Abstract
The dynamics of cellular signaling events are tightly regulated by a diverse set of ubiquitin chains. Recent work has suggested that branched ubiquitin chains composed of Lys11 and Lys48 isopeptide linkages play a critical role in regulating cell cycle progression. Yet, endogenous Lys11/Lys48 branched chains could not be detected. By combining a Lys11 linkage specific antibody with high-resolution middle-down mass spectrometry (an approach termed UbiChEM-MS) we sought to identify endogenous Lys11/Lys48 branched ubiquitin chains in cells. Using asynchronous cells, we find that Lys11-linked branched chains can only be detected upon cotreatment with a proteasome and nonselective deubiquitinase inhibitor. Releasing cells from mitotic arrest results in a marked accumulation of Lys11/Lys48 branched chains in which branch points represent ~3–4% of the total ubiquitin population. This report highlights the utility of UbiChEM-MS in characterizing the architecture of Lys11 Ub chains under various cellular conditions and corroborates the formation of Lys11/Lys48 branched chains during mitosis.
Keywords: polyubiquitin chains, branched ubiquitin chains, mass spectrometry, UbiChEM-MS
Graphical abstract
INTRODUCTION
Ubiquitin (Ub) regulates the dynamics of every biochemical pathway in eukaryotes.1 Most of Ub’s function stems from the formation of an immense array of polymeric Ub chains.2–4 Different chains are recognized by distinct effector proteins to transmit information into a biological outcome.5 How, when, why, and where specific Ub chains are formed, recognized, and removed all remain open and pressing questions in biology.
Ub chains are produced through the concerted actions of E1, E2 and E3 enzymes.6 Once a substrate protein is selected by an E3 and decorated with a single Ub moiety, additional rounds of modification often target Ub itself resulting in the formation of Ub chains. Single linkage (homotypic), mixed linkage (heterotypic), and branched chains can all be formed since there are eight possible linkages (Met1, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) that can be made between Ub molecules.2–4 Quantitative proteomics has shown that all eight linkage types are generated in cells7–10 and there are certain signaling pathways that require the formation of specific linkage types, indicating different Ub chains do not have redundant functions.11–15
The “ubiquitin code” hypothesis is based on the idea that each Ub chain regulates the dynamics of a cellular pathway in a distinct way.2–4 The paradigm is based on Lys48 and Lys63 homotypic chains. The former targets substrate proteins to the proteasome for degradation, while the latter regulates protein trafficking and the assembly of large multiprotein signaling complexes.16,17 With exception of Met1 chains, which are vital for immune signaling,15 much less is known about the other chain types.
The function of Lys11 chains is particularly controversial. The first reports showed Lys11 chains produced by the anaphase-promoting complex/cyclosome (APC/C) are necessary for triggering proteasomal degradation during mitosis.14,18–21 Key to these findings was the advent of a Lys11 linkage-specific antibody.22 Subsequent studies found Lys11-linked chains attached to membrane receptors,23,24 proteins involved in NF-κB signaling,25 autophagy,26 and endoplasmic reticulum-associated degradation (ERAD).27 In none of these cases are Lys11 chains installed by APC/C nor involved in proteasomal degradation. There is also a recent study indicating Lys11 chains on their own are poor proteasome targeting signals and instead require added Lys48 chains to promote efficient degradation.28 In accord, evidence suggests that APC/C marks substrates for degradation by building Lys11/Lys48 branched chains rather than homotypic Lys11 chains.29 Yet, endogenous Lys11/Lys48 branched chains have not been detected at any point in the cell cycle.
We sought to evaluate whether Lys11/Lys48 branched chains accumulate upon proteasome inhibition and whether their presence is cell cycle dependent. However, identifying and characterizing branched chains in cell extracts is rather challenging due to multiple modifications on a single Ub polypeptide and their predicted low frequency. To overcome these issues, we recently described a method called UbiChEM-MS (ubiquitin chain enrichment middle-down mass spectrometry) that enables characterization of Ub chain architecture in enriched populations from cell extracts.30,31 With this approach, Ub chains composed of a specific linkage are isolated using an immobilized Ub-binding domain. Trypsin-mediated minimal digestion is then performed to remove the C-terminal diGly motif of each Ub and afford a mixture of Ub species representative of mono-Ub or the end-caps of a chain (Ub1–74), singly modified Ub or the linear portion of a chain (GGUb1–74), and the doubly modified form or branch points (2×GGUb1–74). The relative intensities of the three species inform on the distribution of the Ub pool as well as the extent of chain branching.
In this study, we combined the utility of the Lys11 linkage specific antibody (herein referred to as α-Lys11-IgG) with our UbiChEM-MS approach to detect branched chains in cells. Much to our surprise, we find that the Lys11 chains isolated from asynchronous cell populations are short with no observable branching. Proteasome inhibition does little to change the distribution. Only upon inhibiting both the proteasome and deubiquitinases do we observe an increase in the abundance of Ub chains with branch points. Releasing cells from nocodazole-induced mitotic arrest also leads to an accumulation of branched chains. Congruent with studies on APC/C, the branched chains produced during mitosis are composed of Lys11 and Lys48 linkages. Our findings demonstrate that UbiChEM-MS coupled with α-Lys11-IgG effectively captures the global dynamics of Lys11 chains in both linear and branched configurations.
