Abstract
Histone methylation is closely associated with the occurrence of cancer. Histone H3 trimethylation at lysine 4 (H3K4me3) has been reported to modulate the expression of tumor-associated expression and be altered during the progression of several human cancers. WD Repeat Domain 82 (Wdr82), a key epigenetics-associated factor, is a component of the H3K4me3 methyltransferase complex. An aim of the present study was to determine H3K4me3 and Wdr82 expression and their clinical significances in colorectal cancer (CRC). Immunohistochemistry results demonstrated that the expression level of the H3K4me3 and Wdr82 were significantly decreased in CRC tissues compared with paired noncancerous tissues from 123 patients with CRC. Furthermore, the negative expression of H3K4me3 and Wdr82 expression were significantly associated with lymph node (n=33, P=0.0001) and liver metastasis (n=30, P=0.0001). Additionally, multivariate Cox regression analysis indicated that the low expression level of H3K4me3 or Wdr82 was associated with reduced overall survival (OS, P<0.05), and patients with a low H3K4me3 and Wdr82 expression had a significantly poorer outcome compared with patients with a high expression of H3K4me3 and Wdr82 (P=0.0001), suggesting that H3K4me3 and Wdr82 expression were independent factors for OS in patients with CRC. In conclusion, the decreased expressions of H3K4me3 and Wdr82 were associated with a poor prognosis in CRC. The combined expression of H3K4me3 and Wdr82 may serve as a novel prognostic marker for CRC.
Keywords: colorectal cancer, immunohistochemistry, clinicopathology, overall survival
Introduction
Colorectal cancer (CRC) is known as a widespread malignant tumor, and represents the third most commonly occurring cancer in men and women worldwide (1). It has been reported that the incidence of colorectal cancer tended to occur in younger individuals (2). While the incidence rate in people ≥50 years has declined, the incidence among people aged 20–49 years has increased (2). By 2020 and 2030, the incidence rate for colorectal cancer are expected to increase by ~44 and 107%, respectively, for patients aged 20–34 years old (2). Approximately one in five patients is diagnosed with metastatic disease, while 30–41% of patients develop metastasis in the process of the evolution of disease (3). Unfortunately only a small fraction of patients with metastatic CRC can undergo curative resection and progress with disease-free survival (4). The inconvenience of endoscopy, the golden criteria of CRC diagnosis and the lack of reliable biomarkers culminate in a late diagnosis of malignancy, distant metastasis and consequently low 5-year survival rates (5). Therefore, the identification of prognostic markers may help to reduce CRC mortality (6,7) and incidence (8,9), and to improve the application of presently available therapies.
Trimethylation of histone H3 at lysine4 (H3K4me3) is a post-translational histone modification (10,11). Abnormal H3K4me3 is able to lead to epigenetic alterations, with these alterations resulting in a change in the expression of cancer-associated proteins (12,13) and the regulation of fundamental cancer-associated functions, including growth and metastasis (14,15). Epigenetic alteration is a hallmark of all cancers, therefore; abnormal H3K4me3 has emerged as a mechanism responsible for cancer-associated changes within the epigenome (10,11). H3K4me3 is well characterized to be enriched in gene promoters and is associated with gene transcription (16–19). Levels of H3K4me3 are commonly altered during different cancer development stages, and can be employed as a predictor for cancer recurrence (20–25). The alteration of H3K4me3 has been reported to be important for the activation of genes associated with proliferation and invasion in breast cancer (26). It has also been demonstrated that promoter-linked H3K4me3 is associated with variant enhancer loci in the colon cancer transcriptome (27). In addition, H3K4me3 was reported to be principally catalyzed by the histone methyltransferase human SETD1A (hSETD1A), which is an important co-activator of gene transcription (28). HSETD1A has been identified to be associated with multiple cell activities such as proliferation, migration and invasion, and serves an important role in the metastasis of breast cancer tumors (29). These data suggested that H3K4me3 may be involved in cell proliferation and metastasis in cancers.
Wdr82, a key epigenetics-associated factor, is an integral component of the SETD1A complex (30). The SETD1A complex is a principal enzyme that is responsible for global histone H3K4 trimethylation in mammalian cells, which is highly associated with transcriptional activation. It is associated with RNAP II containing Ser5-P CTD and is tethered to RNAP II by Wdr82 (31). Furthermore, it has been demonstrated that hSETD1A regulates Wnt signaling and controls cellular growth in CRC (25). Depletion of Wdr82 was demonstrated to decrease SETD1A expression and decrease the occupancy of the SETD1A complex and histone H3K4me3 near the transcription start site of transcribed genes (30), suggesting that Wdr82 and H3K4me3 may serve roles in the biological functions of cancer cells. However, the expression and clinical significance of Wdr82 in the progression of cancers are still unclear, and the clinical and histopathological features of H3K4me3 in CRC have yet to be established.
In the present study, the expression levels of Wdr82 and H3K4me3 and prognosis of patients CRC were investigated in a large cohort, and the differences between different clinical parameters were compared to determine the role of Wdr82 and H3K4me3 in tumorigenesis and tumor development. The present study demonstrated that Wdr82 and H3K4me3 may be a promising predictor of prognosis in patients with CRC.
