Abstract
Osteosarcoma is a common bone tumor and a frequently occuring cancer-associated threat to children. Notably, the prognosis of osteosarcoma is very poor when it is diagnosed with metastasis. A growing number of studies have indicated that various microRNAs (miRs) serve important regulatory roles in the pathogeny of different types of cancer. However, the functions of miR-210 in osteosarcoma need to be elucidated comprehensively. The aim of the present study was to investigate the potential roles of miR-210 in osteosarcoma by targeting fibroblast growth factor receptor-like 1 (FGFRL1). Reverse transcription-quantitative polymerase chain reaction results revealed that the expression of miR-210 was highly elevated while FGFRL1 expression was reduced inversely in osteosarcoma tissues compared with matched normal tissues. The results of Transwell assays showed that miR-210 promoted osteosarcoma cell migration and invasion. Furthermore, the luciferase reporter assay results suggested that miR-210 could directly bind to FGFRL1 in osteosarcoma cells. In addition, the present findings demonstrated that miR-210 could negatively regulate FGFRL1 expression by targeting the 3′untranslated region. In conclusion, the findings of the present study suggested that miR-210 exerted tumor carcinogenic functions in osteosarcoma by targeting FGFRL1. The findings of this study demonstrated that FGFRL1 was a direct target of miR-210 in osteosarcoma involved in the promoting functions mediated by miR-210 in the invasion and migration of osteosarcoma, suggesting that miR-210/FGFRL1 may be promising for discovering diagnostic and prognostic biomarkers for the therapies of osteosarcoma.
Keywords: miR-210, human osteosarcoma, migration, invasion, FGFRL1
Introduction
Osteosarcoma, a kind of lethal malignant bone tumor, predominantly occurs among children as well as adolescents (1). Accumulating studies have showed that morbidity and mortality are both high in osteosarcoma. Currently, the major therapies for osteosarcoma are chemotherapy and surgery (2). With the development of combination including neoadjuvant chemotherapy and limb salvage surgery, the survival rates of osteosarcoma patients have significantly increased (3,4). However, osteosarcoma patients with metastasis often have a poor prognosis, in the meantime, overall clinical outcomes of osteosarcoma remain unsatisfactory (5). Therefore, it is necessary to understand the molecular mechanisms underlying the osteosarcoma development and progression. Moreover, finding out useful novel biomarkers for the diagnosis and treatment of osteosarcoma is one of the most important clinical problems. In recent years, emerging studies have indicated that miRNAs have important roles in different kinds of tumors.
miRNA is a kind of small non-coding RNA and plays crucial role in regulating gene expression, being considered as cancer biomarkers (6,7). Studies have indicated that the expressions of almost 30% human genes were controlled by different miRNAs (8,9). Briefly, miRNAs can regulate the expression of their mRNA targets by complete or partial binding to the 3′untranslated regions (3′-UTRs) (10). Based on the characteristics of their target genes, they can inhibit or promote multiple biological processes of obvious cancers (11). For instance, miR-130b suppresses epithelial ovarian carcinoma by regulating runt-related transcription factor 3 (RUNX3) (12); miR-92b functions as an oncogene in hepatocellular carcinoma via regulating Smad7 (13); miR-143 functions as a suppressor in prostate cancer via targeting matrix metalloproteinase 13 (14). In addition, miR-210 has been reported to modulate the development of many malignancies, including glioblastoma (15), breast carcinoma (16) and pancreatic carcinoma (17). However, the function of miR-210 in osteosarcoma needs to be fully elucidated.
Fibroblast growth factor receptor-like 1 (FGFRL1) is one member of FGFR family and unlike the other members of FGFR, FGFRL1 has no intracellular kinase domain, instead, it contains a short intracellular domain (18,19). Increasing evidence has shown that the FGFR family has vital regulatory roles in proliferation, differentiation and migration of various cancers (20). FGFRL1 has also been reported to exert multiple biological functions in different kinds of cancers. However, the specific functions of FGFRL1 in osteosarcoma still remain unclear. Present study aimed to investigate the correlation between miR-210 and FGFRL1 in osteosarcoma cell migration and invasion.
Materials and methods
Osteosarcoma cell lines and tissue specimens
Two osteosarcoma cell lines (MG63 and Saos-2) and one human normal osteoblast cell line hFOB 1.19 were purchased from Shanghai Institute for Biological Sciences (Shanghai, China). Both the osteosarcoma cells and the hFOB 1.19 were maintained in DMEM medium which contained penicillin-streptomycin and 10% FBS (all from Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in an atmosphere with 5% CO2 at 37°C.
