Abstract
The aim of this study was to investigate whether the miR-184 could regulate the proliferation of the tongue squamous cell carcinoma (TSCC) through sex-determining region Y-box 7 (SOX7) gene. miR-184 expression was upregulated in TSCC cell lines and tissues. MTT assay revealed that overexpression of miR-184 significantly promoted the proliferation of the TSCC cells in vitro. SOX7 was the direct target of miR-184 and luciferase reporter assay confirmed that miR-184 downregulated the expression of SOX7. MTT assay verified that knockdown of SOX7 remarkably promoted the proliferation of TSCC cells in vitro. miR-184 promoted the proliferation of TSCC by targeting SOX7. Taken together, our results provided a new potential therapeutic target for TSCC treatment.
Keywords: luciferase reporter assay, miR-184, sex-determining region Y-box 7, tongue squamous cell carcinoma
Introduction
Tongue squamous cell carcinoma (TSCC) is one of the most common malignant tumors of the head and neck worldwide and it is characterized by its rapid invasion and metastasis (1,2). TSCC has a number of progression stages, which start with moderate dysplasia, proceeds to severe dysplasia and eventually invasive disease (3). TSCC can be cured by surgical treatment with early detection and diagnosis. Many therapeutic treatments and efforts have been proposed to prevent and treat TSCC. However, TSCC mortality rate is still high and increasing in many countries (4). Accordingly, identification of new biomarkers or pathways is of great importance and benefit for the treatment of TSCC.
The microRNAs (miRNAs) are small, 20–24 nucleotides in length, non-coding RNAs, which regulate the gene expression post-transcriptionally and have the potential to regulate ~1/3 of human genome (5–7). The miRNAs modulate the gene expression by binding to the 3′-UTR of the target mRNA. Most miRNA genes are solo, and exhibit their functions under the modulation of their own promoters and regulatory sequences (8).
The miR-184 plays a pivotal role in proliferation in many cancers. Tung et al found that miR-184 induced cisplatin resistance in non-small cell lung cancer (NSCLC) downregulated by E6 oncoprotein (9). The suppression of miR-184 upregulated SND1 and contributed to tumor invasion in malignant glioma (10). Foley et al found that the knockdown of miR-184 could promote neuroblastoma cell proliferation, increasing the levels of AKT2 (11). The miR-184 also played a role in epithelia by maintaining the SH2-containing phosphoinositide 5′-phosphatase 2 (SHIP2) level through inhibiting miR-205 (7). In addition, many studies found that miR-184 also could function as an essential component in nasopharyngeal carcinoma (12), hepatocellular carcinoma (HCC) (13), and TSCC (14).
The Sry-related high-mobility-group box (SOX) genes are transcription factors, in which the high mobility group (HMG) box domain is one of the components (15). Sex-determining region Y-box 7 (SOX7) is grouped into the SOX-F subfamily, which also includes the sex-determining region Y-box 17 (SOX17) and sex-determining region Y-box 18 (SOX18) (16). The SOX proteins have the specific DNA binding sites, which are 5′-(A/T)(A/T)CAA(A/T)G-3′ (17). It is reported that SOX7 played an important role in various developmental processes, such as cardiogenesis, endoderm differentiation, and hematopoiesis (18). The expression of SOX7 was remarkably downregulated in the breast cancer tissues compared with the non-tumorous tissues (16). In HCC, the SOX7 was the direct target of the miR-425, and the overexpression of the SOX7 could significantly suppress migration and invasiveness of HCC cells (19). However, the function of the SOX7 in TSCC progression has not been determined.
In this study, we found that the miR-184 was upregulated in TSCC cell lines and tissues compared with the corresponding adjacent non-tumorous tissues. We further found that overexpression of miR-184 could promote the migration, and metastasis of the TSCC cells. Moreover, we identified that SOX7 was the target of miR-184 and miR-184 could suppress the expression of SOX7. Taken together, our present study provides a new potential therapeutic target for TSCC treatment.
Materials and methods
Patient samples
The TSCC tissues and the corresponding non-tumorous counterparts were obtained from patients of the Affiliated Hospital of Taishan Medical University (Taian, China). The tissues samples were frozen in liquid nitrogen and stored at −80°C immediately after resection from patients who were undergoing surgical treatment of tongue carcinoma. Written consent of tissue donation for research purposes was obtained from patients and the research was approved by the Ethics Committee of the Affiliated Hospital of Taishan Medical University (Taian, China).