EXPERIMENTAL SECTION
Materials
The following materials were used in this study: pFUSEss-CHIg-hG1 (InvivoGen), pFUSE2ss-CLIg-hk (InvivoGen), Xfect transfection reagent (Takara Clontech; 631318), DMEM, high glucose (ThermoFisher Scientific; 11965118), Fetal Bovine Serum, qualified, USDA-approved regions (ThermoFisher Scientific; 10437028), Super Low IgG Fetal Bovine Serum, US Origin (VWR; 95042), GlutaMAX supplement (ThermoFisher Scientific; 35050061), Trypsin-EDTA (0.25%), phenol red (ThermoFisher Scientific; 25200056), Chloroacetamide (Sigma-Aldrich; C0267), N-Ethylmaleimide (Alfa Aesar; AA4052606), PR619 DUB Inhibitor (Fisher Scientific; NC0818370), MG132 (Enzo Life Sciences; BML-PI102), ProBlockTM gold mammalian protease inhibitor cocktail (Goldbio; GB-331–5), Trypsin Gold, mass spectrometry grade (Promega; V5280), Ubiquitin monoclonal antibody, P4D1 (Enzo Life Sciences; BML-PW0930), Goat antimouse IgG antibody, HRP conjugate (EMD Millipore; 12–349), and Goat antihuman IgG antibody, HRP conjugate (EMD-Millipore; AP112P).
Methods
Asynchronous Cell Culture
HEK293 cells were grown in media containing DMEM, 10% (v/v) FBS, 1× glutamax, 100 U/mL penicillin, and 100 mg/mL streptomycin, at 37 °C, 5% CO2. Once cells reached 80–90% confluency, they were washed with cold PBS, and treated with either DMSO, 10 µM MG132, 30 µM PR619, or combination of 10 µM MG132 and 30 µM PR619 for 4 h. Cells were then harvested by first removing the growth media, then scrapping cells in the presence of cold PBS (2 mL). Cells were pelleted at 1000g, 4 °C for 5 min and the supernatant decanted. Cell pellets were resuspended in cold PBS (10 mL) and pelleted again at 1000g, 4 °C for 10 min. To every 10× 150 mm plates of HEK cells, was added 5 mL of cold lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, 0.05% IGEPAL CA-630, 10 mM 2-chloroacetamide, 10 mM N-ethylmaleamide, and 1× protease inhibitor cocktail (Gold Biotechnology)). In addition, lysis buffer was supplemented with 10 µM MG132, 30 µM PR619, or both. Cells were incubated in lysis buffer on ice for 30 min prior to sonication. The resulting lysates were clarified at 16000g, 4 °C for 30 min.
Nocodazole-Induced Mitotic Arrest
To induce mitotic arrest,32 HEK293 cells were grown in media containing nocodazole (100 ng/mL) for 14 h at 37 °C, 5% CO2. Cells were then gently washed with PBS, and allowed to progress through mitosis by adding normal media without nocodazole for 2 h at 37 °C, 5% CO2. Aliquots of cells were stained with Hoechst 33342 and the cell cycle distribution was analyzed using FlowJo version 10.1 and MACSQuant10.
α-Lys11-IgG Production
DNA sequences encoding the variable regions of light and heavy chain of conformational Lys11-linkage specific antibody22 were cloned into the pFUSE plasmids as recommended by InvivoGen. HEK293 cells were grown in media containing super low IgG Fetal Bovine Serum (FBS) to bring down bovine IgG to negligible levels. HEK cells were cotransfected with CHIg-hG1:CLIg-hk plasmids in 2:3 ratio, as recommended, and allowed to grow for 4 h at 37 °C, 5% CO2. The media was changed and the cells were allowed to grow for 38 h at 37 °C, 5% CO2. The media was pipetted out and saved. The cells were gently washed with cold PBS and combined with the saved media. The combined solution was buffer exchanged into PBS using 50K MWCO Amicon Ultra-15 centrifugal filters, and subsequently concentrated and stored in PBS containing 0.01% sodium azide. The selectivity of this IgG was analyzed by ELISA (Figure S1).
Generation of α-Lys11-IgG:ProteinA Resin
Protein A resin (5 mL, Thermo Scientific Pierce) was washed and equilibrated with 5 column volumes (C.V.) of binding buffer (25 mM Tris pH 7.2, 150 mM NaCl, 0.05% IGEPAL CA-630). Approx. 150 mg of α-Lys11-IgG was added to this resin and incubated on a revolver for 16 h at 4 °C. The suspension was then pelleted at 1000g for 5 min; the supernatant was removed, and the resin was washed with 20 C.V. of binding buffer (Figure S2).