Materials and methods
Patients and tissues
The Ethics Committee of The Harbin Medical University (Harbin, China) approved the research project. CRC cases were identified from the pathology files of the Harbin Medical University Cancer Hospital and patients provided written informed consent. A total of 123 cases record materials from the Department of Pathology at the Harbin Medical University Cancer Hospital and the Department of colorectal surgery at Harbin Medical University Cancer Hospital during the period between January 2005 to December 2015. The present study comprised 73 males and 50 females aged between 29–88 years old (mean age was 58.8 years). Of those, in accordance with the postoperative pathological Tumor-Node-Metastasis stage (32), the 123 patients were divided into four groups. The number of patients in each group are shown in Table I. Paraffin-embedded colorectal tissues and adjacent normal colorectal tissues were gathered retrospectively. Histopathological and clinical data, histopathological diagnosis, pathological stage and tumor grade were extracted from archival material. Primary carcinomas were assessed according to the 7th edition American Joint Committee on Cancer (AJCC) staging system (32). The present study used the tissue containing Wdr82 and H3K4me3 including normal mucosa, neoplastic tissue, lymph node and liver metastases to perform immunohistochemical analyses. The inclusion criteria for the present study were as follows: i) All patients with primary tumors underwent complete surgical resection and; ii) No patients received neoadjuvant therapy prior to surgical resection. Tumor characteristics and demographic of the patients are detailed in Table I.
Table I.
Wdr82 and H3k4me3 staining in tumor cells and associations with clinicopathological characteristics.
| Wdr82 | H3k4me3 | |||||||
|---|---|---|---|---|---|---|---|---|
| Clinicopathological parameter | No. | Negative 36 cases (%) | Positive 87 cases (%) | P-value | No. | Negative 59 cases (%) | Positive 64 cases (%) | P-value |
| Age (years) | 0.969 | 0.969 | ||||||
| <60 | 68 | 20 (29.4) | 48 (70.6) | 68 | 31 (45.6) | 37 (54.4) | ||
| ≥60 | 55 | 16 (29.1) | 39 (70.9) | 55 | 28 (50.9) | 27 (49.1) | ||
| Sex | 0.510 | 0.995 | ||||||
| Male | 73 | 23 (31.5) | 50 (68.5) | 73 | 35 (47.9) | 38 (52.1) | ||
| Female | 50 | 13 (26.0) | 37 (74.0) | 50 | 24 (48.0) | 26 (52.0) | ||
| Location | 0.706 | 0.82 | ||||||
| Colon | 45 | 12 (26.7) | 33 (73.3) | 45 | 22 (48.9) | 23 (51.1) | ||
| Rectum | 78 | 23 (30.0) | 55 (70.0) | 78 | 37 (47.4) | 41 (52.6) | ||
| Tumor size (cm) | 0.086 | 0.902 | ||||||
| <5 | 57 | 21 (36.8) | 36 (63.2) | 57 | 27 (47.4) | 30 (52.6) | ||
| ≥5 | 66 | 15 (22.7) | 51 (77.3) | 66 | 32 (48.5) | 34 (51.5) | ||
| Grade | 0.023a | 0.142 | ||||||
| Low (low, middle and low differentiation, mucinous adenocarcinoma) | 77 | 17 (22.1) | 60 (77.9) | 77 | 33 (42.9) | 44 (57.1) | ||
| High (high, high and middle, middle differentiation adenocarcinoma) | 46 | 19 (41.3) | 27 (58.7) | 46 | 26 (56.5) | 20 (43.5) | ||
| pT classification | 0.719 | 0.616 | ||||||
| T1-T3 | 55 | 17 (30.9) | 38 (69.1) | 55 | 25 (45.5) | 30 (54.5) | ||
| T4 | 68 | 19 (27.9) | 49 (72.1) | 68 | 34 (50.0) | 34 (50.0) | ||
| pN classification | 0.492 | 0.181 | ||||||
| N0 | 65 | 18 (27.7) | 47 (72.3) | 65 | 29 (44.6) | 36 (55.4) | ||
| N1-N2 | 58 | 18 (31.0) | 40 (69.0) | 58 | 30 (51.7) | 28 (48.3) | ||
| AJCC stage | 0.716 | 0.133 | ||||||
| I | 27 | 9 (33.3) | 18 (66.7) | 27 | 9 (33.3) | 18 (66.7) | ||
| II | 32 | 7 (21.9) | 25 (78.1) | 32 | 14 (43.8) | 18 (56.2) | ||
| III | 36 | 12 (33.3) | 24 (66.7) | 36 | 18 (50.0) | 18 (50.0) | ||
| IV | 28 | 8 (28.6) | 20 (71.4) | 28 | 18 (64.3) | 10 (35.7) | ||
| Wdr82 expression | 0.023a | |||||||
| ≤5.0 (Negative) | 36 | 36 | 23 (63.9) | 13 (36.1) | ||||
| >5.0 (Positive) | 87 | 87 | 36 (41.4) | 51 (58.6) | ||||
| CEA | 0.656 | 0.415 | ||||||
| ≤5.0 (Negative) | 61 | 19 (31.1) | 42 (68.9) | 61 | 27 (44.3) | 34 (55.7) | ||
| >5.0 (Positive) | 62 | 17 (27.4) | 45 (72.6) | 62 | 32 (51.6) | 30 (48.4) | ||
| Total | 123 | 36 (29.3) | 87 (70.7) | 123 | 59 (48.0) | 64 (52.0) | ||
P<0.05. Wdr82, WD repeat domain 82; H3K4me3, Histone H3 trimethylation at lysine 4; AJCC, American Joint Committee on Cancer; pN primary nodes; pT, primary tumor; CEA, carcinoembryonic antigen.