A total of 54 osteosarcoma tissue specimens and the corresponding para-carcinoma tissues were collected from the People's Hospital of Qingdao West Coast New District between 2015 and 2017 and snap-frozen at −80°C. The clinical characteristics of osteosarcoma patients were shown in Table I. All the patients involved in current study signed written informed consent. The present study was approved by the Ethics Committee of the People's Hospital of Qingdao West Coast New Area (Shandong, China).
Table I.
Correlation between miR-210 expression and the clinicopathological characteristics of the osteosarcoma patients.
| miR-210a expression | ||||
|---|---|---|---|---|
| Clinicopathological features | Cases (n=54) | High (n=39) | Low (n=15) | P-value |
| Age (years) | 0.5768 | |||
| >60 | 26 | 20 | 6 | |
| ≤60 | 28 | 19 | 9 | |
| Sex | 0.4937 | |||
| Male | 28 | 18 | 8 | |
| Female | 26 | 21 | 7 | |
| Tumor size (cm) | 0.3864 | |||
| ≥5.0 | 30 | 25 | 8 | |
| <5.0 | 24 | 14 | 7 | |
| TNM stage | 0.0066b | |||
| I–II | 22 | 11 | 11 | |
| III | 32 | 28 | 4 | |
| Lymph-node metastasis | 0.0042b | |||
| Yes | 33 | 31 | 4 | |
| No | 21 | 8 | 11 | |
| Distant metastasis | 0.5204 | |||
| Yes | 28 | 18 | 9 | |
| No | 26 | 21 | 6 | |
The mean expression level of miR-210 was used as the cut-off
statistically significant. miR, microRNA; TNM, tumor-node-metastasis.
Cell transfection
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect miR-210 mimics, inhibitor or scrambled negative miR-control (NC) as well as FGFRL1 overexpression plasmid and the negative control vector (GenePharma Co., Ltd., Shanghai, China) into osteosarcoma cells. Plasmids were transfected into osteosarcoma cell lines by X-treme GENE HP DNA Transfection Reagent (Roche Diagnostics, Basel, Switzerland).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was conducted to measure the miR-210 expressions and FGFRL1 expressions in osteosarcoma cell lines and tissues. Firstly, the total RNA from osteosarcoma tissues and cells as well as normal tissues was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's recommendation. Reverse transcription assay was conducted to obtain the complementary DNA (cDNA) by PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) and qPCR assay was conducted by SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd.) on the system of ABI 7900 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The miR-210 expression was normalized to U6 while the FGFRL1 was normalized to GAPDH. The relative expression levels of genes were calculated by the 2−ΔΔCq method (21). The primers used were shown in Table II.
Table II.
Primer sequences for reverse transcription-quantitative polymerase chain reaction.
| Primer and direction | Sequence |
|---|---|
| miR-210 forward | 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-210 reverse | 5′-CTGTGCGTGTGACAGCGGCTGA-3′ |
| U6 forward | 5′-CTCGCTTCGGCAGCACA-3′ |
| U6 reverse | 5′-AACGCTTCACGAATTTGCGT-3′ |
| FGFRL1 forward | 5′-TGTGAACACAACGGTGGACT-3′ |
| FGFRL1 reverse | 5′-GGGCAACACCACAAACTTCT-3′ |
| GAPDH forward | 5′-TAATCTTCGCCTTAATACTT-3′ |
| GAPDH reverse | 5′-AGCCTTCATACATCTCAA-3′ |
miR, microRNA; FGFRL1, fibroblast growth factor receptor-like 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western blot analysis
The total proteins were extracted by lysing cells with RIPA buffer which contained protease and phosphatase inhibitors (Beyotime Institute of Biotechnology, Haimen, China). Put the lysates onto ice for half an hour, then centrifuged them for 20 min at 12,000 × g. The protein concentrations were quantified with the BCA kit (Pierce; Thermo Fisher Scientific, Inc.). The protein lysates were separated by SDS-PAGE and subsequently transferred onto PVDF membrane which was blocked with 5% non-fat milk and incubated with the primary antibodies at 4°C overnight. Then, a secondary incubation step was performed with appropriate secondary antibody at room temperature for one hour. The proteins were detected by Chemoluminescene method. GAPDH (Abcam, Cambridge, UK) was used as an internal loading control.