Ubiquitin Chain Enrichment from Cell Lysate
500 µL of α-Lys11-IgG:proteinA resin was incubated with 150 mg of cell lysate on a revolver for 16 h at 4 °C. The resin was pelleted at 1000g for 5 min and then washed with 25 C.V. of a minimal buffer (25 mM Tris, 50 mM NaCl, pH 8) containing a lower concentration of salt than previously used.31 The resin was then resuspended in minimal buffer to bring the final volume to 800 µL. 6× Laemmli loading buffer was added to aliquots of each step, and the enriched ubiquitinated proteins were separated on a 15% SDS-PAGE gel, and analyzed by Western blot with anti-Ub antibody (Figure S3).
On-Resin Minimal Trypsinolysis of Isolated Ub Chains
To a slurry of resin-bound ubiquitinated proteins (800 µL), 7.5 µg of trypsin was added and the mixture was incubated on a revolver for 16 h at room temperature. The on-resin minimal trypsinolysis samples were then acidified to pH 2 with acetic acid to deactivate trypsin. These acidified solutions were incubated for 20 min at 4 °C, and centrifuged for 2 min at 1000g, 4 °C. The supernatant was removed and saved. The resin was resuspended in 500 µL of 0.1% TFA in water, incubated for 20 min at 4 °C, and then centrifuged for 5 min at 1000g, 4 °C. The supernatants were combined and concentrated 8–10 fold using a speedvac. Samples were centrifuged for at least 30 min at 16 000g, 4 °C, prior to injection into the mass spectrometer.
Middle-Down Mass Spectrometry and Quantitative Analysis
Minimally digested samples were separated using a nanoAcquity LC system (Waters) equipped with home-packed PLRP-S column. A multistep gradient over 60 min with solvent A: 0.1% formic acid in water, and solvent B: 0.1% formic acid in 1:1 solution of acetonitrile and ethanol was used for each sample. The column equilibrated in 5% B was brought to 30% B in 5 min and kept at 30% B for another 20 min. The gradient from 30% B to 41% B over next 18 min was critical for removing unwanted peptides/impurities and reproducibly yield quantifiable Ub species. The gradient was increased to 95% B by 43 min and then brought back to 5% B for the last 8 min. The LC system was coupled online with a Bruker Impact II Q-TOF mass spectrometer, where the resolving power of the mass analyzer was set at 50000. All spectra were processed with Compass DataAnalysis 4.3 software using a signal-to-noise (S/N) threshold of 3 and a quality factor of 70%. Maximum Entropy was used as the deconvolution algorithm. Quantitative analysis of the relative abundance of the three Ub1–74 species was performed as described previously.30,31 The percent distribution was calculated from three biological replicates, each with three technical replicates; and data is represented as the mean ± standard error of the mean (SEM).
Tandem Mass Spectrometry (MS/MS) Analysis of Ub Chain Linkages
Minimally digested products were separated using a nanoAcquity LC system (Waters) equipped with home-packed PLRP-S column and multistep gradients over 60 min with solvents A and B aforementioned. The LC system was coupled online with a Bruker maXis II Q-TOF mass spectrometer. For MS/MS experiments, individual charge states of protein molecular ions were isolated then the ions were subjected to electron transfer dissociation (ETD).33 The ranges for accumulation time, reagent time, and reaction time for ETD fragmentation were 2500–4000 ms, 5–20 ms and 1–10 ms, respectively. All spectra were processed with Compass DataAnalysis 4.3 software to generate mass lists which were supplied to MSAlign34 to identify possible sites of diglycine (GG) modification (data shown in Supporting Information). The spectra were also processed with the MASH suite software35,36 using a S/N threshold of 3 and a fit factor of 70%. All reported calculated (calc.) and experimental (expt.) values correspond to the monoisotopic molecular weight.