Cell culture
TSCC cell lines Cal27, Tca-8113, and SCC-9 were purchased from the American Type Culture Collection. All the cell lines were cultured in RPMI-1640 with 10% fetal bovine serum.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted and purified from the TSCC tissues and corresponding non-tumorous tissues using the TRIzol reagent (Invitrogen: Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) following the manufacturer's instructions. The concentration and purity of RNA were determined using the NanoDrop 2000 instrument (Thermo Fisher Scientific, Inc., Waltham, MA, USA). cDNAs were synthesized using the PrimeScript 1st Strand cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China). The RT-qPCR was carried out using the PrimeScript RT Reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd.) on the LightCycler 480 Instrument II (Roche Diagnostics, Basel, Switzerland). The GAPDH was used as the control for the normalization of expression levels of genes. The snRNA U6 was used as the internal control for miRNA. The RT-qPCR data were calculated by the 2−ΔΔCq method and relative to SnRNA U6 or GAPDH. The conditions for PCR were as follows: 95°C for 5 min, 40 cycles of denaturation at 95°C (15 sec), 50°C (30 sec) and 72°C (30 sec). The primers were as follows: miR-184 forward, 5′-TGGACGGAGAACTGATAAGGGT-3′ and reverse, 5′-CCTTATCAGTTCTCCGTCCATT-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; SOX7 forward, 5′-GGGGCCGCCGCCCGAAGCTGATA-3′ and reverse, 5′-CGGGATCCAGAAGGGACCTTGGCTATCA-3′; GAPDH forward, 5′-TTGCCGACAGGATGCAGAAGGA-3′ and reverse, 5′-AGGTGGACAGCGAGGCCAGGAT-3′.
Plasmid construction and small interfering RNA (siRNA) assay
The miR-184 mimic or inhibitor was constructed for the overexpression or knockdown of the miR-184. The siRNA was synthesized and purchased from Takara Biotechnology Co., Ltd., which was used for the knockdown of the SOX7. The miR-184 mimic or inhibitor and siRNA were transfected into Cal27 and Tca-8113 cell lines using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The sequences of miR-184 mimics/inhibitor, negative control (NC) were the following: miR-184 mimic 5′-UGGACGGAGAACUGAUAAGGGU-3′, miR-184 inhibitor 5′-CUGGAGGAAGGGCCCCAGAGG-3′, and the NC is a scrambled oligonucleotide: 5′-UUCUCCGAACGUGUCACGUTT-3′. miR-184 mimics/inhibitor, NC were designed and synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). For SOX7 depletion, siRNA was synthesized and purified by RiboBio Co., Ltd. (Guangzhou, China). SOX7-siRNA sequence was as follows: 5′-ACGCCGAGCTGTCGGATGG-3′.
Western blotting
Total protein was extracted from cells using the radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China). Protein concentration was determined using the BCA method. The proteins were separated by the SDS-PAGE using the Mini-PROTEAN Tetra instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA), then transfered onto PVDF membranes (Bio-Rad Laboratories, Inc.). The membranes were blocked by 5% BSA and incubated overnight at 4°C with specific primary antibody rabbit polyclonal anti-SOX7 antibody (ab220293, 1:1,000; Abcam, Cambridge, UK). After that, the membrane was incubated in the secondary antibody goat polyclonal anti-rabbit IgG H&L secondary antibody (ab150077, 1:2,000; Abcam) at room temperature for 1 h. Anti-GAPDH antibody (sc-25778; 1:3,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was used as a loading control at 4°C overnight. Signal was measured using the GelDoc 2000 instrument (Bio-Rad Laboratories, Inc.).
Luciferase reporter assay
A sequence of 3′-UTR of SOX7 containing the predicted miR-184 binding site was synthesized and inserted into the p-MIR-reporter plasmid (p-MIR-SOX7-3′-UTR-WT) (Ambion: Thermo Fisher Scientific, Inc., Foster City, CA, USA). Simultaneously, a sequence that contains seven mutant nucleotides of the 3′-UTR of SOX7 was inserted into the the p-MIR-reporter plasmid (p-MIR-SOX7-3′-UTR-MU). For the luciferase assay, the Cal27 and Tca-8113 cells were transfected with firefly luciferase reporter plasmid, miR-184 mimic, or the NC using the Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). The pMIR-REPORT β-galactosidase vector was used as the control. The luciferase activity was measured using the ONE-Glo Luciferase Assay instrument (Promega Corp., Madison, WI, USA).