Migration and invasion assays
The invasion and migration abilities of the treated osteosarcoma cells were assessed by Transwell assays using Transwell chambers (Corning Incorporated, Corning, NY, USA) with or without Matrigel (Clontech Laboratories, Inc., Mountainview, CA, USA). For the invasion assay, the Transwell chambers were pretreated with Matrigel. Firstly, osteosarcoma cell transfections were suspended in serum-free medium and seeded into the upper chamber. At the same time, DMEM including 10% FBS was joined into the lower chamber. Being cultivated for 48 h, cells on the upper chambers were removed with cotton swabs carefully, while the invasive cells on the lower chamber were subsequently fixed and stained with 4% formaldehyde and 0.1% crystal violet, respectively. The difference between the migration assay and the invasion assay was that there was no Matrigel in the Transwell chambers. An inverted microscope (Olympus Corporation, Tokyo, Japan) was used to measure and count the invasive and migratory cells in five randomly selected fields.
Luciferase reporter assay
The amplified FGFRL1-3′-UTR-WT and corresponding FGFRL1-3′-UTR-MUT were respectively cloned into pGL3 luciferase vector (Promega Corporation, Madison, WI, USA). Osteosarcoma cells were added into a 96-well plate, and after 24 h, FGFRL1-3′UTR-WT and miR-210 mimics or FGFRL1-3′UTR-MUT and miR-210 mimics were transfected into the treated osteosarcoma cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, the Dual Luciferase Reporter Assay kit (Promega Corporation) was used to detect the relative luciferase activities 48 h after the transfections.
Statistical analysis
All the above assays were performed three times. Statistical analysis was performed using SPSS statistical software, version 17.0 (SPSS, Inc., Chicago, IL, USA) with Student's t-test and one-way ANOVA with Scheffe's post hoc test. Correlation between mRNA and miRNA were estimated using the Spearman's correlation method. Kaplan-Meier method with log-rank test were applied to estimate the survival rates and compare the survival curves respectively. The data was indicated as means ± SD. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-210 expression is upregulated and FGFRL1 expression is downregulated in osteosarcoma
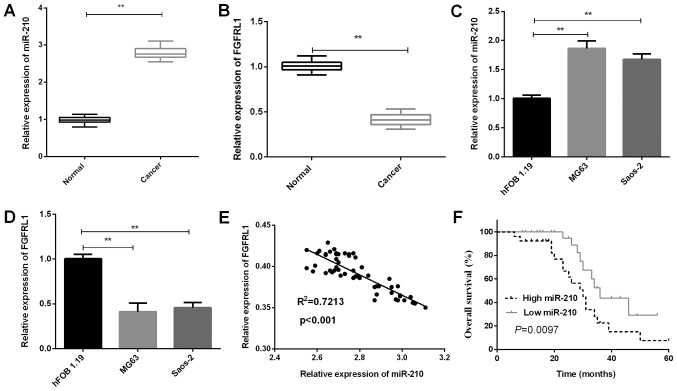
In this current study, the expressions of miR-210 and FGFRL1 in 54 paired osteosarcoma tissue specimens and cell lines were measured. The results of RT-qPCR revealed that the miR-210 expressions in osteosarcoma tissues were significantly increased in contrast with that in the matched para-carcinoma tissues (P=0.0015; Fig. 1A). On the contrary, FGFRL1 expressions in osteosarcoma tissues were found to be prominently downregulated (P=0.0023; Fig. 1B). Additionally, the same result was also found in osteosarcoma cell lines. From the results of RT-qPCR, we could also easily find a higher miR-210 expression both in MG63 (P=0.0064) and Saos-2 cells (P=0.0060) than that in hFOB 1.19 (P<0.01; Fig. 1C). Subsequently, we also measured the FGFRL1 mRNA expression in osteosarcoma cell lines, the results demonstrated a significant decrease in MG63 (P=0.0035) and Saos-2 (P=0.0039) cells compared to that in hFOB 1.19 (Fig. 1D). Moreover, to better understand the relationship between miR-210 and FGFRL1, we analyzed the correlation of miR-210 and FGFRL1 expression and found a negative correlation between them (P<0.001; Fig. 1E). To address the clinical significance of miR-210 in osteosarcoma, the mean expression level of miR-210 was defined as cutoff value to divide osteosarcoma patients into miR-210 low group and high group. Subsequently, the results indicated that osteosarcoma patients who had low miR-210 expression levels showed higher overall survival rates than those with high miR-210 expression levels (P=0.0097; Fig. 1F). Clinical association analysis demonstrated that the high miR-210 expression was notably correlated with advanced TNM stage (P=0.0066) and lymph node metastasis (P=0.0042; Table I).