RESULTS AND DISCUSSION
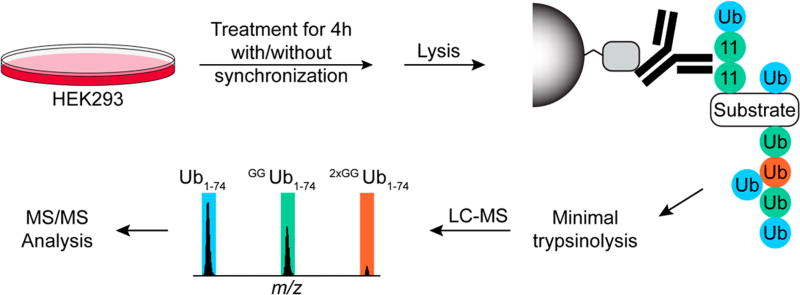
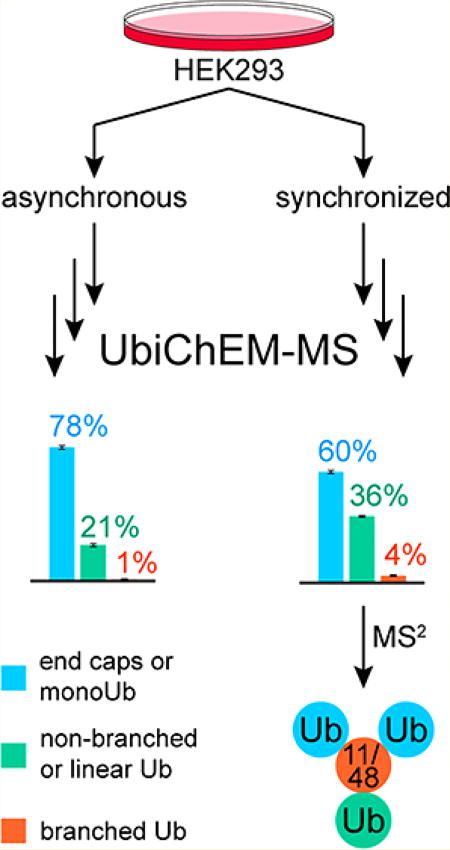
Dynamic changes in Lys11 linkages have been observed during the cell cycle and upon proteasome inhibition, implying that chain comprised of these linkages regulate cell cycle progression by stimulating the degradation of certain cell cycle-mediators.22 More recently, data has suggested that the principal drivers of proteasome-mediated cell cycle progression are not Lys11 homotypic chains but instead heterotypic or branched Lys11/Lys48 chains.28,29 Testing this supposition has been difficult due to the inability to detect endogenous Lys11/Lys48 branched chains using standard bottom-up proteomic approaches. We sought to address this problem using a UbiChEM-MS approach, in which the total population of Lys11 chains is enriched and chain architecture is evaluated using middle-down mass spectrometry (Figure 1).
Figure 1.
General workflow for α-Lys11-IgG based UbiChEM-MS analysis. Batches of cells are grown under different conditions and lysed. The clarified lysates are then incubated with α-Lys11-IgG:Protein A resin to enrich for ubiquitin chains. The isolated chains are subjected to minimal trypsinolysis yielding the three Ub1–74 species. These samples are then characterized by high-resolution mass spectrometry coupled with reverse phase chromatography.
Integrating α-Lys11-IgG into the UbiChEM-MS Workflow
Previously, we demonstrated that two different Ub-binding domains (UBDs)–the nonselective tandem Ub-binding elements (TUBEs) and the Lys29 specific NZF1 domain–could be used to pull-down specific populations of Ub chains to facilitate the analysis of chain architecture.31 Building on this work, we sought to exploit the utility of Ub chain linkage specific antibodies, specifically the Lys11 linkage specific antibody (α-Lys11-IgG). Data has shown α-Lys11-IgG exhibits a high affinity (Kd ~ 12 nM) for Lys11-linked chains.22
We generated α-Lys11-IgG and tested it against a panel of Ub dimers to confirm selectivity toward Lys11 linkages. As previously described,37 the synthetic bivalent Fab antibody fragment specific for Lys11 linkages was converted into a full-length immunoglobulin (IgG) in HEK293 cells. Using an enzyme-linked immunosorbent assay (ELISA) with four different Ub dimers along with monomeric Ub immobilized on a plate, the isolated α-Lys11-IgG was indeed found to be selective for Lys11 Ub dimers (Figure S1). These results are consistent with previously published reports on the selectivity of α-Lys11-IgG.
With the α-Lys11-IgG antibody in hand, we sought to apply it in our UbiChEM-MS workflow (Figure 1). During the initial stages of method development, we encountered issues that were unique in comparison to the previously evaluated UBDs. Under all circumstances, UbiChEM-MS requires minimal trypsinolysis under nondenaturing conditions. In the case of α-Lys11-IgG, Ub species could not be detected by LC–MS after on-resin minimal trypsinolysis. We reasoned that this could be due to a high abundance of coeluting antibody fragments. To overcome this, we modified the minimal trypsinolysis conditions and LC gradient (see Methods). Adhering to the revised method led to reproducible MS detection levels of Ub species.