Immunohistochemical (IHC) analysis of paraffin-embedded colorectal tissue
Wdr82 and H3K4me3 expression were evaluated by IHC staining using 4-µm-thick sections. The paraffin-embedded sections were dewaxed in xylene and then rehydrated in 100, 95, 90, 85 and 75% graded series of ethanol solutions for 5 min. Subsequently, antigen retrieval in a steam pressure cooker for 4 min in citrate buffer at pH 6.0 at 120°C. Endogenous peroxidase activity was blocked with 3% H2O2 for 30 min at room temperature. The sections were incubated with primary antibodies against Wdr82 (#AP20978b, 1:25, Abgent Biotech Co., Ltd., Suzhou, China) and H3K4me3 (CI1038, 1:500, Boster Biological Technology, Pleasanton, CA, USA) overnight at 4°C and then incubated with Polink-1 horseradish peroxidase-conjugated secondary antibody (ab150113; 1:500; Abcam, Cambridge, UK) at room temperature for 30 min. 3,3′-diaminobenzidine tetrahydrochloride (DAB) was used for development and the slides were counterstained using Mayer's hematoxylin (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) for 15 sec at room temperature.
Two experienced pathologists blinded to the clinicopathological information scored the H3k4me3 and Wdr82 level in tumor cells by assessing: i) The proportion of positively stained cells: (0, <5%; 1, 6 to 20%; 2, 21 to 50%; 3, 51 to 75%; 4, >75%) and ii) The signal intensity: (0, no signal; 1, weak; 2, moderate; 3, strong). The score was determined by multiplying i and ii (31). The expression level of H3k4me3 and Wdr82 was obtained by counting the positively and negatively stained cells in 5–10 separate −100 or −400 magnified high-powered microscopic fields and calculating the mean percentage of positively stained cells among the total cells per field by an optical microscope. (Olympus Corporation, Tokyo, Japan). A final scores obtained from the extent scores multiplied by intensity scores were used to identify expression levels. Scores of 0–4 were defined as low expression, and 5–12 were defined as high expression.
Western blot analysis
Cells were washed twice using cold PBS, and then lysed in RIPA cell lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) containing a protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA) at 4°C for 15 min. Following centrifugation at 4°C at 12,000 × g for 10 min, the supernatant was collected and quantified using a bicinchoninic acid quantification kit (Beyotime Institute of Biotechnology). The proteins (30 µg) were separated by 10% SDS-PAGE (Beijing Solarbio Science & Technology Co., Ltd.) and transferred to polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA). PVDF membranes were blocked with 5% non-fat dried milk in TBST for 1 h at room temperature, and incubated overnight at 4°C with specific primary antibodies: Wdr82 antibody (1:1,000) and H3K4me3 antibody (1:500) and β-actin antibody (1:500) (TA-09; OriGene Technologies., Inc., Beijing, China). HRP-conjugated secondary antibodies: Goat anti-mouse (1:2,000; cat. no. sc-2005, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) were used for incubation at room temperature for 2 h. Development was performed using ECL-detecting reagent (Tanon Science and Technology Co., Ltd., Shanghai, China) and then was detected using an chemiluminescence gel imaging system (FluorChem HD2, AlphaView software, version 3.4.0.0729, Alpha Innotech, CA, USA).
Reagents and antibodies
Peroxidase blocking solution (S2023, Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) was used. The following antibodies were used in the present study: Rabbit polyclonal anti-Wdr82 (1:200, ab175071, Abcam), rabbit monoclonal anti-H3K4me3 antibody (1:200, OriGene Technologies, Inc., Beijing, China) and Polink-1 HRP-conjugated Goat anti-rabbit IgG secondary antibody (1:1,000, L3012-2, Signalway Antibody LLC, College Park, MD, USA).
Statistical analysis
Data are presented as mean ± standard deviation or standard error of the mean. Statistical significance of the differences between treated samples was determined by using either a two-tailed Student's t-test or analysis of variance followed by the Mann-Whitney U test. Differences were considered statistically significant at P<0.05. A χ2 test was used to analyze the association between the expression of Wdr82 and H3K4me3 and various clinicopathological parameters. Cumulative overall survival (OS) was plotted by the Kaplan-Meier method and the relationship between each of the variables and survival was assessed by log-rank test in a univariate analysis. The parameters were then tested by multivariate Cox proportional hazards model, which was performed to identify independent variables for predicting survival. P<0.05 was considered to indicate a statistically significant difference.