Figure 1.
miR-210 expression elevated and FGFRL1 expression reduced in osteosarcoma. (A) RT-qPCR analysis was applied to measure the miR-210 expression in osteosarcoma tissues (n=54) and matched normal tissues (n=54). (B) FGFRL1 expressions in osteosarcoma tissues (n=54) and matched normal tissues (n=54) were measured by RT-qPCR. (C) miR-210 expression in osteosarcoma cells was detected by RT-qPCR. (D) The FGFRL1 expression was measured using RT-qPCR in MG63 and Saos-2 cells (**P<0.01 as indicated). (E) Correlation between miR-210 and FGFRL1 expressions. (F) miR-210 expression level in osteosarcoma was associated with overall survival. miR, microRNA; FGFRL1, fibroblast growth factor receptor-like 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-210 accelerates osteosarcoma cell invasion and migration
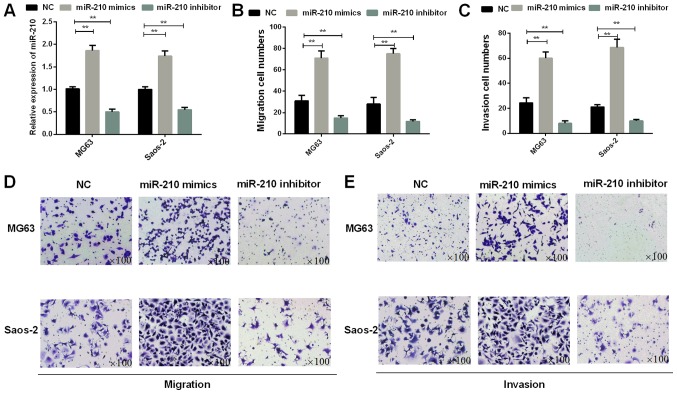
Subsequently, we investigated the invasion and migration abilities of osteosarcoma cells which were transfected with miR-210 mimics or inhibitor. The osteosarcoma cells (MG63 and Saos-2) transfected with miR-210 mimics or inhibitor were used to detect the roles of miR-210 in osteosarcoma cell invasion and migration. The results of RT-qPCR assays demonstrated that the expression of miR-210 mimics was high in MG63 (P=0.0035) and Saos-2 (P=0.0041) cells (P<0.01; Fig. 2A). The results of Transwell assays indicated that miR-210 overexpression facilitated migration ability of osteosarcoma cells (P=0.0042 for MG63 cells and 0.0030 for Saos-2 cells). On the contrary, the transwell results also demonstrated a significant decrease in the migration of MG63 (P=0.0048) and Saos-2 (P=0.0035) cells transfected with miR-210 inhibitor in contrast to the control group (Fig. 2B and D). In addition, according to the Transwell assays, the consequences also demonstrated that miR-210 could promote osteosarcoma invasion ability (P=0.0021 for MG63 cells and 0.0019 for Saos-2 cells) (Fig. 2C and E).
Figure 2.
