Abstract
Karst caves are obviously characterized by darkness, constantly low temperature, high humidity, and oligotrophy. Previous studies revealed that Karst caves have a high and specific bio-diversity. A large number of troglobiont animals had been discovered and their evolution and speciation have been well investigated. However, the origin and evolution of cave fungi remain unknown. In a previous study, we have identified 20 new species, which accounted for 49% of the total number of new species of fungi ever described from caves. In this study, we inferred the divergence times of these 20 new species and compared to the cave formation geologic age. The fossil-calibrated molecular clock showed that the divergence times of these 20 suspected troglobitic fungi are between late Miocene (7.2 Mya for Metapochonia variabilis) and late Jurassic (158 Mya for Gymnoascus exasperates). While based on the historical geological movement and the paleoclimate of Guizhou, it has been estimated that the development of caves in this area was later than middle Pliocene (3.5–4 Mya). It is therefore concluded that the new species described from these caves are unlikely troglobitic fungi but travelers from other environments. The geographic history of caves appeared to be too short for fungal speciation.
Keywords: BEAST, geologic age, molecular clock, obligate troglobitic fungi, speciation
Introduction
Caves are strongly zonal environment, with unique characteristics determined by the surrounding rock, subterranean water, and karst morphology (Kuzmina et al., 2012; Gabriel and Northup, 2013). It significantly differs from the land surface environment in the darkness, constantly low temperature, high humidity, and oligotrophy (Gabriel and Northup, 2013). As a relatively closed space, cave environment can be affected by various factors, such as the air currents, chemolithoautotrophy, visitors, and water movement (streams or water seeps; Hose et al., 2000; Barton and Jurado, 2007a; Gabriel and Northup, 2013; Ortiz et al., 2014). All these above factors contribute to the biota in caves (Ogórek et al., 2013; Ortiz et al., 2014) and caves were suspected to encompass a high, specific biodiversity (Culver and Holsinger, 1992; Gabriel and Northup, 2013; Vanderwolf et al., 2013; Jiang et al., 2017).
Cave fauna can be classified into four major categories: troglobiont, eutroglophile, subtroglophile, and trogloxene. Troglobiont is a species or population that bound to a subterranean habitat strictly, in other words, true cave life (Sket, 2008). The earliest description of fungi in caves was published by Humboldt in 1794, as described in Dobat (1967), and the first ecological literature of caves was by Megušar (1914). Most of the previous studies were focused on cave fauna and it has been suspected that there might be 50,000–100,000 species of cave animals on the earth (Culver and Holsinger, 1992). Quite many troglobitic animals had been discovered, such as Typhlobarbus nudiventris (Chu and Chen, 1982), Typhlocaridina semityphlata (Cai, 1995), Angulifemur tridigitis (Zhang, 1995), and several species of Macrochlamys and Kaliella (Li, 2003). A number of studies on the evolution of cave fauna had been published, and several hypotheses for the evolution and origin of cave animals have been proposed, such as adaptive shift hypothesis (Rouch and Danielopol, 1987; Desutter-Grandcolas and Grandcolas, 1996) and climatic relict hypothesis (Barr and Holsinger, 1985; Peck and Finston, 1993).
Microbes play important roles in cave (Barton and Northup, 2007b; Gabriel and Northup, 2013). More than 1150 fungal species in 550 genera have been discovered in caves and mines worldwide by 2017 (Vanderwolf et al., 2013; Zhang et al., 2017) and some suspected obligate troglobitic fungi have been reported, such as Acaulium caviariforme, Aspergillus baeticus, and Aspergillus thesauricus (Vanderwolf et al., 2013). Recently, Zhang et al. (2017) described 20 new species from caves, accounting for 49% new fungal species ever described from caves. However, the origin of cave microbiology was unknown and there is no hypothesis on their evolution and origin. The answer to the question “whether if there is real obligate troglobitic fungus” is important for understanding the origin and evolution of cave fungi. The factors affecting the materials and energy in caves, such as water movement, air currents, and visitors, can also act as pathways of microorganism exchange between subterrane and surface environment (Gabriel and Northup, 2013). Meanwhile, most of the currently discovered fungi from caves were known from other environments (Vanderwolf et al., 2013; Zhang et al., 2017). Zhang et al. (2017) suggested that cave fungi may have an origin from outside environment, as all the recorded genera and most of the identified species are known from other environments.
Molecular clock hypothesis is useful in estimating divergence times (Boutin and Coineau, 2000; Bromham and Penny, 2003). To reveal the origin of fungi in caves, a comparison was made between the divergence times of suspected obligate troglobitic fungi with the independent estimation of cave formation geologic age (Juan and Emerson, 2010). In the present study, six nucleotide sequences of the 20 suspected obligate troglobitic fungi described by Zhang et al. (2017), and some aligned representative species in the phylum Ascomycota were combined to infer a fossil-calibrated phylogeny and estimate divergence times for fungi in caves.
Materials and Methods
Cave Information
Suiyang county, located in Guizhou, China, has a typical subtropical monsoon climate. The annual mean temperature is 13.5°C, and the annual rainfall is 1116–1350 mm (Jiang et al., 2012).
Two unnamed Karst caves in Wangcao town, Suiyang county, herein named Cave 1 (28° 12′ 629″ N, 107° 13′ 639″ E) and Cave 2 (28° 12′ 599″ N, 107° 13′ 661″ E), are located at the southern edge of Kuankuoshui National Natural Reserve. Both caves are horizontal and zonal and have one entrance hiding in the forest of the hillside (Figure 1, Zhang et al., 2017). These two caves are 500 m away from each other, and might belong to the same cave system with subterranean river connection. The elevation of Cave 1 is 908 m; the length is 400 m; the humidity is 75–80%; and the temperature is 21–22°C. The elevation of Cave 2 is 930 m; the length is 750 m; the humidity is 75–85%; and the temperature is 20–23°C (Zhang et al., 2017).