Results
Wdr82 and H3K4me3 expression decreased during carcinogenesis
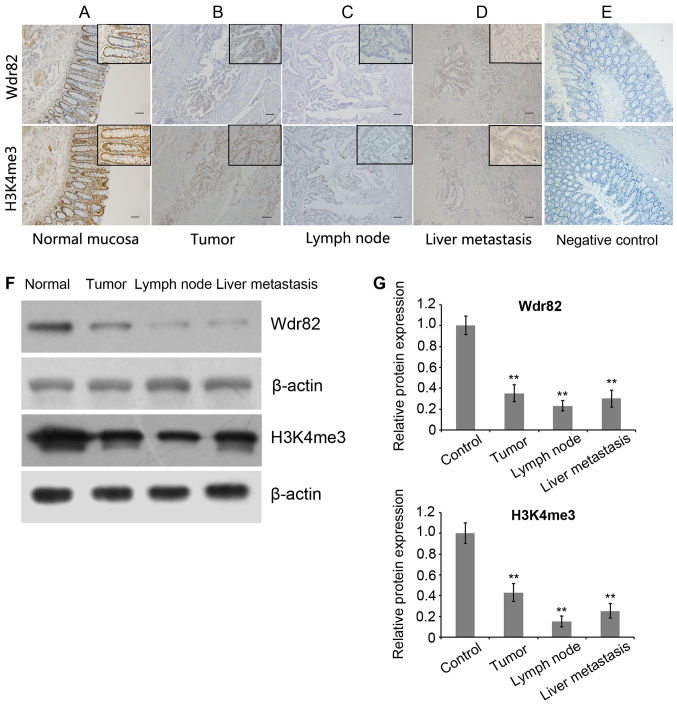
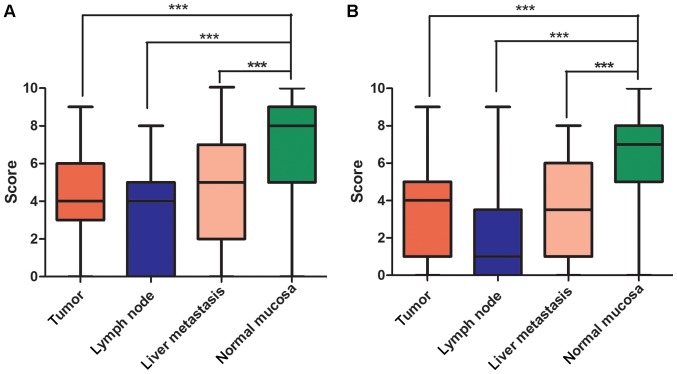
To validate whether Wdr82 and H3K4me3 are involved in the carcinogenesis of CRC, Wdr82 and H3K4me3 expression levels were examined in paired tumors, lymph nodes, liver metastasis and normal mucosa tissues from patients with CRC patients (n=123) by immunohistochemical staining (Fig. 1A-E). As predicted from previous analyses of Wdr82 and H3K4me3 intracellular localization, Wdr82 immunohistochemical staining displayed cytoplasmic, while H3K4me3 appeared as brown particles, primarily localizing within the nuclei of colorectal epithelial cells (Fig. 1). Western blot analysis was used to detect Wdr82 and H3K4me3 protein levels in patient tissues and showed the identical results to immunohistochemical staining (Fig. 1F and G). The Wdr82 (Fig. 2A) and H3K4me3 (Fig. 2B) expressions significantly decreased in carcinomas compared with that in non-cancerous mucosa (n=70, P=0.0007; n=70, P=0.0008). Similarly, the Wdr82 and H3K4me3 expressions in lymph node (n=35, P=0.0006; n=33, P<0.0004) and liver metastasis tissues (n=30, P=0.0009; n=30, P=0.0007) markedly reduced relative to non-cancerous mucosa, respectively (Figs. 1 and 2). These results suggest that Wdr82 and H3K4me3 are involved in tumorigenesis and metastasis of CRC and Wdr82 and H3K4me3 are negatively associated with the progression of CRC.
Figure 1.
Immunohistochemical staining for H3K4me3 and Wdr82 in human colorectal cancer. (A) Expression levels of H3K4me3 and Wdr82 in normal mucosa (n=123) from patients with CRC were detected (magnification, −100 and −400). (B) Expression levels of H3K4me3 and Wdr82 in primary carcinoma tissues (n=70) were detected (−10 and −40 magnification). (C) Expression levels of H3K4me3 and Wdr82 in lymphnode metastasis tissues (n=33) were detected (magnification, −10 and −40). (D) Expression levels of H3K4me3 (n=30) and Wdr82 in liver metastasis tissues (n=30) were examined (magnification, −10 and −40). (E) Negative controls with no primary antibody (magnification, −100 and −400). (F) Western blot analysis for H3K4me3 and Wdr82 in human CRC tissues was performed and. β-actin was detected as an internal reference. (G) Quantification of the western blot analysis showed the amount of interest proteins normalized to the amount of β-actin. **P<0.01 vs. normal mucosa. Data are presented as mean ± standard error of the mean. Wdr82, WD repeat domain 82; H3K4me3, Histone H3 trimethylation at lysine 4.
Figure 2.