Cell proliferation assays
In this study, we used the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) as the monitor of the cell proliferation. After transfection, the cells were cultured for 1–4 days. The cells were incubated for 4 h at 37°C after adding the MTT. The supernatant was removed and dissolved the formazan crystals using DMSO (150 µl/well). The absorbance at 490 nm for each sample was measured using the Multilabel Plate Reader (PerkinElmer, Inc., Waltham, MA, USA).
Clone formation assay
Cal27 and Tca-8113 cells were seeded on a 24-well plate at a density of 300 cells/well. Then the cells were transfected with miR-184 mimic, NC, miR-184 inhibitor using Lipofectamine 2000. Subsequently, the cells were cultured in a humidified chamber at 37°C containing 5% CO2 for 14 days. When clones were macroscopic, the culture process was terminated. Then the cells were fixed by applying neutral methyl alcohol 4% for 15 min and stained with Wright-Giemsa compound stain for 30 min. Finally, the number of clones was counted under a microscope (Olympus, Tokyo, Japan).
Statistical analysis
Student's t-test and Tukey's post hoc test after ANOVA was used to determine the significant difference between the groups. All the experiments were performed in three independent replicates. The results are presented as the mean ± SD and the differences were considered statistically significant at P<0.05. Correlation between mRNA and miRNA were estimated using the Spearman's correlation method. SPSS v.19.0 software (IBM Corp., Armonk, NY, USA) was used to perform statistical analyses and GraphPad Prism 5.02 (GraphPad Software, Inc., La Jolla, CA, USA) to complete graph presentation.
Results
miR-184 expression is upregulated and SOX7 expression is downregulated in TSCC cell lines and tissues
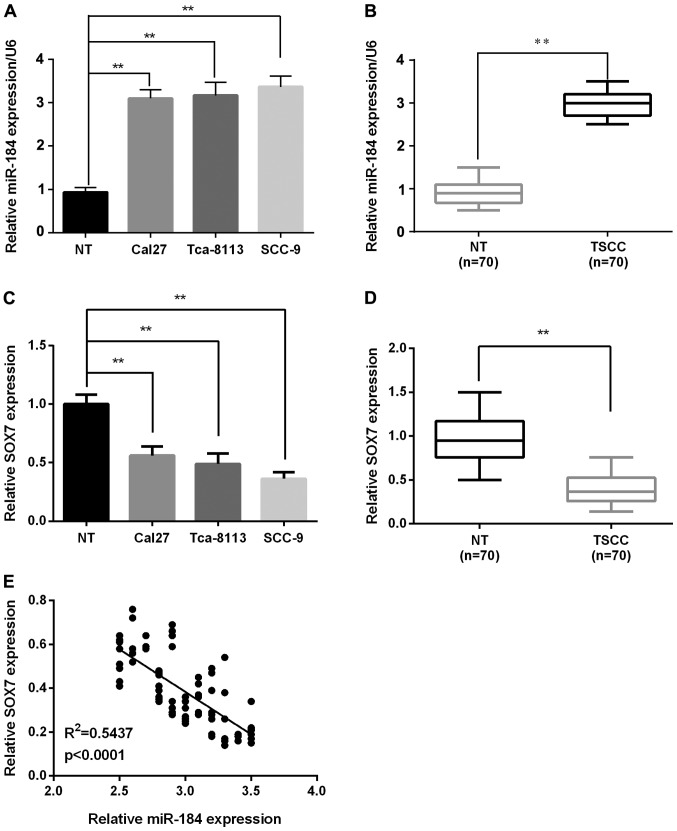
RT-qPCR analysis showed that the miR-184 expression was significantly increased in all cell lines (Cal27, Tca-8113, and SCC-9) compared with the normal cell line (Fig. 1A). In TSCC tumors, miR-184 expression was remarkably upregulated in 70 pairs of TSCC tissues compared with the corresponding adjacent non-tumorous tissues (Fig. 1B). However, SOX7 expression in TSCC cells and tissues was decreased significantly compared to normal tissues by RT-qPCR (Fig. 1C and D). Furthermore, to evaluate miR-184 modulation of SOX7 at the gene expression level, the association between the expression of miR-184 and SOX7 was investigated in these clinical specimens. It was demonstrated that miR-184 expression was negatively correlated with SOX7 expression in TSCC (R2=0.5437; P<0.0001; Fig. 1E), indicating that miR-184 regulates the expression of SOX7.