FIGURE 1.

Visited caves. (a) Location of caves in Suiyang, Guizhou. (b) Entrance to Cave 1. (c) Stalactite. (d) Pool at the end of Cave 1. (e) Sampled rocks. (f) Bat guano colonized by fungal mycelia. (g) Pellucid tadpole (Zhang et al., 2017).
According to the geological features and the historically tectonic movement nearby, the development periods of these two investigated caves were estimated as later than middle Pliocene (3.5–4 Mya), which fitted well with the estimation of the formation age of most caves in China (late Pliocene to early Pleistocene, ca. 3 Mya; Xiong and Zhu, 1994; Qin and He, 2004; Ping et al., 2011; Zhang and Zhu, 2012).
Taxon Sampling, Molecular Data, and Phylogenetic Analysis
We included representative species of most lineages in Ascomycota with a total number of 167 species (Supplementary Table S1), including the 20 suspected obligate troglobitic fungi described by Zhang et al. (2017), and two species of Basidiomycota, Boletus edulis and Rubroboletus sinicus, as outgroup. Six loci were used for analysis, i.e., ITS, LSU, SSU, TEF, RPB1, and RPB2. Two new loci, SSU and RPB1, of these 20 species were amplified using primers NS1/NS4 (White et al., 1990) and RPB1-Cr/RPB1-Af (Matheny et al., 2002), respectively. Amplification reactions were performed according to the program of Zhang et al. (2017), except the annealing temperature, 54°C for SSU and RPB1.
All sequences of different loci were aligned using MAFFT1 (Katoh and Toh, 2010) and modified manually in MEGA v. 7 (Kumar et al., 2016) separately. Then, individual alignments were concatenated and used for phylogenetic analysis next step. Ambiguously aligned regions were excluded from all analyses.
Maximum likelihood (ML) and Bayesian inference (BI) methods were used to reveal the phylogenetic relationship. The ML analyses were executed using RAxML-HPC v. 8.2.7 (Stamatakis, 2014) with 1,000 replicates under the GTR-GAMMY model. The robustness of branches was assessed by bootstrap analysis with 1,000 replicates. For Bayesian analysis, the best model of evolution was estimated using jModelTest v. 2.1.7 (Guindon and Gascuel, 2003; Darriba et al., 2012). Posterior probabilities (PPs) (Rannala and Yang, 1996; Zhaxybayeva and Gogarten, 2002) were calculated by Markov chain Monte Carlo (MCMC) sampling in MrBayes v. 3.2.1 (Huelsenbeck and Ronquist, 2001), using the estimated evolutionary models. Six simultaneous Markov chains were run for 10,000,000 generations, and trees were sampled every 1,000th generations (totally resulting 10,000 trees). The first 2,000 trees, representing the burn-in phase of the analyses, were discarded and the remaining 8,000 trees were used to calculate PPs in the majority rule consensus tree. The final trees were visualized in TreeView (Page, 1996). All the sequences generated in this study were deposited in GenBank (Supplementary Table S1).
Molecular Clock Analysis
We used five fossils with reliable age and accurate identification to estimate the divergence times. Each fossil age served as a minimum constraint. Paleopyrenomycites devonicus was discovered in thin-section preparations of lower Devonian [400 Mya (million years)] and considered to be the oldest ascomycete fossil (Taylor et al., 1999). Here it was used as the minimum divergence time of Pezizomycotina (Prieto and Wedin, 2013; Garnica et al., 2016). Paleoophiocordyceps coccophagus was described as the oldest fossil of animal parasitic fungi (99–105 Mya) with a striking morphological similarity to the asexual states of Ophiocordyceps (Sung et al., 2008). Fossils of Calicium and Chaenotheca were reported from Baltic amber dating back to 35–55 Mya (Rikkinen, 2003). The Alectoria fossil from the Baltic amber (35–40 Mya) was used as the calibration point for Alectoria clade (Magdefrau, 1957; De Paz et al., 2011; Prieto and Wedin, 2013).
Bayes MCMC algorithm was implemented to estimate divergence times using the data from multi-locus and according multiple fossil calibration nodes Supplementary Datasheet S1. The analysis was performed using BEAST v. 2.4.5 software package (Bouckaert et al., 2014). The tree topology was estimated by RAxML in the last step. We partitioned the data by gene using the general time reversible (GTR) substitution model for each partition, as estimated by jModelTest v. 2.1.7. We used the relaxed clock log normal model, specifying an exponential distribution for the ucld.mean parameter with a mean of 10.0 and offset 0. A birth-death tree prior was implemented (Divergence Time Estimation using BEAST2). All groups containing calibration points were regarded as monophyletic in beast analysis. Beast analyses were run for 50,000,000 generation; meanwhile, parameters and trees were logged every 1000 generations. Convergence, mixing, and effective sample sizes (ESS) of parameters were checked in Tracer v. 1.6.5 (Rambaut et al., 2014). Three repeat analyses were performed for accuracy and LogCombiner (Bouckaert et al., 2014) was used to combine the runs. The first 20% trees representing the starting and unreliable results were removed from the analysis and a maximum clade tree was created with TreeAnnotator v. 2.4.5 (Bouckaert et al., 2014). Age estimation is followed by their highest posterior density (HPD) in parentheses, which based on 95% confidence interval of all sample values.
Divergence time estimation calculates stem and node ages for each clade. Subsequently, the stem ages are considered as the time a group originated, or differentiated from its sister clade, and the node ages as a group started diversification.