Expression of Wdr82 and H3K4me3 were analyzed by immunohistochemistry scores. Wdr82 and H3K4me3 expression decreases from non-cancerous mucosa to carcinomas. (A) Wdr82 and (B) H3K4me3 expressions in normal mucosa, primary carcinoma tissues, lymph node metastasis tissues and liver metastasis tissues were quantified. ***P<0.001. Wdr82, WD repeat domain 82; H3K4me3, Histone H3 trimethylation at lysine 4.
Association of Wdr82 and H3K4me3 with clinicopathological findings
The association between Wdr82 and H3K4me3 expression and clinicopathological features in tissue samples from 123 patients with CRC was investigated. Wdr82 and H3K4me3 staining were also compared in carcinomas according to histological subtype, and the differences between Wdr82 and H3K4me3 expression are illustrated in Table I. Positive expression of Wdr82 was negatively associated with tumor grade (P=0.023) and expression of H3K4me3 was significantly positively correlated with Wdr82 expression (P=0.023). However, there were no significant associations between Wdr82 or H3K4me3 expression and primary tumor location (P=0.820), tumor status (pathological assessment of the primary tumor (pT, P=0.719, P=0.616); pathological assessment of regional lymph nodes (pN, P=0.492, P=0.181)) and AJCC stage (P=0.716, P=0.133). In addition, the tumor marker CEA was not associated with Wdr82 or H3K4me3 expression (P=0.656, P=0.415).
Univariate and multivariate analysis of prognostic factors for overall survival (OS)
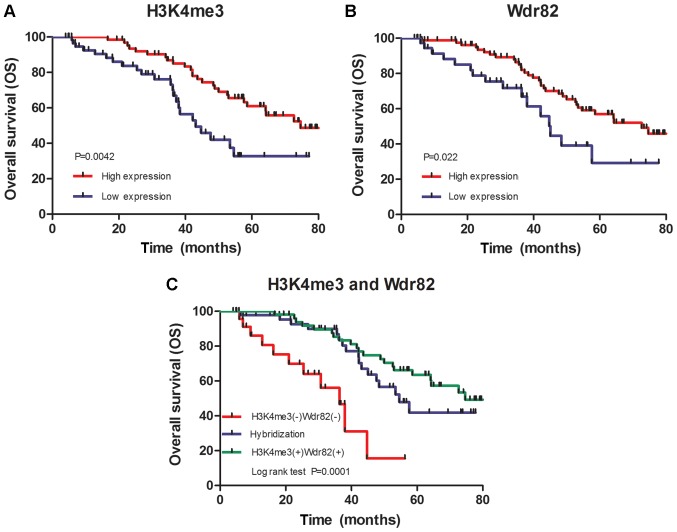
To examine the clinical significance of H3K4me3 and Wdr82 expression, survival analyses were conducted using the Kaplan-Meier curves with a log-rank test. The median OS for the all patients (n = 123) was 45.24 months and the 5-year OS rate was 33.3%. The results demonstrated that patients with a high expression of H3K4me3 or Wdr82 had a significantly improved OS relative to the patients with a low expression of H3K4me3 (50.72 vs. 39.29 months, χ2 = 9.440, P=0.0042) (Fig. 3A) or Wdr82 (47.51 vs. 44.30 months, χ2 = 0.4365, P=0.0224) (Fig. 3B). Especially, patients with a positive expression of H3K4me3 and Wdr82 exhibited a markedly improved OS compared with the patients with a negative expression of H3K4me3 and Wdr82 (P=0.0001, Fig. 3C), suggesting that the expression levels of H3K4me3 and Wdr82 were positively associated with the OS of patients with CRC.
Figure 3.
Kaplan-Meier survival analysis in patients with colorectal cancer. (A) OS curves for patients according to the high and low expression levels of H3K4me3 of immunohistochemical variables in tumor cells. (B) OS curves for patients according to the high and low expression levels of Wdr82 of immunohistochemical variables in tumor cells. (C) OS curves for patients according to the positive and negative expression of H3K4me3 and Wdr82 of immunohistochemical variables in tumor cells. OS, overall survival; Wdr82, WD repeat domain 82; H3K4me3, Histone H3 trimethylation at lysine 4.
Univariate analysis indicated that clinicopathological parameters such as pN classification status, AJCC stage and tumor grade are also of prognostic value, and the expression of H3K4me3 or Wdr82 in tumor cells were potential predictors of OS (Table II). In addition, multivariate Cox model analysis demonstrated that patients with positive H3K4me3 and Wdr82 expression [Hazard ratio (HR) (95% confidence interval CI)], 2.988 (1.591–5.612), P=0.001 and HR (95% CI), 1.980 (1.064–3.847), P=0.032, respectively) had an improved OS in comparison to negative H3K4me3 and Wdr82 expression (Table II). The data suggests that H3K4me3 and Wdr82 expression are independent prognostic indicators of OS in patients with CRC (Table II).
Table II.