miR-210 accelerates osteosarcoma cell invasion and migration. (A) The miR-210 expressions in transfected MG63 and Saos-2 cells were detected using RT-qPCR (**P<0.01). (B) The migration cell numbers of osteosarcoma cells were counted (**P<0.01). (C) The invasion cell numbers of osteosarcoma cells were counted (**P<0.01). (D) Cell invasion was observed by the Transwell assay in transfected osteosarcoma cells (magnification, ×100). (E) The Transwell assay was conducted to detect cell migration in transfected osteosarcoma cells (magnification, ×100). miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-210 suppresses FGFRL1 gene transcription in osteosarcoma by targeting its 3′-UTR
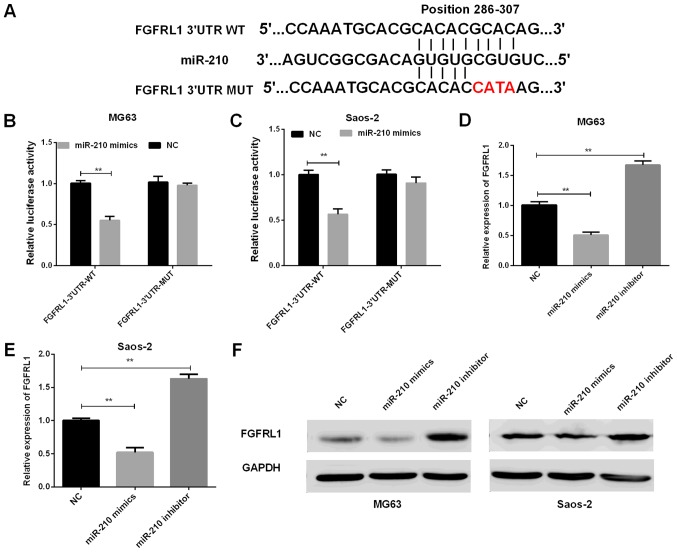
To investigate whether FGFRL1 expression was associated with the expression of miR-210 and to better understand the mechanisms of miR-210 in osteosarcoma, Target Scan was used to find out the target sites in the FGFRL1 sequence of miR-210 (Fig. 3A). To confirm the results, the luciferase reporter vector contained the wide-type (WT) or mutant (Mut) FGFRL1 3′-UTR was constructed. Then, the luciferase reporter assays were conducted in osteosarcoma cells. miR-210 mimics and FGFRL1-3′UTR-WT vector or FGFRL1-3′UTR-Mut vector were cotransfected into osteosarcoma cells to investigate whether FGFRL1 was the target of miR-210. Then, we detected the roles of miR-210 in the regulation of FGFRL1 mRNA and protein expression. The results of luciferase reporter assays indicated that miR-210 remarkably reduced the FGFRL1 3′-UTR-WT luciferase activities in both MG63 and Saos-2 cells. However, miR-210 had no significant influence on the FGFRL1 3′-UTR-Mut luciferase activities (P=0.0039 for MG63 cells and 0.0027 for Saos-2 cells) (Fig. 3B and C), suggesting that FGFRL1 was direct target of miR-210. Furthermore, we also detected the FGFRL1 expressions at mRNA level as well as at protein level in osteosarcoma cell lines transfected with miR-210 mimics or inhibitor. The results of RT-qPCR and western blot both demonstrated that miR-210 could inhibit the expression of FGFRL1 in MG63 (P=0.0044 for miR-210 mimics and 0.0032 for miR-210 inhibitor) and Saos-2 cells (P=0.0051 for miR-210 mimics and 0.0023 for miR-210 inhibitor (Fig. 3D-F).
Figure 3.
miR-210 de-regulated FGFRL1 expression via binding to the 3′-UTR of FGFRL1 directly. (A) The miR-210 binding sequence in the 3′-UTR of FGFRL1. (B and C) The luciferase reporter gene assays were performed to detect the fluorescence activities of the FGFRL1 3′UTR in MG63 cells (B) and Saos-2 cells (C) which were cotransfected with wild-type FGFRL1 3′UTR or mutational type FGFRL1 3′UTR and miR-210, respectively (**P<0.01). (D and E) RT-qPCR results of the FGFRL1 mRNA level in MG63 cells (D) and Saos-2 cells (E) with different transfections (**P<0.01 as indicated). (F) Western blot results of the FGFRL1 expression in osteosarcoma cells with different transfections. miR, microRNA; FGFRL1, fibroblast growth factor receptor-like 1; 3′-UTR, 3′untranslated region; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
The roles of FGFRL1 in regulating miR-210 effects in osteosarcoma cell migration and invasion
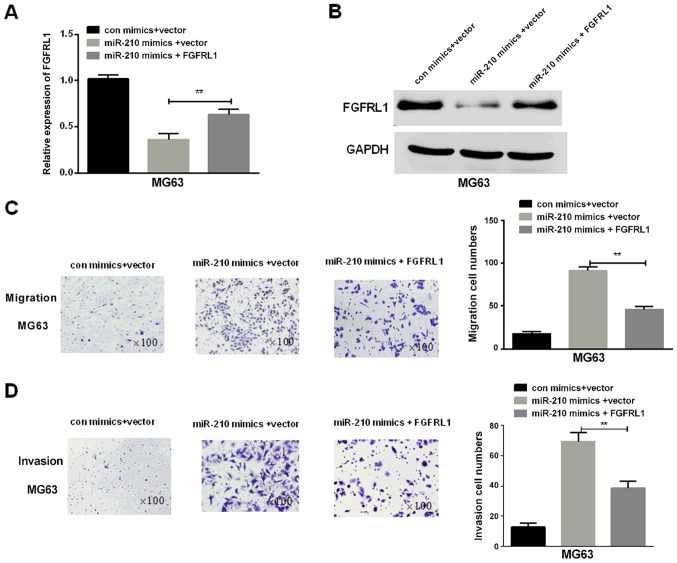
We continued to investigate whether FGFRL1 was needed in regulation of the promoting functions mediated by miR-210 in osteosarcoma cell invasion and migration. Firstly, FGFRL1 overexpression vector and miR-210 mimics were transfected into osteosarcoma cells. Then, RT-qPCR and western blotting were performed to measure the mRNA and protein expression levels respectively and the results indicated that the FGFRL1 expression in osteosarcoma cells transfected with FGFRL1 was significantly increased in contrast to the control group (P=0.0056; Fig. 4A and B). The results of Transwell assays indicated that FGFRL1 deprived the promotion effect of miR-210 on cell migration and invasion, suggesting that FGFRL1 played important roles in the miR-210-mediated biological functions in osteosarcoma cells (P=0.0043 for migration and 0.0036 for invasion) (Fig. 4C and D).