Results
Phylogenetic Relationships
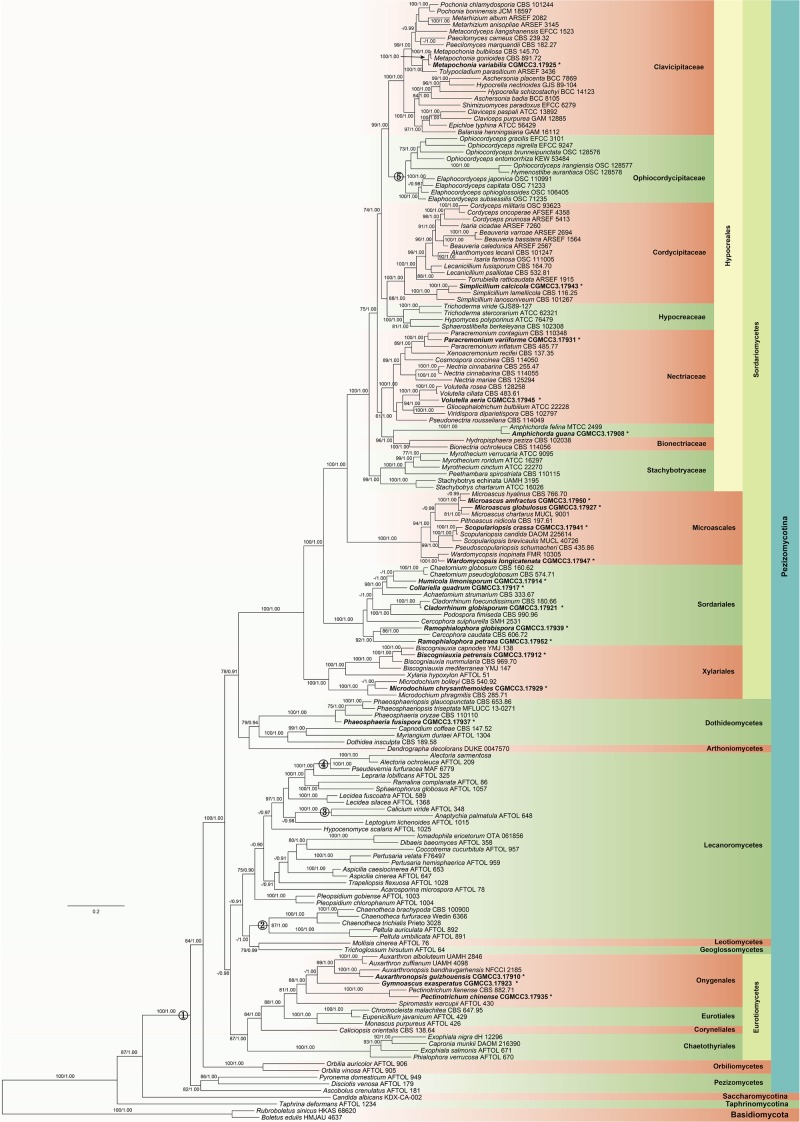
All of the major clades in Ascomycota were strongly supported with ML bootstrap proportion and Bayesian PPs (Figure 2) and the overall topology of the best tree generated in ML analysis is basically congruent with previous phylogenetic studies of Ascomycota (Hibbett et al., 2007; Sung et al., 2007; Schoch et al., 2009; Blackwell, 2010; Gazis et al., 2012; Prieto and Wedin, 2013). These 20 fungal species clustered into three classes and six orders which were well supported.
FIGURE 2.
Maximum likelihood (ML) tree based on ITS, LSU, SSU, TEF, RPB1, and RPB2 sequences. ML bootstrap values (≥70%) and Bayesian posterior probability (≥90%) are indicated along branches (ML/PP). The tree is rooted with Rubroboletus sinicus and Boletus edulis (Basidiomycota). Numbers in the circles indicate the nodes used for fossil calibration: (1) Paleopyrenomycites devonicus; (2) Chaenotheca sp.; (3) Calicium sp.; (4) Alectoria succinica; and (5) Paleoophiocordyceps coccophagus. The suspected obligated fungal species are indicated in bold font and marked with “∗”. The major clades were marked with different colors and names are shown on the right side.