UbiChEM-MS Analysis Reveals the Dynamics of Lys11 Branched Chains in Asynchronous Cell Populations
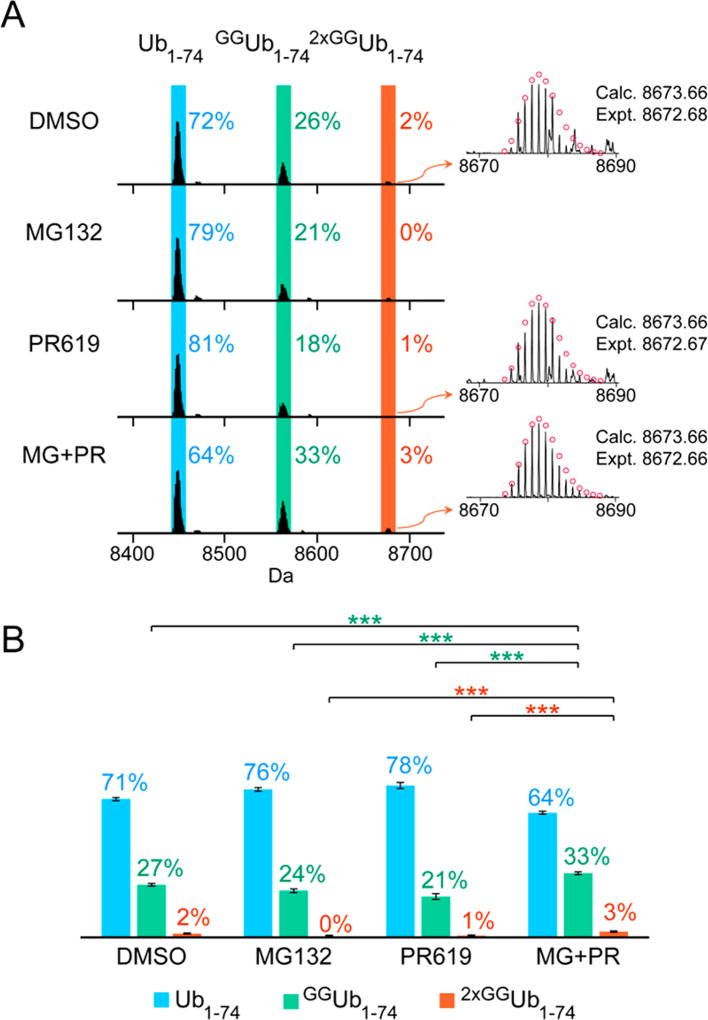
Once we optimized UbiChEM-MS with α-Lys11-IgG, we wanted to first analyze the Lys11 chain population in asynchronously growing HEK293 cells. Consistent with previous characterization data on α-Lys11-IgG, we observe an enrichment of high molecular weight Ub species by Western blot analysis (Figure S5). When we subjected these conjugates to minimal trypsinolysis and high-resolution MS, we were surprised to find the predominant Ub species (72–78%) is Ub1–74, which represents the end-caps of chains or mono-Ub marks (Figures 2 and 3; untreated and DMSO treated). Only 21–26% of the enriched Ub population corresponds to singly modified GGUb1–74 and little (~1%), of the doubly modified 2×GGUb1–74 could be detected. These results suggest α-Lys11-IgG enriches a subpopulation of ubiquitinated proteins that, on average, are tagged with mono-Ub or multiple mono-Ub modifications along with short Lys11 chains. This is reminiscent of APC/C-mediated ubiquitination, as studies examining in vitro reactions have shown that Ub modifications are built in stages.38,39 In the initial phase, APC/C conjugates mono-Ub to several substrate lysines, and then in the second phase, short chains are assembled.
Figure 2.
UbiChEM-MS analysis of Ub chains enriched from asynchronous cells. (A) Representative deconvoluted MS spectra for all the conditions tested highlighting the three minimal digestion products: Ub1–74 (blue), GGUb1–74 (green) and 2×GGUb1–74 (orange). (B) The distribution was calculated by averaging relative abundance of each Ub1–74 species to total abundance of all three Ub1–74 species from three biological replicates for each condition (Table S1). Error bars represent standard error of the mean (SEM) for each data set; * p < 0.05, ** p < 0.01, and *** p < 0.001 (Student’s t test). The reported calculated (calc.) and experimental (expt.) values correspond to the monoisotopic molecular weight.
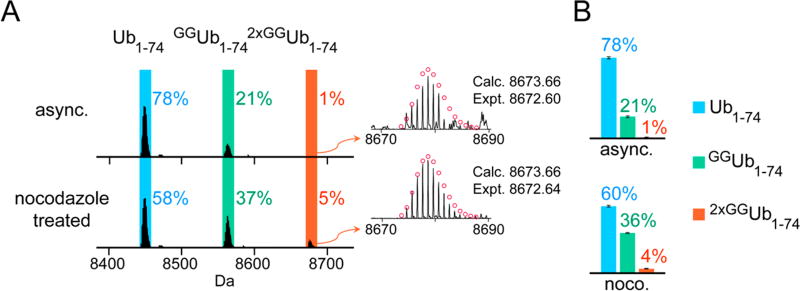
Figure 3.
UbiChEM-MS analysis of Ub chains enriched from untreated asynchronous cells and nocodazole treated cells. (A) Representative deconvoluted MS spectra for all the conditions tested highlighting the three minimal digestion products: Ub1–74 (blue), GGUb1–74 (green) and 2×GGUb1–74 (orange). (B) The distribution was calculated by averaging relative abundance of each Ub1–74 species to total abundance of all three Ub1–74 species from three biological replicates for each condition (Table S2). Error bars represent standard error of the mean (SEM) for each data set. The reported calculated (calc.) and experimental (expt.) values correspond to the monoisotopic molecular weight.