Figure 1.
miR-184 is significantly increased and SOX7 was decreased in the TSCC cell lines and tissues. (A) miR-184 expression levels in TSCC cell lines and normal cell line were tested using RT-qPCR. The expression levels were calculated using miR-184/U6 expression ratio (2−ΔΔCq). (B) The expression levels of miR-184 in TSCC and the corresponding non-tumorous tissues were identified using RT-qPCR. (C) SOX7 expression levels in TSCC cell lines and normal cell line were tested using the RT-qPCR. (D) The expression levels of SOX7 in TSCC and the corresponding non-tumorous tissues were identified using RT-qPCR. (E) The correlation between miR-184 and SOX7 expression was negative (r=−0.7373, p<0.0001). **P<0.01. SOX7, sex-determining region Y-box 7; TSCC, tongue squamous cell carcinoma.
miR-184 promotes proliferation of the TSCC cells in vitro
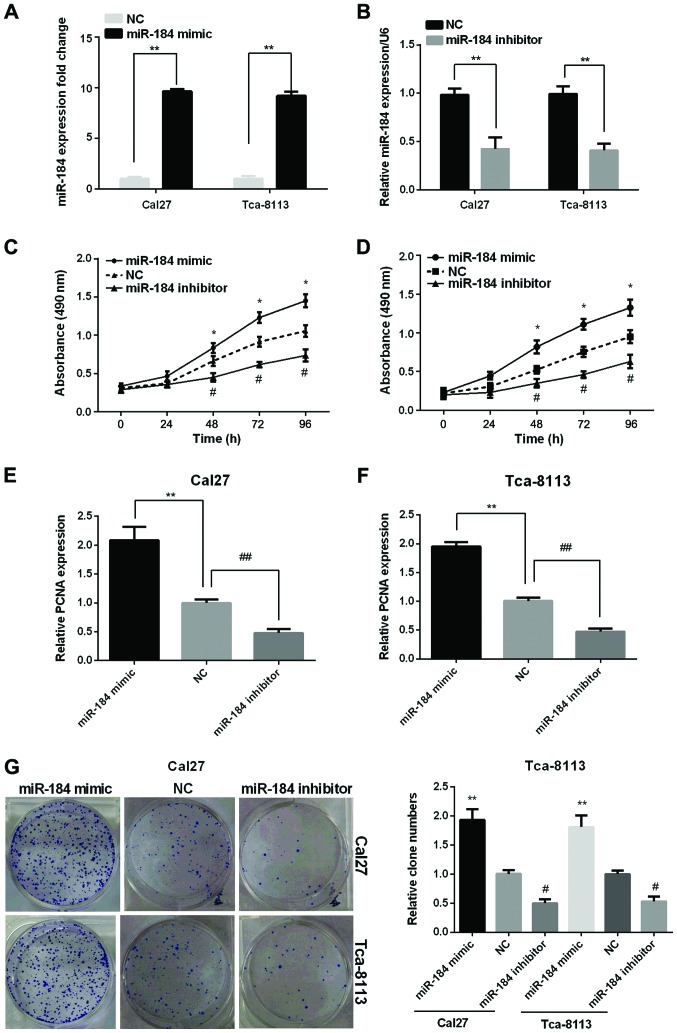
To better understand the function of miR-184 in the TSCC tissues, we transfected the miR-184 mimic or inhibitor to the TSCC cell lines (Cal27, Tca-8113). The successful overexpression or silencing of miR-184 was confirmed by the RT-qPCR (Fig. 2A and B). The results demonstrated that the miR-184 expression levels were remarkably increased in the two TSCC cell lines after transfected with miR-184 mimic while decreased after transfected with miR-184 inhibitor compared with the NC.
Figure 2.
miR-184 promotes the proliferation of TSCC cells in vitro. The expression of miR-184 was detected using RT-qPCR in Cal27 and Tca-8113 cells after transfected with miR-184 (A) mimic and (B) inhibitor. Cell viability of (C) Cal27 and (D) Tca-8113 cells was tested using MTT assay after transfected with miR-184 mimic or inhibitor. PCNA expression was detected by RT-qPCR in (E) Cal27 and (F) Tca-8113 cells after transfected with miR-184 mimic or inhibitor. (G) Clone number of Cal27 and Tca-8113 cells after transfection with miR-184 mimic or inhibitor was detected by clone formation assay. *p<0.05, **p<0.01; #p<0.05, ##p<0.01. PCNA, proliferating cell nuclear antigen.
The influence of miR-184 on the proliferation of the TSCC cell lines was investigated using the MTT, RT-qPCR and clone formation assays. MTT results showed that the cell viability in the two cell lines was significantly increased by miR-184 mimic, but reduced by miR-184 inhibitor (Fig. 2C and D). RT-qPCR analysis showed that the proliferating cell nuclear antigen (PCNA) expression was raised remarkably in the two cell lines after transfected with miR-184 mimic while declined after transfection with miR-184 inhibitor (Fig. 2E and F). As expected, clone formation assay showed an increased clone number in miR-184 mimic group while decreased clone number in miR-184 inhibitor group compared to control group (Fig. 2G).