Divergence Times
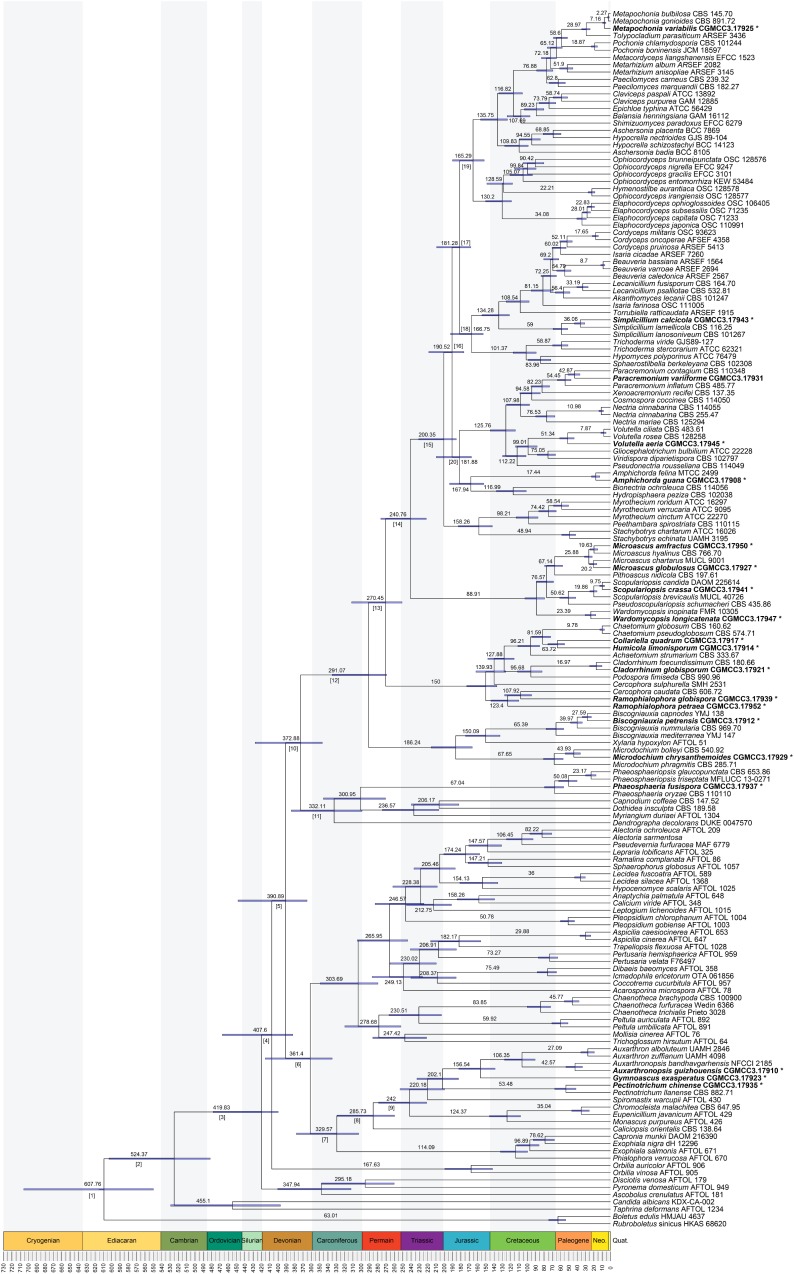
The divergence of Basidiomycota and Ascomycota linage occurred in the Sinian to Early Cambrian (node 1; 608 Mya, 549–701.5 Mya for 95% HPD; Figure 3 and Table 1). The first divergence in Ascomycota, i.e., the origin of Saccharomycotina, Taphrinomycotina, and Pezizomycotina, took place in Early Cambrian (node 2; 524 Mya, 481–601 Mya for 95% HPD). The earliest split in Pezizomycotina took place in Silurian (node 3; 420 Mya, 400–476 Mya for 95% HPD), resulting in Pezizomycetes and Pezizomycotina crown groups; in Silurian to Devonian (node 4; 408 Mya, 382–464 Mya for 95% HPD), resulting in Orbiliomycetes; in Carboniforous (node 6; 361 Mya, 334–413 Mya for 95% HPD), resulting in Eurotiomycetes and Lecanoromycetes; in Carboniferous (node 10; 373 Mya, 346–425 Mya for 95% HPD), resulting in Sordariomycetes crown (Figure 3 and Table 1).
FIGURE 3.
Maximum clade credibility chronogram of Ascomycota evolution. The chronogram is the result from the beast analysis based on the topology of ML tree. Each node represents the mean divergence time estimate (written above the nodes) and bars show the 95% HPD. Numbers corresponding to dated groups shown in Table 1 are written above the nodes with square brackets. The suspected obligated fungal species are indicated in bold font and marked with “∗”.
Table 1.
Mean and range (95% HPD) divergence time estimations (Mya) of the major Ascomycota lineages.
| Node label | Mean | 95% HPD | Node label | Mean | 95% HPD |
|---|---|---|---|---|---|
| 1 | 607.76 | 548.91–701.46 | 11 | 332.11 | 297.24–382.13 |
| 2 | 524.37 | 481.31–600.56 | 12 | 291.07 | 268.62–332.4 |
| 3 | 419.83 | 400.00–476.09 | 13 | 270.45 | 249.23–308.75 |
| 4 | 407.6 | 381.57–464.21 | 14 | 240.76 | 221.41–275.18 |
| 5 | 390.89 | 364.87–445.37 | 15 | 200.35 | 184.91–229.36 |
| 6 | 361.4 | 333.77–412.68 | 16 | 190.52 | 175.80–217.62 |
| 7 | 329.57 | 302.47–376.62 | 17 | 181.28 | 166.71–206.89 |
| 8 | 285.73 | 259.31–327.66 | 18 | 166.75 | 152.34–191.03 |
| 9 | 242.00 | 220.81–278.22 | 19 | 165.29 | 151.72–189.06 |
| 10 | 372.88 | 346.03–424.85 | 20 | 181.88 | 166.44–207.94 |
Divergence times of the suspected obligate troglobitic fungi were different (Figure 3 and Table 2). The most recent divergence is Metapochonia variabilis, occurred in Miocene (7.2 Mya, 5.3–9.2 Mya for 95% HPD), and the oldest is Gymnoascus exasperates in late Jurassic (157 Mya, 139–181 Mya for 95% HPD). While these two caves were developed within 4 Mya, clearly younger than the suspected obligate troglobitic fungi. In other words, the fungal speciation did not occurred in the caves, and cave fungi were most likely exogenetic. The estimated evolutionary rates of different loci and combined are detailed in Table 3.
Table 2.