Univariate and multivariate analysis of prognostic factors for overall survival.
| Variable | Univariate HR (95% CI) | P-value | Multivariate HR (95% CI) | P-value |
|---|---|---|---|---|
| Sex | ||||
| Male | 1 | |||
| Female | 0.672 (0.370–1.222) | 0.190 | ||
| Age (years) | ||||
| <60 | 1 | |||
| ≥60 | 1.193 (0.681–2.091) | 0.536 | ||
| Location | ||||
| Colon | 1 | |||
| Rectum | 1.257 (0.701–2.253) | 0.441 | ||
| Tumor size (cm) | ||||
| <5 | 1 | |||
| ≥5 | 0.915 (0.522–1.604) | 0.756 | ||
| pT classification | ||||
| T1-T3 | 1 | 1 | ||
| T4 | 2.257 (1.264–4.030) | 0.549 | 0.549 (0.236–1.280) | 0.165 |
| pN classification | ||||
| N0 | 1 | 1 | ||
| N1-N2 | 2.035 (1.155–3.585) | 0.012a | 0.480 (0.098–2.357) | 0.366 |
| AJCC stage | 0.004b | 0.007b | ||
| I | 1 | 1 | ||
| II | 0.210 (0.086–0.513) | 0.001b | 0.895 (0.096–8.308) | 0.922 |
| III | 0.274 (0.116–0.649) | 0.003b | 0.923 (0.114–7.495) | 0.940 |
| IV | 0.378 (0.168–0.848) | 0.018a | 0.191 (0.093–0.604) | 0.001b |
| Grade | ||||
| High | 1 | 1 | ||
| Low | 2.150 (1.209–3.825) | 0.008b | 0.614 (0.325–1.163) | 0.134 |
| Wdr82 expression | ||||
| Negative | 1 | 1 | ||
| Positive | 0.499 (0.272–0.916) | 0.032a | 1.980 (1.064–3.847) | 0.032a |
| H3K4me3 expression | ||||
| Negative | 1 | 1 | ||
| Positive | 2.283 (1.280–4.072) | 0.005b | 2.988 (1.591–5.612) | 0.001b |
| CEA | ||||
| Negative | 1 | 1 | ||
| Positive | 0.488 (0.695–2.140) | 0.487 | 0.206 (0.025–1.678) | 0.140 |
P<0.05
P>0.001. Wdr82, WD repeat domain 82; H3K4me3, Histone H3 trimethylation at lysine 4; HR, hazard ratio; CI, confidence interval; AJCC, American Joint Committee on Cancer; pN, primary nodes; pT, primary tumor; CEA, carcinoembryonic antigen.
Discussion
The prevalence of CRC means it is one of the most commonly occurring cancer types worldwide (33). Several biomarkers, including CEA, CA199 and CA125, are associated with clinical significances and have been explored in scientific research or used in clinical practice; however, its diagnostic applications have thus far been limited (33). The inconvenience of endoscopy, the golden criteria of CRC diagnosis and the lack of reliable biomarkers lead to a late diagnosis of malignancy, distant metastasis and consequently poor survival rates (5). Therefore, the identification of prognostic markers may be significant in the reduction of CRC mortality and improving the presently available treatments.
Epigenetics serves an important role in a variety of cellular biological processes, such as proliferation, migration and invasion (29). H3K4 methylation is associated with transcriptional activation, altered in several human cancer types (29) and H3K4me3, which can be catalyzed to the histone methyltransferase SETD1A (28), has been reported to regulate the activation of cancer-associated genes and be associated with growth and invasion in breast cancer (26). Wdr82, is a component of the H3K4me3 methyltransferase complex, which has been demonstrated to affect the levels of Setd1A expression and histone H3K4me3 near the transcription start site of transcribed genes (30). However, the associations between Wdr82 and cancer progression and the physiological functions of H3K4me3 and Wdr82 in cancer remain unclear. In the present study, the results indicated, to the best of our knowledge for the first time that Wdr82 expression level in tumor cells is significantly and independently associated with the OS of patients with CRC. An association between H3K4me3 and Wdr82 expression and cancer progression and prognosis in CRC was established, and the expressions of H3Ke3 and WdrR82 in tumor, lymph node or liver metastasis in tissues were identified to be significantly decreased compared with the normal mucosa in patients with CRC. The present study also demonstrated that the increased expressions of H3K4me3 and Wdr82 were associated with an improved prognosis in CRC, suggesting that H3K4me3 and functioned as suppressors for CRC.
Post-translational modifications of histones, including phosphorylation, ubiquitination, acetylation and methylation, have been associated with the regulation of gene expression (34). Histone methylation has been found for over 30 years (35) and site-specific histone methylation and the corresponding methyltransferases have also been identified by previous studies (36,37). The catalytic core for the majority of histone lysine methyltransferases is located in the SET domain (36). The SET domain is an evolutionarily conserved motif, with homologues present in organisms ranging from yeast to humans (38). Set1 methyltransferase is responsible for histone H3 lysine 4 (H3K4) methylation and catalyzes the addition of up to three methyl groups to the substrate (39–42). Setd1A belongs to the Set1 family of H3K4 methyltransferases (43). H3K4me3 is relative to the transcription start site of transcribed genes and the human Setd1A histone H3-Lys4 methyltransferase complex was identified to be relative to the RNA polymerase II large subunit by binding to the Ser5-phosphorylated C-terminal domain (CTD) of RNA polymerase II (17). Wdr82, a C-terminal domain-binding protein, quickly identifies Ser5-P CTD (30) and recruits the Setd1A histone H3-Lys4 methyltransferase complex through the RNA recognition motif domain of Setd1A to transcription start sites of transcribed human genes, therefore regulating gene expression (30).