SOX7 is a direct target of miR-184 and miR-184 regulates the expression of SOX7
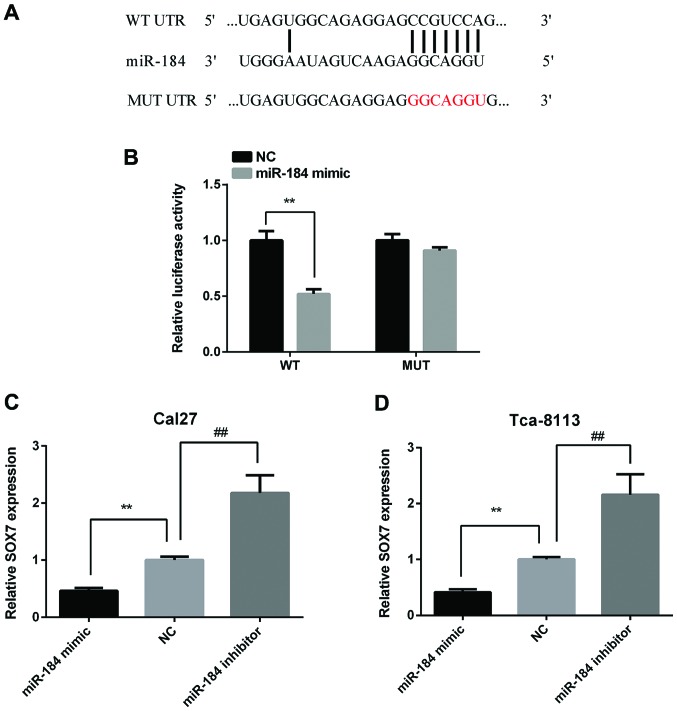
In order to discover the target gene of miR-184, bioinformatical prediction was performed using the TargeScan website (http://www.targetscan.org/mamm_31/). As Fig. 3A shows, SOX7 has a binding site at the 36–42 bp of the 3′-UTR for miR-184. To verify this prediction, Cal27 cells were transfected with the p-MIR-SOX7-3′-UTR-WT or p-MIR-SOX7-3′-UTR-MT luciferase reporter and miR-184 mimic, respectively. The relative luciferase activity was significantly reduced in Cal27 cells co-transfected with p-MIR-SOX7-3′-UTR-WT luciferase reporter and miR-184 mimic than the NC cells. However, there were no significance between Cal27 cells co-transfected with p-MIR-SOX7-3′-UTR-MU luciferase reporter and miR-184 mimic and the NC cells (Fig. 3B). Then, we detected SOX7 expression in Cal27 and Tca-8113 cells after transfected with miR-184 mimic or inhibitor. The results showed that miR-184 mimic reduced SOX7 expression and miR-184 inhibitor increased SOX7 expression in both two cell lines (Fig. 3C and D).
Figure 3.
SOX7 is the direct target of miR-184 and miR-184 regulates the expression of SOX7. (A) The binding site of SOX7 and miR-184 was predicted using the TargetScan tool. (B) Cal27 cells were transfected with the p-MIR-SOX7-3′-UTR-WT or p-MIR-SOX7-3′-UTR-MT luciferase reporter and miR-184 mimic. The luciferase activity was measured using the ONE-Glo Luciferase Assay instrument. Relative SOX7 expression was tested in (C) Cal27 and (D) Tca-8113 cells after transfected with miR-184 mimic or inhibitor. **p<0.01; ##p<0.01. SOX7, sex-determining region Y-box 7.