Mean and range (95% HPD) divergence time estimations (Mya) of the suspected obligate troglobitic fungi.
| Node label | Strains No. | Source | Mean | 95% HPD |
|---|---|---|---|---|
| Amphichorda guana | CGMCC3.17908T | Bat guano | 17.44 | 12.76–22.96 |
| Auxarthronopsis guizhouensis | CGMCC3.17910T | Air | 42.57 | 33.11–53.19 |
| Biscogniauxia petrensis | CGMCC3.17912T | Rock | 27.59 | 22.63–33.73 |
| Cladorrhinum globisporum | CGMCC3.17921T | Water | 16.97 | 9.82–25.24 |
| Collariella quadrum | CGMCC3.17917T | Soil | 63.72 | 55.16–74.58 |
| Gymnoascus exasperates | CGMCC3.17923T | Bat guano | 156.54 | 139.07–181.04 |
| Humicola limonisporum | CGMCC3.17914T | Soil | 63.72 | 55.16–74.58 |
| Metapochonia variabilis | CGMCC3.17925T | Soil | 7.16 | 5.33–9.21 |
| Microascus amfractus | CGMCC3.17950T | Plant debris | 19.63 | 15.08–24.61 |
| Microascus globulosus | CGMCC3.17927T | Bat guano | 20.2 | 15.92–24.83 |
| Microdochium chrysanthemoides | CGMCC3.17929T | Air | 43.93 | 35.81–53.43 |
| Paracremonium variiforme | CGMCC3.17931T | Water | 42.87 | 36.23–50.75 |
| Pectinotrichum chinense | CGMCC3.17935T | Soil | 53.48 | 41.61–67.1 |
| Phaeosphaeria fusispora | CGMCC3.17937T | Air | 50.08 | 39.96–61.8 |
| Ramophialophora globispora | CGMCC3.17939T | Plant debris | 107.92 | 93.94–125.78 |
| Ramophialophora petraea | CGMCC3.17952T | Rock | 123.4 | 109.64–142.64 |
| Scopulariopsis crassa | CGMCC3.17941T | Soil | 9.75 | 7.07–12.75 |
| Simplicillium calcicola | CGMCC3.17943T | Rock | 36.06 | 30.29–42.77 |
| Volutella aeria | CGMCC3.17945T | Air | 51.34 | 43.73–61.07 |
| Wardomycopsis longicatenata | CGMCC3.17947T | Air | 23.39 | 17.28–30.31 |
T indicates ex-type strains.
Table 3.
Range of estimated evolutionary rates with confidence intervals (site/Mya).
| Gene | Mean | Min | Max | SD |
|---|---|---|---|---|
| Combined | 0.0033 | 0.0025 | 0.0041 | 0.0005 |
| ITS | 0.0020 | 0.0016 | 0.0024 | 0.0003 |
| LSU | 0.0009 | 0.0007 | 0.0010 | 0.0001 |
| RPB1 | 0.0034 | 0.0026 | 0.0041 | 0.0004 |
| RPB2 | 0.0039 | 0.0029 | 0.0051 | 0.0006 |
| SSU | 0.0003 | 0.0002 | 0.00035 | 0.00003 |
| TEF | 0.0061 | 0.0035 | 0.0092 | 0.0016 |
Discussion
Hypothesis for the Origin of the Subterranean Fauna
The adaptation of organisms to caves or groundwater environments has been subject of many studies (Leys et al., 2003). Adaptive shift hypothesis and climatic relict hypothesis are two general hypotheses on the origin of the subterranean fauna. According to the climatic relict hypothesis, an epigean species preadapted to the underground life may survive in the subterranean refuge when the surface environment becomes unfavorable due to climate change. While under the adaptive shift hypothesis, a preadapted epigen species may actively enter the subterranean habitats to exploit new resources once they become accessible (Bellés, 1992; Leys et al., 2003; Juan and Emerson, 2010; Ribera et al., 2010). Leys et al. (2003) studied the evolution of subterranean diving beetles in Australia based on phylogeny and molecular clock method and found that the diving beetles were preadapted to the underground life, and when the drought occurred in the Early Pliocene, they survived and adapted to cave or groundwater life (climatic relict hypothesis). Ribera et al. (2010) investigated the origin and evolution of subterranean beetles of the tribe Leptodirini in Western Mediterranean based on mitochondrial and nuclear sequences. Their results suggested that the main lineages of Leptodirini had developed the necessary modifications for the hypogean life by the early-mild Oligocene. In another word, they had preadapted to the underground life. In contrast to most current assumptions on the evolution of the underground fauna, there was no evidence for the extinction of epigean populations (adaptive shift hypothesis). Johnson et al. (2011) reported a new eel-like fish, Protanguilla palau, from an undersea cave in Palau and divergence time estimation showed that Protanguilla lineage diverged about 200 Mya, much earlier than the formation age of Palau-Kyushu Ridge (60–70 Mya). Accordingly, it has been suggested that the P. palau possibly had a considerably broader distribution than previous assumption.
Similar to the cave fauna, to determine the origin of cave fungi, it is important to determine if the true troglobitic fungi exist. Currently, there are 36 suspected troglobitic fungi reported, including the troglobitic fungi summarized by Vanderwolf et al. (2013) and the new cave fungi published by Zhang et al. (2017). Although these species might be discovered from non-cave environment in subsequent studies, they do stand as the currently most reasonable suspect of troglobitic fungi. Geomyces destructans, the most famous “cave fungus” previously, was isolated from bats outside caves by Dobony in 2012 (Vanderwolf et al., 2013). Comparing the divergence time of suspected troglobitic fungi and cave development geologic age is one of the best ways to infer the origin of cave fungi.