Human Setd1A has been demonstrated to be upregulated in multiple metastatic breast cancer cell lines and clinical tumor specimens (29). A high expression of H3K4me3 was associated with a poor prognosis in hepatocellular carcinoma (22). However, in the present study, the expression of Wdr82 and H3K4me3 were identified to be inversely associated with lymph node and liver metastasis and predicted an improved prognosis in CRC. Additionally, it has also been reported that patients with low levels of H3K4me1-3 exhibited a shorter overall survival time in renal cell carcinoma (20). The above results seemed to be conflicting; however, considering that each cancer is unique, protein expression and clinical outcomes differ between different tumor types (44). Setd1A has been reported to regulate Wnt target genes and controls the tumor growth of colorectal cancer cells (25). Furthermore, Setd1A modulates cell cycle progression and tumorigenesis through regulating p53 target genes in mouse xenograft models, suggesting that the markedly specific genetic consequences are associated with alterations in chromatin modulators in cancer. However, the underlying mechanism of regulation of Wdr82 and H3K4me3 in CRC tumorigenesis remains unclear. Whether these cancer-associated genes are involved in epigenetic modulation in CRC requires further investigation.
To conclude, the present study determined the association between H3K4me3 and Wdr82 expression and prognosis in CRC, and identified that increased expressions of H3K4me3 and Wdr82 were associated with an improved prognosis in CRC. The combined expression of H3K4me3 and Wdr82 may represent a novel and promising prognostic marker for CRC.
Acknowledgements
The authors would like to thank the Key Laboratory of Myocardial Ischemia Ministry of Education (Harbin, China) for their expert technical assistance.
Funding
This project was supported by the National Natural Science Foundation of China (grant nos. 1272704 and 81402367), the Science and Technology Bureau of Harbin (grant nos. 2014RFQGJ and 2015RAXYJ063) and Research Fund for the Doctoral Program of Higher Education (grant no. 20132307120012), the Heilongjiang postdoctoral fund (grant no. LBH-Z12155) and the Harbin Municipal Science and Technology Committee of Harbin outstanding academic leaders plan (grant no. 2015RAXYJ063).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Author's contributions
YaL and BC designed the study and analyzed the data. HL and YoL wrote the manuscript. HL, YoL, JL, YaL and BC conducted the experiments. HL and YoL performed statistical analyses. HL, YoL and JL provided clinical samples. HL, YoL, JL, YaL and BC were involved in discussions regarding experimental protocols and data. YaL and BC provided logistic and budget support.
Ethics approval and consent to participate
This retrospective study was approved by the ethics committee of Harbin Medical University Cancer Hospital (Harbin, China).
Consent for publication
All patients provided written informed consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bandyopadhyay A, Wang L, Agyin J, Tang Y, Lin S, Yeh IT, De K, Sun LZ. Doxorubicin in combination with a small TGFbeta inhibitor: A potential novel therapy for metastatic breast cancer in mouse models. PLoS One. 2010;5:e10365. doi: 10.1371/journal.pone.0010365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey CE, Hu CY, You N. Increase in incidence of colorectal cancer in young adults, rates expected to rise. JAMA Surg. 2014. (early release online)
- 3.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. IARC Cancer Base No. 11 [Internet] Lyon, France; International Agency for Research on Cancer; 2013. GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide. [Google Scholar]
- 4.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics. 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 5.Das V, Kalita J, Pal M. Predictive and prognostic biomarkers in colorectal cancer: A systematic review of recent advances and challenges. Biomed Pharmacother. 2017;87:8–19. doi: 10.1016/j.biopha.2016.12.064. [DOI] [PubMed] [Google Scholar]
- 6.Hardcastle JD, Chamberlain JO, Robinson MH, Moss SM, Amar SS, Balfour TW, James PD, Mangham CM. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. Lancet. 1996;348:1472–1477. doi: 10.1016/S0140-6736(96)03386-7. [DOI] [PubMed] [Google Scholar]
- 7.Kronborg O, Jørgensen OD, Fenger C, Rasmussen M. Randomized study of biennial screening witha faecal occult blood test: Results after nine screening rounds. Scand J Gastroenterol. 2004;39:846–851. doi: 10.1080/00365520410003182. [DOI] [PubMed] [Google Scholar]
- 8.Mandel JS, Church TR, Ederer F, Bond JH. Colorectal cancer mortality: Effectiveness of biennialscreening for fecal occult blood. J Natl Cancer Inst. 1999;91:434–437. doi: 10.1093/jnci/91.5.434. [DOI] [PubMed] [Google Scholar]
- 9.Hewitson P, Glasziou P, Watson E, Towler B, Irwig L. Cochrane systematic review of colorectalcancer screening using the fecal occult blood test (Hemoccult): An update. Am J Gastroenterol. 2008;103:1541–1549. doi: 10.1111/j.1572-0241.2008.01875.x. [DOI] [PubMed] [Google Scholar]