Next, we wanted to test whether proteasome inhibition alters the distribution of Ub species pulled-down by α-Lys11-IgG. Cells were treated with the inhibitor MG132 for 4 h. Previous studies have shown that proteasome inhibition increases the abundance of Lys11 linkages in a number of asynchronous cell populations.7–10,40 UbiChEM-MS analysis reveals the distribution of Ub species—Ub1–74, GGUb1–74, and 2×GGUb1–74 —remains similar to as that observed with untreated or DMSO treated cells (Figures 2 and 3). Thus, while the total amount of Ub chains increases upon proteasome impairment, the overall distribution of α-Lys11-IgG-enriched chains does not significantly change. We reasoned that perhaps by compromising proteasome function there is an accumulation of Ub chains that serve as substrates of deubiquitinases. According to the principles of steady-state kinetics, we would expect that increasing substrate concentrations would lead to faster chain disassembly, which could obscure changes in the distribution of Ub species pulled-down from MG132-treated cells. On the basis of this logic, we surmised the addition of a nonselective deubiquitinase inhibitor, e.g., PR619, along with MG132 would reveal the architecture of Lys11-linked Ub chains that are recruited to the proteasome.41
To test this, we first treated cells with PR619 alone as a control. The MS data shows the distribution of Ub species is again, quite similar to previous samples (Figures 2A and 2B). Upon addition of PR619 and MG132, however, we observe a redistribution of Ub species (Figures 2A and 2B). There is a decrease on mono-Ub/end-caps and a concomitant increase in singly (from ~23% to 33%) and doubly modified (from ~1% to 3%) Ub. These data are based on three biological replicates and three technical replicates, indicating the changes are indeed significant (Figure 2B). Thus, these results suggest the steady-state population of Lys11-linked chains, which is what we observe when cells are not treated with a combination of MG132 and PR619, differs from the population involved in proteasomal degradation. The latter involves long chains with branch points that are rather short-lived.
ETD fragmentation was used to identify the linkages in the branch points. Accounting for all seven possible linkages that could accompany Lys11, the fragmentation pattern best fit Lys11/Lys48 branch point, with 11 c and 14 z• ions identified. The presence of c11 and z•28 ions supports the presence of modifications on Lys11 and Lys48 (Figures S6 and S7). We also analyzed the ETD data using MSAlign, a database search tool used to identify proteins from top-down tandem MS.34 As opposed to manual analysis of ETD data, MSAlign is unbiased, thus, enabling the analysis of all possible combinations of 2×GG modifications. Congruent with our manual approach, MSAlign revealed the formation of Lys11/Lys48 branch points (Figure S8). On the basis of these results, we contend that short-lived chains involved in proteasomal degradation have Lys11 and Lys48 linkages.
Mitotic Arrest Leads to the Accumulation of Lys11/Lys48 Branched Chains
Having established the transient nature of Lys11 branched chains in asynchronous cells, we wanted to evaluate the architecture of Lys11 chains during the cell cycle. Previous work has shown that Lys11 chains increase in abundance as cells advance through mitosis.22 Higher levels of Lys11 linkages correlate precisely with the activity of APC/C. Thus, we surmised that if APC/C is responsible for building branched chains,29 we should be able to detect an accumulation of these chains during mitosis.
To test this, we used the spindle toxin nocodazole to arrest HEK293 cells in mitosis. Cells were treated with nocodazole for 14 h, released for 2 h and then analyzed by flow cytometry. The DNA content is consistent with cells in the G2/M phase of the cell cycle (Figure S4). Subjecting this cell population to the UbiChEM-MS workflow using α-Lys11-IgG reveals a Ub population comprised of 58% Ub1–74, 37% GGUb1–74, and 5% 2×GGUb1–74 (Figures 3A and 3B). This distribution is similar to that observed with the asynchronous cells cotreated with MG132 and PR619; however, there is a slight increase in singly and doubly modified forms of Ub in the synchronized cell population. Our data therefore shows that branched chains accumulate in cells released from nocodazole-induced mitotic arrest, providing unambiguous support for their formation during mitosis.