Origin of Cave Fungi
The divergence time of Ascomycota and Basidiomycota estimated in our analysis (Figure 3) is similar to that inferred from previous analyses (Berbee and Taylor, 2001; Dhanasekaran et al., 2006; Taylor and Berbee, 2006; Floudas et al., 2012; Prieto and Wedin, 2013; Garnica et al., 2016). Divergence of our 20 suspected troglobitic fungi was estimated to be no later than Miocene (7.2 Mya), which is much earlier than cave development age (3.5–4.0 Mya). The speciation of these cave fungi was thus unlikely to occur in caves. This provided reasonably support for the speculation of Zhang et al. (2017) that cave fungi may have an origin of external environment.
Considering the features of subterranean habitat and outside environment, allopatric speciation appears to be a reasonable explanation for cave fauna (Mayr, 1966; Barr and Holsinger, 1985; Giraud et al., 2008). This, however, cannot explain cave fungi. The geographic age of caves is obviously too short for a most fungal speciation processes. Apparently, the constantly lower temperature (Rohde, 1992; Gillman and Wright, 2006; Gillman et al., 2009; Goldie et al., 2010; Wright et al., 2010) and extremely limited energy resources also lead to a limited microevolution in caves (Goldie et al., 2010). Unfortunately, nowadays, studies of the evolution speed were mainly on plants and animals.
The data obtained from this study well demonstrated the non-cave origin of cave fungi, as the caves were generally believed to be formed later than Pliocene period (3.5–4.0 ma), which is shown to be much shorter than the most recently speciated species M. variabilis (late Miocene, ca. 7.2 Mya). It is therefore concluded that the new species described from cave are unlikely troglobitic fungi but travelers from other environments, although they have not been reported from a terrestrial environment.
Although some new species have been discovered from caves, hitherto no new genus was described from caves. Even within the described new species, none of the any two were shown to be sister lineages (Zhang et al., 2017), indicating no evidence of fungal evolutionary divergence in caves. Within the currently descried new species from caves, we could not recognize any distinct morphological and physiological feature that might be potentially troglobitic. Future studies employing metagenomics techniques could possibly bring broader and more conclusive data to answer above questions.
Author Contributions
Z-FZ designed the work that led to the submission, analyzed the data, and drafted most part of the manuscript. PZ revised our manuscript. LC conceived the work, drafted part of the manuscript, and revised our manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Yuan-hai Zhang of the Institute of Karst Geology, Chinese Academy of Geological Sciences, for providing the information of the investigated caves. We thank all of the individuals in our lab for help with technological supporting, valuable, and constructive suggestions.
Footnotes
Funding. This study was financially supported by NSFC 31725001 and the Project for Fundamental Research on Science and Technology, MOST (2014FY120100). Z-FZ acknowledged CAS project for supporting his studentship.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01407/full#supplementary-material
Specimens used in this study with GenBank accession numbers (xlsx).
Information about the priors and parameters used in BEAST to obtain the divergence time.
References
- Barr T. C., Holsinger J. R. (1985). Speciation in cave faunas. Ann. Rev. Ecol. Syst. 16 313–337. 10.1146/annurev.es.16.110185.001525 [DOI] [Google Scholar]
- Barton H. A., Jurado V. (2007a). What’s up down there? Microbial diversity in caves. Microbe 2 132–138. [Google Scholar]
- Barton H. A., Northup D. E. (2007b). Geomicrobiology in cave environments: past, current and future perspectives. J. Cave Karst Stud. 69 163–178. [Google Scholar]
- Bellés X. (1992). Survival, opportunism and convenience in the processes of cave colonization by terrestrial faunas. Oecol. Aquat. 10 325–335. [Google Scholar]
- Berbee M. L., Taylor J. W. (2001). “Fungal molecular evolution: gene trees and geologic time,” in Systematics and Evolution, eds McLaughlin D. J., Spatafora J. W. (New York, NY: Springer Press; ), 229–245. [Google Scholar]
- Blackwell M. (2010). Fungal evolution and taxonomy. BioControl 55 7–16. 10.1007/s10526-009-9243-8 [DOI] [Google Scholar]
- Bouckaert R., Heled J., Kühnert D., Vaughan T., Wu C. H., Xie D., et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin C., Coineau N. (2000). “Evolutionary rates and phylogenetic age of some stygobiontic species,” in Subterranean Ecosystems – Ecosystems of the World 30, eds Wilkens H., Culver D. C., Humphreys W. F. (Amsterdam: Elsevier; ), 433–451. [Google Scholar]
- Bromham L., Penny D. (2003). The modern molecular clock. Nat. Rev. Genet. 4 216–224. 10.1038/nrg1020 [DOI] [PubMed] [Google Scholar]
- Cai Y. X. (1995). A new troglobitic shrimp from China (Decapoda: Atyidae). Acta Zootaxonom. Sin. 20 157–160. 10.11646/zootaxa.4399.3.2 [DOI] [PubMed] [Google Scholar]
- Chu X., Chen Y. (1982). A new genus and species of blind cyprinid fish from China with special reference to its relationships. Acta Zool Sin. 28 383–388. [Google Scholar]
- Culver D. C., Holsinger J. R. (1992). How many species of troglobites are there. Natl. Speleol. Soc. Bull. 54 79–80. [Google Scholar]
