen I. T., Saier M. H., Jr. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera I. C., Grove A. (2010). Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell Biol. 2 243–254. 10.1093/jmcb/mjq021 [DOI] [PubMed] [Google Scholar]
- Romero D., Traxler M. F., Lopez D., Kolter R. (2011). Antibiotics as signal molecules. Chem. Rev. 111 5492–5505. 10.1021/cr2000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Rodríguez A., Robledo-Casados I., Sanchez S. (2015). An overview on transcriptional regulators in Streptomyces. Biochim. Biophys. Acta 1849 1017–1039. 10.1016/j.bbagrm.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Rutledge P. J., Challis G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13 509–523. 10.1038/nrmicro3496 [DOI] [PubMed] [Google Scholar]
- Sun D., Wang Q., Chen Z., Li J., Wen Y. (2017). An alternative σ factor, σ8, controls avermectin production and multiple stress responses in Streptomyces avermitilis. Front. Microbiol. 8:736 10.3389/fmicb.2017.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Zhu J., Chen Z., Li J., Wen Y. (2016). SAV742, a novel AraC-family regulator from Streptomyces avermitilis, controls avermectin biosynthesis, cell growth and development. Sci. Rep. 6:36915. 10.1038/srep36915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urem M., Swiatek-Polatynska M. A., Rigali S., van Wezel G. P. (2016). Intertwining nutrient-sensory networks and the control of antibiotic production in Streptomyces. Mol. Microbiol. 102 183–195. 10.1111/mmi.13464 [DOI] [PubMed] [Google Scholar]
- van Wezel G. P., McDowall K. J. (2011). The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat. Prod. Rep. 28 1311–1333. 10.1039/c1np00003a [DOI] [PubMed] [Google Scholar]
- Wang W., Ji J., Li X., Wang J., Li S., Pan G., et al. (2014). Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 111 5688–5693. 10.1073/pnas.1324253111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Liu X., Wen Y., Song Y., Chen Z., Li J. (2015). The PhoP transcription factor negatively regulates avermectin biosynthesis in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 99 10547–10557. 10.1007/s00253-015-6921-6 [DOI] [PubMed] [Google Scholar]
- Yu Q., Bai L., Zhou X., Deng Z. (2012). Inactivation of the positive LuxR-type oligomycin biosynthesis regulators OlmRI and OlmRII increases avermectin production in Streptomyces avermitilis. Chin. Sci. Bull. 57 869–876. 10.1007/s11434-011-4865-5 [DOI] [Google Scholar]
- Zhang Q., Chen Q., Zhuang S., Chen Z., Wen Y., Li J. (2015). A MarR family transcriptional regulator, DptR3, activates daptomycin biosynthesis and morphological differentiation in Streptomyces roseosporus. Appl. Environ. Microbiol. 81 3753–3765. 10.1128/AEM.00057-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wen Y., Chen Z., Song Y., Li J. (2007). An adpA homologue in Streptomyces avermitilis is involved in regulation of morphogenesis and melanogenesis. Chin. Sci. Bull. 52 623–630. 10.1007/s11434-007-0105-4 [DOI] [Google Scholar]
- Zhu D., Wang Y., Zhang M., Ikeda H., Deng Z., Cane D. E. (2013). Product-mediated regulation of pentalenolactone biosynthesis in Streptomyces species by the MarR/SlyA family activators PenR and PntR. J. Bacteriol. 195 1255–1266. 10.1128/JB.02079-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Chen Z., Li J., Wen Y. (2017). AvaR1, a butenolide-type autoregulator receptor in Streptomyces avermitilis, directly represses avenolide and avermectin biosynthesis and multiple physiological responses. Front. Microbiol. 8:2577. 10.3389/fmicb.2017.02577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Sun D., Liu W., Chen Z., Li J., Wen Y. (2016). AvaR2, a pseudo gamma-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol. Microbiol. 102 562–578. 10.1111/mmi.13479 [DOI] [PubMed] [Google Scholar]
- Zianni M., Tessanne K., Merighi M., Laguna R., Tabita F. R. (2006). Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17 103–113. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.