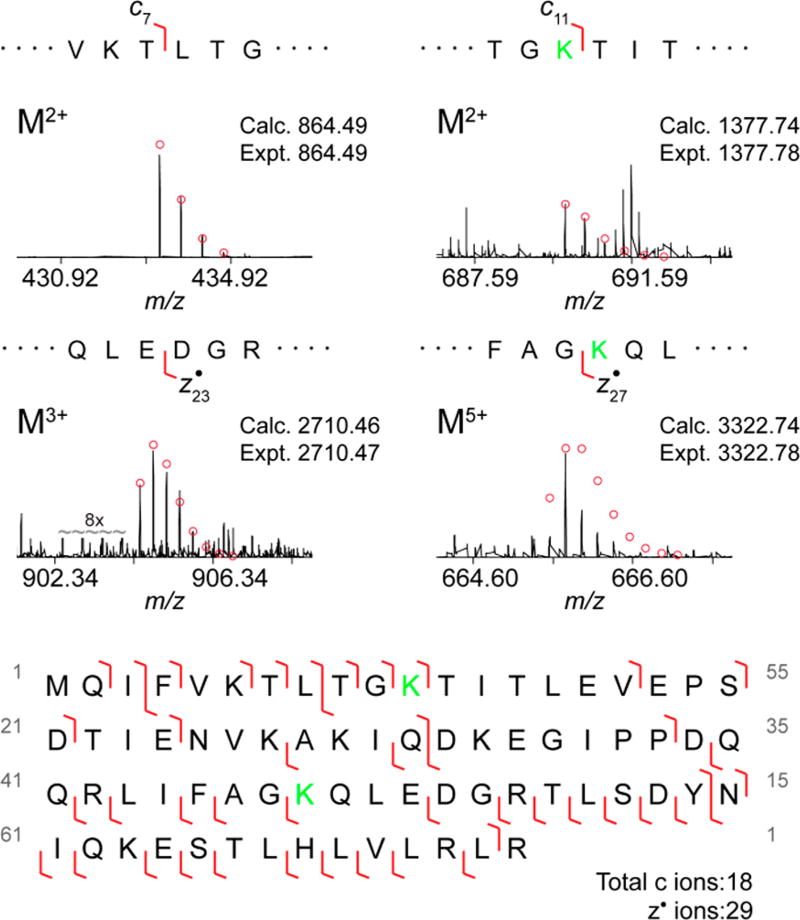
To identify the linkages in branch points, we again employed ETD fragmentation. We focused on the M12+ charge state of the 2×GGUb1–74 species as it provided the most extensive fragmentation. Assuming the presence of a Lys11 linkage, we varied the position of the other linkage to find the best fit to the fragmentation data. With 18 c and 29 z• ions, the presence of both Lys11 and Lys48 provided the best match (Figures 4 and S9). The key fragments are c7, c11, z•23 and z•27 (Figure 4). Both c7 and c11 ions reveal that a diGly modification resides within residues 8 and 11, i.e., Lys11. And the z•23 and z•27 ion shows the other diGly modification is on Lys48. MSAlign confirmed the presence of Lys11 and Lys48 linkages in the branch point (Figure S10). Collectively, these data support the notion that Lys11/Lys48 branched chains are formed in a cell cycle-dependent manner.
Figure 4.
ETD analysis of 2×GGUb1–74 species isolated from synchronized cells. ETD fragments used to deduce the site for the two diGly modifications to be at Lys11 and Lys48. The position of modified lysine is highlighted in green (top). Observed ETD fragments (c and z• ions) mapped onto the sequence of Ub containing GG modifications at Lys11 and Lys48 (bottom).
CONCLUSION
Branched Ub chains are thought to play critical roles in signaling pathways and protein turnover. Detecting branched chains, however, is difficult. It has been proposed that Lys11/Lys48 branched chains are formed by the E3 APC/C to drive proteasome-mediated degradation of cell cycle regulators.29 In this case, the detection of Lys11/Lys48 branched chains relied on overexpressing an engineered form of Ub in cells. Thus, it has remained unclear whether and to what extent Lys11/Lys48 branched chains are generated in a cell cycle-dependent manner.
In this study, we integrated the Lys11 linkage specific antibody, α-Lys11-IgG, into our UbiChEM-MS approach to examine the formation of branched conjugates under different cellular conditions. In asynchronous cells, we uncovered a transient population of Lys11 Ub chains that contain a substantial amount of branching. We propose that this population is involved in proteasomal degradation. When proteasome function is impaired, long Lys11 branched chains begin to accumulate, increasing their susceptibility to the activity of DUBs. Our results demonstrate how the fate of a protein is tightly balanced by the proteasome and DUBs. Counter to the low abundance of branched chains in asynchronous cells, we find that branched conjugates are prevalent in cells released from nocodazole-induced mitotic arrest. These conjugates are comprised of Lys11 and Lys48 linkages, corroborating the formation of Lys11/Lys48 branched chains during mitosis. In future work, it will be interesting to identify DUBs that regulate the abundance of branched conjugates with Lys11 linkages.
Supplementary Material
Acknowledgments
This work was supported by research grant RO1GM110543 from the National Institutes of Health (NIH). Flow cytometry data was collected at UW Carbone Cancer Center, Madison, WI. Y.G. would like to acknowledge NIH grant R01GM117058 and high-end instrument grant S10OD018475.
Footnotes
ASSOCIATED CONTENT
Author Contributions
A.S.J.B.R. designed and conducted all the experiments. A.S.J.B.R., Y.G., and E.R.S. analyzed the results and wrote the manuscript.