- Darriba D., Taboada G. L., Doallo R., Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paz G. A., Cubas P., Divakar P. K., Lumbsch H. T., Crespo A. (2011). Origin and diversification of major clades in parmelioid lichens (Parmeliaceae, Ascomycota) during the Paleogene inferred by Bayesian analysis. PLoS One 6:e28161. 10.1371/journal.pone.0028161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desutter-Grandcolas L., Grandcolas P. (1996). The evolution toward troglobitic life: a phylogenetic reappraisal of climatic relict and local habitat shift hypotheses. Mém. Biospéol. 23 57–63. [Google Scholar]
- Dhanasekaran V., Jeewon R., Hyde K. D. (2006). Molecular taxonomy, origins and evolution of freshwater ascomycetes. Fungal Divers. 23 351–390. [Google Scholar]
- Dobat K. (1967). Ein bisher unveroffentlichtes botanisches manuskript Alexander von Humboldts: Plantae subterranae Europ. 1794 cum Iconibus. Akademie Der Wissenschaften und der Literatur. 6 16–19. [Google Scholar]
- Floudas D., Binder M., Riley R., Barry K., Blanchette R. A., Henrissat B., et al. (2012). The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336 1715–1719. 10.1126/science.1221748 [DOI] [PubMed] [Google Scholar]
- Gabriel C. R., Northup D. E. (2013). “Microbial ecology: caves as an extreme habitat,” in Cave Microbiomes: A Novel Resource for Drug Discovery, ed. Cheeptham N. (New York, NY: Springer Press; ), 85–108. 10.1007/978-1-4614-5206-5_5 [DOI] [Google Scholar]
- Garnica S., Riess K., Schön M. E., Oberwinkler F., Setaro S. D. (2016). Divergence times and phylogenetic patterns of sebacinales, a highly diverse and widespread fungal lineage. PLoS One 11:e0149531. 10.1371/journal.pone.0149531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazis R., Miadlikowska J., Lutzoni F., Arnold A. E., Chaverri P. (2012). Culture-based study of endophytes associated with rubber trees in Peru reveals a new class of Pezizomycotina: xylonomycetes. Mol. Phylogenet. Evol. 65 294–304. 10.1016/j.ympev.2012.06.019 [DOI] [PubMed] [Google Scholar]
- Gillman L. N., Keeling D. J., Ross H. A., Wright S. D. (2009). Latitude, elevation and the tempo of molecular evolution in mammals. Proc. R. Soc. B Biol. Sci. 276 3353–3359. 10.1098/rspb.2009.0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman L. N., Wright S. D. (2006). The influence of productivity on the species richness of plants: a critical assessment. Ecology 87 1234–1243. 10.1890/0012-9658(2006)87[1234:TIOPOT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Giraud T., Refrégier G., Le Gac M., de Vienne D. M., Hood M. E. (2008). Speciation in fungi. Fungal Genet. Biol. 45 791–802. 10.1016/j.fgb.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Goldie X., Gillman L., Crisp M., Wright S. (2010). Evolutionary speed limited by water in arid Australia. Proc. R. Soc. B Biol. Sci. 277 2645–2653. 10.1098/rspb.2010.0439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 52 696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hibbett D. S., Binder M., Bischoff J. F., Blackwell M., Cannon P. F., Eriksson O. E., et al. (2007). A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111 509–547. 10.1016/j.mycres.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Hose L. D., Palmer A. N., Palmer M. V., Northup D. E., Boston P. J., DuChene H. R. (2000). Microbiology and geochemistry in a hydrogen-sulphide-rich Karst environment. Chem. Geol. 169 399–423. 10.1016/S0009-2541(00)00217-5 [DOI] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. (2001). Mrbayes: bayesian inference of phylogenetic trees. Bioinformatics 17 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jiang J.-R., Cai L., Liu F. (2017). Oligotrophic fungi from a carbonate cave, with three new species of Cephalotrichum. Mycology 8 164–177. 10.1080/21501203.2017.1366370 [DOI] [Google Scholar]
- Jiang X. Y., He Y. Q., Shen C. C., Kong X. G., Li Z. Z., Chang Y. W. (2012). Stalagmite-inferred Holocene precipitation in northern Guizhou Province, China, and asynchronous termination of the Climatic Optimum in the Asian monsoon territory. Chin. Sci. Bull. 57 795–801. 10.1007/s11434-011-4848-6 [DOI] [Google Scholar]
- Johnson G. D., Ida H., Sakaue J., Sado T., Asahida T., Miya M. (2011). A ‘living fossil’ eel (Anguilliformes: Protanguillidae, fam. nov.) from an undersea cave in Palau. Proc. R. Soc. B Biol. Sci. 279 934–943. 10.1098/rspb.2011.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan C., Emerson B. C. (2010). Evolution underground: shedding light on the diversification of subterranean insects. J. Biol. 9:17. 10.1186/jbiol227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Toh H. (2010). Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmina L. Y., Galimzianova N. F., Abdullin S. R., Ryabova A. S. (2012). Microbiota of the Kinderlinskaya Cave (South Urals, Russia). Microbiology 81 251–258. 10.1134/S0026261712010109 [DOI] [Google Scholar]
- Leys R., Watts C. H., Cooper S. J., Humphreys W. F. (2003). Evolution of subterranean diving beetles (Coleoptera: dytiscidae: hydroporini, Bidessini) in the arid zone of Australia. Evolution 57 2819–2834. 10.1111/j.0014-3820.2003.tb01523.x [DOI] [PubMed] [Google Scholar]
- Li D. H. (2003). A preliminary study on species diversity and distribution of snails in caves of Guizhou Province. Carsol. Sin. 22 213–218. [Google Scholar]
- Magdefrau K. (1957). Flechten und Moose in baltischen Bernstein. Ber Deut Bot Ges. 70 433–435. [Google Scholar]
- Matheny P. B., Liu Y. J., Ammirati J. F., Hall B. D. (2002). Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe. Agaricales). Am. J. Bot. 89 688–698. 10.3732/ajb.89.4.688 [DOI] [PubMed] [Google Scholar]