- 10.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Ling JQ, Hoffman AR. Epigenetics of long-range chromatin interactions. Pediatr Res. 2007;61:R11–R16. doi: 10.1203/pdr.0b013e31804575db. [DOI] [PubMed] [Google Scholar]
- 13.Low D, Mizoguchi A, Mizoguchi E. DNA methylation in inflammatory bowel disease and beyond. World J Gastroenterol. 2013;19:5238–5249. doi: 10.3748/wjg.v19.i32.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SK, Jang HR, Kim JH, Noh SM, Song KS, Kim MR, Kim SY, Yeom YI, Kim NS, Yoo HS, Kim YS. The epigenetic silencing of LIMS2 in gastric cancer and its inhibitory effect on cell migration. Biochem Biophys Res Commun. 2006;349:1032–1040. doi: 10.1016/j.bbrc.2006.08.128. [DOI] [PubMed] [Google Scholar]
- 15.Dai Y, Duan H, Duan C, Zhou R, He Y, Tu Q, Shen L. Down-regulation of TCF21 by hypermethylation induces cell proliferation, migration andinvasion in colorectal cancer. Biochem Biophys Res Commun. 2016;469:430–436. doi: 10.1016/j.bbrc.2015.09.109. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 18.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellinger J, Kahl P, Mertens C, Rogenhofer S, Hauser S, Hartmann W, Bastian PJ, Büttner R, Müller SC, von Ruecker A. Prognostic relevance of global histone H3 lysine 4 (H3K4) methylation in renal cell carcinoma. Int J Cancer. 2010;127:2360–2366. doi: 10.1002/ijc.25250. [DOI] [PubMed] [Google Scholar]
- 20.Ellinger J, Kahl P, von der Gathen J, Rogenhofer S, Heukamp LC, Gütgemann I, Walter B, Hofstädter F, Büttner R, Müller SC, et al. Global levels of histone modifications predict prostate cancer recurrence. Prostate. 2010;70:61–69. doi: 10.1002/pros.21038. [DOI] [PubMed] [Google Scholar]
- 21.He C, Xu J, Zhang J, Xie D, Ye H, Xiao Z, Cai M, Xu K, Zeng Y, Li H, Wang J. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum Pathol. 2012;43:1425–1435. doi: 10.1016/j.humpath.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Ke XS, Qu Y, Rostad K, Li WC, Lin B, Halvorsen OJ, Haukaas SA, Jonassen I, Petersen K, Goldfinger N, et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 2009;4:e4687. doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18:867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salz T, Li G, Kaye FJ, Zhou L, Qiu Y, Huang S. hSETD1A regulates Wnt target genes and controls tumor growth of colorectal cancer cells. Cancer Res. 2014;74:775–786. doi: 10.1158/0008-5472.CAN-13-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JH, Sharma A, Dhar SS, Lee SH, Gu B, Chan CH, Lin HK, Lee MG. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336:736–739. doi: 10.1126/science.1217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shilatifard A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282:13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Skalnik DG. WDR82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–618. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Ye SB, Ma G, Tang XF, Chen SP, He J, Liu WL, Xie D, Zeng YX, Li J. The expressions of MIF and CXCR4 protein in tumor microenvironment are adverseprognostic factors in patients with esophageal squamous cell carcinoma. J Transl Med. 2013;11:60. doi: 10.1186/1479-5876-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 33.Feng B, Zheng MH, Zheng YF, Lu AG, Li JW, Wang ML, Ma JJ, Xu GW, Liu BY, Zhu ZG. Normal and modified urinary nucleosides represent novel biomarkers for colorectal cancer diagnosis and surgery monitoring. J Gastroenterol Hepatol. 2005;20:1913–1919. doi: 10.1111/j.1440-1746.2005.03888.x. [DOI] [PubMed] [Google Scholar]
- 34.Murray K. The occurrence of iε-N-Methyl lysine in histones. Biochemistry 127: 10–15, 1964. Lachner M, O'Sullivan RJ, Jenuwein T, editors. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 35.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol 14: R546–R551, 2004. Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T, editors. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 36.Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 38.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 40.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roguev A, Schaft D, Shevchenko A, Pijnappel WWMP, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alicea-Velázquez NL, Shinsky SA, Loh DM, Lee JH, Skalnik DG, Cosgrove MS. Targeted disruption of the interaction between WD-40 repeat protein 5 (WDR5) and mixed lineage leukemia (MLL)/SET1 family proteins specifically inhibits MLL1 and SETd1A Methyltransferase complexes. J Biol Chem. 2016;291:22357–22372. doi: 10.1074/jbc.M116.752626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tajima K, Yae T, Javaid S, Tam O, Comaills V, Morris R, Wittner BS, Liu M, Engstrom A, Takahashi F, et al. SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat Commun. 2015;6:8257. doi: 10.1038/ncomms9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahlberg SH, Spiegelberg D, Glimelius B, Stenerlöw B, Nestor M. Evaluation of cancer stem cell markers CD133, CD44, CD24: Association with AKT isoforms and radiation resistance in colon cancer cells. PLoS One. 2014;9:e94621. doi: 10.1371/journal.pone.0094621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.