Overexpression of SOX7 inhibits proliferation of TSCC cells in vitro
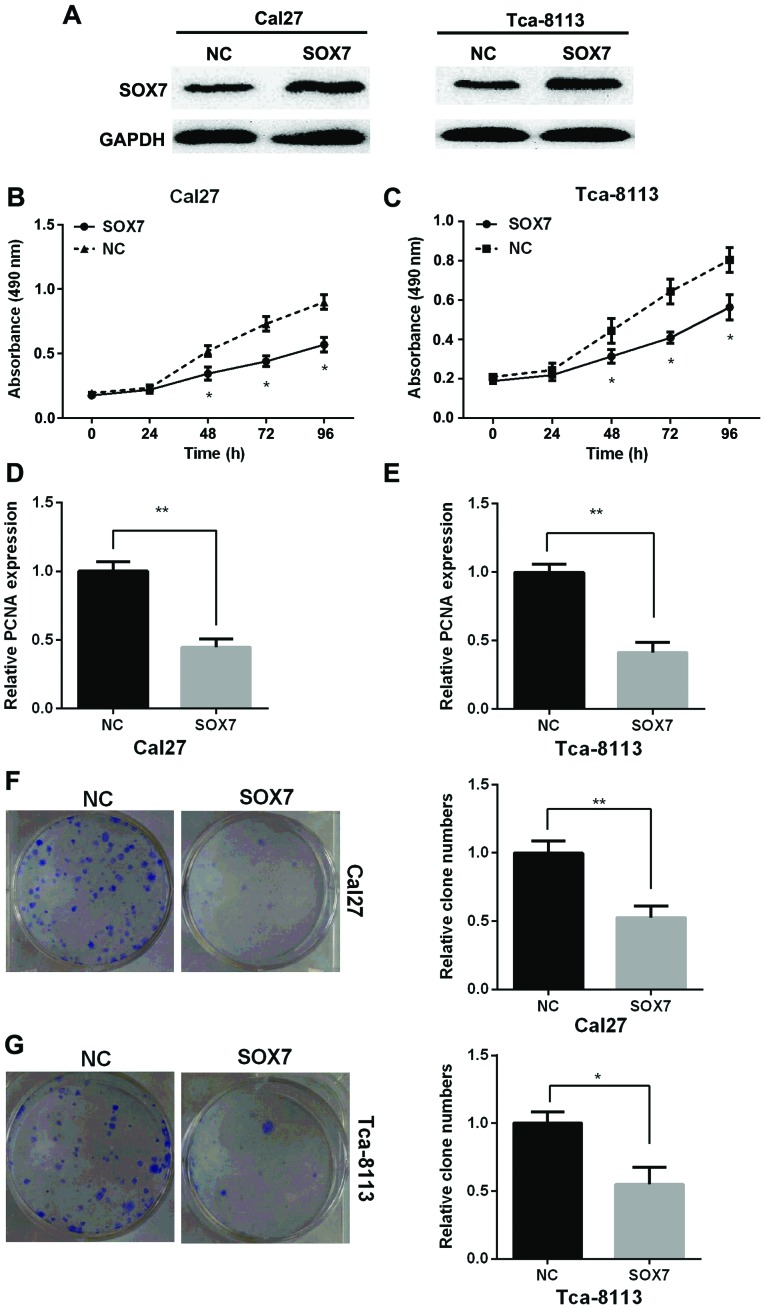
To detect the role of SOX7 on the proliferation of TSCC, Cal27 and Tca-8113 cell lines were transfected with SOX7 vector or the NC to overexpress of SOX7. SOX7 expression in protein level was measured by western blotting assay in Cal27 and Tca-8113 cells and the results showed that SOX7 expression was increased after overexpression of SOX7 compared with NC (Fig. 4A). MTT assay showed that overexpression of SOX7 inhibited the proliferation of Cal27 and Tca-8113 cells (Fig. 4B and C). PCNA expression was decreased after cells were transfected with SOX7 vector in the two TSCC cell lines by RT-qPCR (Fig. 4D and E). Clone formation assay showed that overexpression of SOX7 could suppress the relative clone number in the two TSCC cell lines (Fig. 4F). Taken together, overexpression of SOX7 inhibited proliferation of TSCC cells in vitro.
Figure 4.
Overexpression of SOX7 inhibits the proliferation of TSCC cells in vitro. (A) The alteration of SOX7 expression in protein level was identified using western blotting in Cal27 and Tca-8113 cells after transfected with SOX7 vector or NC. (B) Cal27 and (C) Tca-8113 cell viability was detected using MTT assay after overexpression of SOX7. Relative PCNA expression was measured by RT-qPCR in (D) Cal27 and (E) Tca-8113 cells after overexpression of SOX7. Relative clone numbers were measured in (F) Cal27 and (G) Tca-8113 cells by clone formation assay after overexpression of SOX7. SOX7, sex-determining region Y-box 7; TSCC, tongue squamous cell carcinoma; NC, negative control; PCNA, proliferating cell nuclear antigen.*P<0.05, **P<0.01.