- Mayr E. (1966). Animal Species and Evolution. Cambridge, MA: Harvard University Press. [Google Scholar]
- Megušar F. (1914). Oekologischen Studien an Höhlentieren. Carniola 5 63–83. [Google Scholar]
- Ogórek R., Lejman A., Matkowski K. (2013). Fungi isolated from Niedźwiedzia Cave in Kletno (Lower Silesia. Poland). Int. J. Speleol. 42 161–166. 10.5038/1827-806X.42.2.9 [DOI] [Google Scholar]
- Ortiz M., Legatzki A., Neilson J. W., Fryslie B., Nelson W. M., Wing R. A., et al. (2014). Making a living while starving in the dark: metagenomic insights into the energy dynamics of a carbonate cave. ISME J. 8 478–491. 10.1038/ismej.2013.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. D. M. (1996). TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Peck S. B., Finston T. L. (1993). Galapagos island troglobites: the questions of tropical troglobites, parapatric distributions with eyed-sister-species, and their origin by parapatric speciation. Mémoires de Biospéologie. 20 19–37. [Google Scholar]
- Ping Y. M., Yang G. F., Zhang X. J., Tian M. Z., Chen A. Z., Eg Z. L., et al. (2011). Timing of Zhangjiajie sandstone landforms: evidence from fluvial terraces and karst caves. Geol. Rev. 57 118–124. 10.16509/j.georeview.2011.01.003 [DOI] [Google Scholar]
- Prieto M., Wedin M. (2013). Dating the diversification of the major lineages of Ascomycota (Fungi). PLoS One 8:e65576. 10.1371/journal.pone.0065576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q. W., He C. J. (2004). Morphological accumulation of karst cave and landform development in Guizhou. J. Guizhou Norm. Univ. 22 24–26. 10.16614/j.cnki.issn1004-5570.2004.02.007 [DOI] [Google Scholar]
- Rambaut A., Suchard M. A., Xie D., Drummond A. J. (2014). Tracer v1.6. Available at: http://beast.bio.ed.ac.uk/Tracer [Google Scholar]
- Rannala B., Yang Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Ribera I., Fresneda J., Bucur R., Izquierdo A., Vogler A. P., Salgado J. M., et al. (2010). Ancient origin of a Western Mediterranean radiation of subterranean beetles. BMC Evol. Biol. 10:29. 10.1186/1471-2148-10-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikkinen J. (2003). Calicioid lichens from European Tertiary amber. Mycologia 95 1032–1036. 10.2307/3761911 [DOI] [PubMed] [Google Scholar]
- Rohde K. (1992). Latitudinal gradients in species diversity: the search for the primary cause. Oikos 65 514–527. 10.2307/3545569 [DOI] [Google Scholar]
- Rouch R., Danielopol D. L. (1987). L’origine de la faune aquatique souterraine, entre le paradigme du refuge et le modèle de la colonization active. Stygologia 3 345–372. [Google Scholar]
- Schoch C. L., Sung G. H., López-Giráldez F., Townsend J. P., Miadlikowska J., Hofstetter V., et al. (2009). The Ascomycota tree of life: a phylum wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 58 224–239. 10.1093/sysbio/syp020 [DOI] [PubMed] [Google Scholar]
- Sket B. (2008). Can we agree on an ecological classification of subterranean animals? J. Nat. Hist. 42 1549–1563. 10.1080/00222930801995762 17923422 [DOI] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung G. H., Hywel-Jones N. L., Sung J. M., Luangsa-ard J. J., Shrestha B., Spatafora J. W. (2007). Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57 5–59. 10.3114/sim.2007.57.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung G. H., Poinar G. O., Spatafora J. W. (2008). The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal-arthropod symbioses. Mol. Phylogenet. Evol. 49 495–502. 10.1016/j.ympev.2008.08.028 [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Berbee M. L. (2006). Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia 98 838–849. 10.3852/mycologia.98.6.838 [DOI] [PubMed] [Google Scholar]
- Taylor T. N., Hass H., Kerp H. (1999). The oldest fossil ascomycetes. Nature 399 648–648. 10.1038/21349 [DOI] [PubMed] [Google Scholar]
- Vanderwolf K. J., Malloch D., McAlpine D. F., Forbes G. J. (2013). A world review of fungi, yeasts, and slime molds in caves. Int. J. Speleol. 42 77–96. 10.5038/1827-806X.42.1.9 [DOI] [Google Scholar]
- White T. J., Bruns T. D., Lee S. B., Taylor J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (San Diego, CA: Academic Press; ), 315–322. [Google Scholar]
- Wright S. D., Gillman L. N., Ross H. A., Keeling D. J. (2010). Energy and the tempo of evolution in amphibians. Glob. Ecol. Biogeogr. 19 733–740. 10.1111/j.1466-8238.2010.00549.x 28035890 [DOI] [Google Scholar]
- Xiong K. N., Zhu W. X. (1994). Karst geomorphology and speleogenesis of the Zhijin Cave area. Caisol. Sin. 3 281–292. [Google Scholar]
- Zhang C. Z. (1995). Small Myriapoda in soil from China III: a new species of the millipede genus Nanocondylodesmus Zhang (Diplopoda: Polydesmida: Doratodesmidae). Acta Zootaxonom. Sin. 20 416–419. [Google Scholar]
- Zhang Y. H., Zhu D. H. (2012). Large Karst Caves Distribution and Development in China. J. Guilin Univ. Technol. 32 20–28. 10.3969/j.issn.1674-9057.2012.01.003 [DOI] [Google Scholar]
- Zhang Z. F., Liu F., Zhou X., Liu X. Z., Liu S. J., Cai L. (2017). Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia 39 1–31. 10.3767/persoonia.2017.39.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O., Gogarten J. P. (2002). Bootstrap, bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3:4. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specimens used in this study with GenBank accession numbers (xlsx).
Information about the priors and parameters used in BEAST to obtain the divergence time.