The authors declare no competing financial interest.
References
- 1.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 3.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016;129:875–880. doi: 10.1242/jcs.183954. [DOI] [PubMed] [Google Scholar]
- 5.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 6.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 7.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose CM, Isasa M, Ordureau A, Prado MA, Beausoleil SA, Jedrychowski MP, Finley DJ, Harper JW, Gygi SP. Highly Multiplexed Quantitative Mass Spectrometry Analysis of Ubiquitylomes. Cell Syst. 3. 2016;3:395–403. doi: 10.1016/j.cels.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.013284. M111 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol. Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan WC, Lee YR, Lin SY, Chang LY, Tan YP, Hung CC, Kuo JC, Liu CH, Lin MY, Xu M, Chen ZJ, Chen RH. K33-Linked Polyubiquitination of Coronin 7 by Cul3-KLHL20 Ubiquitin E3 Ligase Regulates Protein Trafficking. Mol. Cell. 2014;54:586–600. doi: 10.1016/j.molcel.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 14.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damgaard RB, Walker JA, Marco-Casanova P, Morgan NV, Titheradge HL, Elliott PR, McHale D, Maher ER, McKenzie AN, Komander D. The Deubiquitinase OTULIN Is an Essential Negative Regulator of Inflammation and Autoimmunity. Cell. 2016;166:1215–1230. doi: 10.1016/j.cell.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 18.Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min M, Mevissen TE, De Luca M, Komander D, Lindon C. Efficient APC/C substrate degradation in cells undergoing mitotic exit depends on K11 ubiquitin linkages. Mol. Biol. Cell. 2015;26:4325–4332. doi: 10.1091/mbc.E15-02-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, Kelley RF, Dixit VM. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Goto E, Yamanaka Y, Ishikawa A, Aoki-Kawasumi M, Mito-Yoshida M, Ohmura-Hoshino M, Matsuki Y, Kajikawa M, Hirano H, Ishido S. Contribution of lysine 11-linked ubiquitination to MIR2-mediated major histocompatibility complex class I internalization. J. Biol. Chem. 2010;285:35311–35319. doi: 10.1074/jbc.M110.112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 2010;11:210–220. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, Vucic D. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platta HW, Abrahamsen H, Thoresen SB, Stenmark H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 2012;441:399–406. doi: 10.1042/BJ20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locke M, Toth JI, Petroski MD. Lys11- and Lys48-linked ubiquitin chains interact with p97 during endoplasmic-reticulum-associated degradation. Biochem. J. 2014;459:205–216. doi: 10.1042/BJ20120662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grice GL, Lobb IT, Weekes MP, Gygi SP, Antrobus R, Nathan JA. The Proteasome Distinguishes between Heterotypic and Homotypic Lysine-11-Linked Polyubiquitin Chains. Cell Rep. 2015;12:545–553. doi: 10.1016/j.celrep.2015.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valkevich EM, Sanchez NA, Ge Y, Strieter ER. Middle-down mass spectrometry enables characterization of branched ubiquitin chains. Biochemistry. 2014;53:4979–4989. doi: 10.1021/bi5006305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crowe SO, Rana A, Deol KK, Ge Y, Strieter ER. Ubiquitin Chain Enrichment Middle-Down Mass Spectrometry Enables Characterization of Branched Ubiquitin Chains in Cellulo. Anal. Chem. 2017;89:4428–4434. doi: 10.1021/acs.analchem.6b03675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zieve GW, Turnbull D, Mullins JM, McIntosh JR. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp. Cell Res. 1980;126:397–405. doi: 10.1016/0014-4827(80)90279-7. [DOI] [PubMed] [Google Scholar]
- 33.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Sirotkin Y, Shen Y, Anderson G, Tsai YS, Ting YS, Goodlett DR, Smith RD, Bafna V, Pevzner PA. Protein identification using top-down. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.008524. M111 008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai W, Guner H, Gregorich ZR, Chen AJ, Ayaz-Guner S, Peng Y, Valeja SG, Liu X, Ge Y. MASH Suite Pro: A Comprehensive Software Tool for Top-Down Proteomics. Mol. Cell. Proteomics. 2016;15:703–714. doi: 10.1074/mcp.O115.054387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guner H, Close PL, Cai W, Zhang H, Peng Y, Gregorich ZR, Ge Y. MASH Suite: a user-friendly and versatile software interface for high-resolution mass spectrometry data interpretation and visualization. J. Am. Soc. Mass Spectrom. 2014;25:464–470. doi: 10.1007/s13361-013-0789-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 38.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y, Wang W, Kirschner MW. Specificity of the anaphase-promoting complex: a single-molecule study. Science. 2015;348:1248737. doi: 10.1126/science.1248737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Udeshi ND, Mani DR, Eisenhaure T, Mertins P, Jaffe JD, Clauser KR, Hacohen N, Carr SA. Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteomics. 2012;11:148–159. doi: 10.1074/mcp.M111.016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.