Discussion
TSCC is the most common type of oral squamous cell carcinoma and it is well known for its high rate of lymph nodal metastasis. Although huge efforts have been made to prevent and cure TSCC, the number of deaths increased during the past 5 years (20). It is meaningful to study the related gene expression and miRNA alteration. Dysregulation of miRNAs is a distinct feature of cancers, which derive from dysregulated gene expression (14). It is reported that miRNA alteration could induce carcinogenesis (21) and miR-184 played an important role in many cancers, such as NSCLC (9), neuroblastoma (11), nasopharyngeal carcinoma (12), and HCC (13). In our study, we found that miR-184 was significantly upregulated in the TSCC tissues compared with the adjacent non-tumorous tissues and promoted the proliferation of TSCC cells. PCNA is a DNA polymerase sliding clamp that serves in DNA replication and repair (22). A previous study demonstrated that a high PCNA index resulted in an increase in cyclin E2 and proliferative activity in pancreatic cancer cells (23). The results of the current study demonstrated that the PCNA expression was raised remarkably in two cell lines after transfected with miR-184 mimic while declined after transfected with miR-184 inhibitor. This further indicates that miR-184 promoted the proliferation by dysregulating the expression of PCNA, moreover, the absence of cell cycle analysis may be a limitation of our study. In human HCC, miR-184 was upregulated in HCC cell lines and overexpression of miR-184 could increase cell proliferation through targeting SOX7 (13). In contrast to our research, miR-184 expression was downregulated in human malignant glioma tumor tissues compared with their normal counterparts (10). This exhibits the complicated functions of miR-184 in the regulation of gene expression. Using the TargetScan and the luciferase reporter assay, we predicted and confirmed that miR-184 bound to the 3′-UTR of SOX7. The luciferase activity was reduced significantly in cell lines co-transfected with p-MIR-SOX7-3′-UTR-WT luciferase reporter and miR-184 mimic than the NC cells.
SOX7 has been found to be involved in regulation of many cancers and to functioned as a suppressor. In the human breast cancer cells, miR-492 promoted the cell cycle and proliferation through inhibiting SOX7 (24). Liu et al found that SOX7 expression was remarkably reduced in the ovarian cancer tissues compared with the non-cancerous tissues and played this role through Wnt/β-catenin signaling pathway (25). In the gastric cancer, miR-935 increased tumorigenesis and cell proliferation by targeting SOX7 (26). Also, SOX7 played an important role in acute myeloid leukemia, and functioned as a tumor suppressor (27). SOX7 contains HMG box, and other important functional domains at the carboxyl and amino terminal regions. The HMG box plays an important role in DNA bending and binding, protein transportation and interaction (15,28). These important domains may explain why the SOX7 could function as tumor suppressor in a number of cancers. Consistent with these identifications, our study showed that overexpression of the SOX7 inhibited the proliferation of TSCC cells and miR-184 could suppress the expression of SOX7 using the luciferase reporter assays.
In conclusion, our results demonstrated that miR-184 was significantly upregulated in the TSCC tissues compared with the non-tumorous tissues and miR-184 promoted TSCC cell proliferation. We found that miR-184 bound to the 3′-UTR of SOX7 and suppressed SOX7 expression. Therefore, our results demonstrated that the miR-184/SOX7 axis might provide a promising therapeutic target for TSCC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
DC and JL contributed to the conception of the study. SL and PH contributed significantly to the data analysis and study preparation. YW performed the data analyses and wrote the study. SD, PH and NL helped perform the analysis with constructive discussions and conducted the experiment. All authors have read and approved the final study.
Ethics approval and consent to participate
Written consent of tissue donation for research purposes was obtained from patients and the research was approved by the Ethics Committee of the Affiliated Hospital of Taishan Medical University (Taian, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wang W, Liu Z, Zhao L, Sun J, He Q, Yan W, Lu Z, Wang A. Hexokinase 2 enhances the metastatic potential of tongue squamous cell carcinoma via the SOD2-H2O2pathway. Oncotarget. 2017;8:3344–3354. doi: 10.18632/oncotarget.22907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosebush MS, Rao SK, Samant S, Gu W, Handorf CR, Pfeffer LM, Nosrat CA. Oral cancer: Enduring characteristics and emerging trends. J Mich Dent Assoc. 2012;94:64–68. [PubMed] [Google Scholar]
- 3.Chen C, Méndez E, Houck J, Fan W, Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG, et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:2152–2162. doi: 10.1158/1055-9965.EPI-07-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang Z, Wu L, Wang L, Yang Y, Meng Y, Yang H. Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas: A possible correlation with cancer metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117:89–95. doi: 10.1016/j.oooo.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 6.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci USA. 2008;105:19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Tung MC, Lin PL, Cheng YW, Wu DW, Yeh SD, Chen CY, Lee H. Reduction of microRNA-184 by E6 oncoprotein confers cisplatin resistance in lung cancer via increasing Bcl-2. Oncotarget. 2016;7:32362–32374. doi: 10.18632/oncotarget.8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emdad L, Janjic A, Alzubi MA, Hu B, Santhekadur PK, Menezes ME, Shen XN, Das SK, Sarkar D, Fisher PB. Suppression of miR-184 in malignant gliomas upregulates SND1 and promotes tumor aggressiveness. Neuro Oncol. 2015;17:419–429. doi: 10.1093/neuonc/nou220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foley NH, Bray IM, Tivnan A, Bryan K, Murphy DM, Buckley PG, Ryan J, O'Meara A, O'Sullivan M, Stallings RL. MicroRNA-184 inhibits neuroblastoma cell survival through targeting the serine/threonine kinase AKT2. Mol Cancer. 2010;9:83. doi: 10.1186/1476-4598-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhen Y, Liu Z, Yang H, Yu X, Wu Q, Hua S, Long X, Jiang Q, Song Y, Cheng C, et al. Tumor suppressor PDCD4 modulates miR-184-mediated direct suppression of C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal carcinoma. Cell Death Dis. 2013;4:e872. doi: 10.1038/cddis.2013.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu GG, Li WH, He WG, Jiang N, Zhang GX, Chen W, Yang HF, Liu QL, Huang YN, Zhang L, et al. Mir-184 post-transcriptionally regulates SOX7 expression and promotes cell proliferation in human hepatocellular carcinoma. PLoS One. 2014;9:e88796. doi: 10.1371/journal.pone.0088796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 15.Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Mastriani E, Yan ZQ, Yin SY, Zeng Z, Wang H, Li QH, Liu HY, Wang X, Bao HX, et al. SOX7 co-regulates Wnt/β-catenin signaling with Axin-2: Both expressed at low levels in breast cancer. Sci Rep. 2016;6:26136. doi: 10.1038/srep26136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harley VR, Lovell-Badge R, Goodfellow PN. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stovall DB, Cao P, Sui G. SOX7: From a developmental regulator to an emerging tumor suppressor. Histol Histopathol. 2014;29:439–445. doi: 10.14670/hh-29.10.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Z, Liu J, Yang Z, Wu L, Xie H, Jiang C, Lin B, Chen T, Xing C, Liu Z, et al. MicroRNA-452 promotes stem-like cells of hepatocellular carcinoma by inhibiting Sox7 involving Wnt/β-catenin signaling pathway. Oncotarget. 2016;7:28000–28012. doi: 10.18632/oncotarget.8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X, Li Z. MicroRNA expression and its implications for diagnosis and therapy of tongue squamous cell carcinoma. J Cell Mol Med. 2016;20:10–16. doi: 10.1111/jcmm.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 22.Shiomi N, Mori M, Tsuji H, Imai T, Inoue H, Tateishi S, Yamaizumi M, Shiomi T. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2007;35:e9. doi: 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng J, He M, Chen L, Chen C, Zheng J, Cai Z. The loss of miR-26a-mediated post-transcriptional regulation of cyclin E2 in pancreatic cancer cell proliferation and decreased patient survival. PLoS One. 2013;8:e76450. doi: 10.1371/journal.pone.0076450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen F, Cai WS, Feng Z, Li JL, Chen JW, Cao J, Xu B. MiR-492 contributes to cell proliferation and cell cycle of human breast cancer cells by suppressing SOX7 expression. Tumour Biol. 2015;36:1913–1921. doi: 10.1007/s13277-014-2794-z. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Yan ZQ, Li B, Yin SY, Sun Q, Kou JJ, Ye D, Ferns K, Liu HY, Liu SL. Reduced expression of SOX7 in ovarian cancer: A novel tumor suppressor through the Wnt/β-catenin signaling pathway. J Ovarian Res. 2014;7:87. doi: 10.1186/s13048-014-0087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang M, Cui G, Ding M, Yang W, Liu Y, Dai D, Chen L. miR-935 promotes gastric cancer cell proliferation by targeting SOX7. Biomed Pharmacother. 2016;79:153–158. doi: 10.1016/j.biopha.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Man CH, Fung TK, Wan H, Cher CY, Fan A, Ng N, Ho C, Wan TS, Tanaka T, So CW, et al. Suppression of SOX7 by DNA methylation and its tumor suppressor function in acute myeloid leukemia. Blood. 2015;125:3928–3936. doi: 10.1182/blood-2014-06-580993. [DOI] [PubMed] [Google Scholar]
- 28.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: Extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/S1534-5807(02)00223-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.