Figure 4.
Restoration of FGFRL1 could reverse partial function of miR-210 in osteosarcoma cells. (A) RT-qPCR and western blot analyses of the (B) FGFRL1 expressions in osteosarcoma cells with different transfections (**P<0.01). (C) Transwell assay was performed to observe migration of MG63 cells with different transfection treatments (magnification, ×100) (**P<0.01). (D) Transwell assay was performed to observe invasion of MG63 cells with different transfection treatments (magnification, ×100) (**P<0.01). One-way ANOVA and Scheffe's post-hoc test were utilized to analyze the data. FGFRL1, fibroblast growth factor receptor-like 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
Osteosarcoma is one common bone tumor which affects more and more children globally. At present, the primary treatments for osteosarcoma are radiotherapy, chemotherapy and surgical resection (22). Although great efforts have been made to explore the underlying mechanisms of osteosarcoma carcinogenesis, the prognosis is still unsatisfied due to extremely-high metastatic rate as well as aggressive invasion into local tissues (23,24). Therefore, it is urgent to elucidate the molecular mechanism of osteosarcoma to explore the novel therapeutic approaches, such as diagnostic biomarkers and therapeutic targets. During the past decades, increasing studies have demonstrated that various miRNAs are involved in osteosarcoma proliferation, apoptosis and metastasis (25).
Growing evidence has demonstrated that miR-210 is aberrantly expressed in multiple tumors. Additionally, miR-210 can modulate multiple physiological processes, including cell differentiation, apoptosis and survival proliferation via regulating its target genes (26). For instance, miR-210 has been reported to accelerate proliferation, autophagy and angiogenesis of schwannoma cells (27); miR-210 has also been reported to induce endothelial cell apoptosis by directly targeting PDK1 in the setting of atherosclerosis (28); in addition, miR-210-3p can inhibit the bladder cancer growth and metastasis through regulating FGFRL1 (29). As we all know, miR-210 has been demonstrated to play important roles in many tumors, but its role in osteosaroma remains unclear. This study aimed to explore the function of miR-210 in osteosaroma. The results in this study revealed that miR-210 was significantly upregulated in osteosaroma. Recent studies have indicated that hypoxia is one of the hallmarks of cancer and cells within the tumor become hypoxic as the tumor mass increases, resulting in activation of HIF-1α to induce various malignant phenotypes (30,31). The potential mechanism underlying the miR-210 overexpression in osteosaroma is that miR-210 is a hypoxia regulated miRNA and the induction of miR-210 is a consistent characteristic of the hypoxic response (32). FGFRL1, the fifth member of the FGFR family, plays important roles in the development of virtually all cell types such as the proliferation, differentiation, migration and apoptosis (33). FGFRL1 is newly described and usually expressed in adult pancreas and embryonic bone (34). A study has demonstrated that FGFRL1 overexpression in HEK 293 Tet-On cells suppresses the development of tumor in nude mice (35). For another example, FGFRL1 can't enhance cell proliferation but induce cell adhesion (36). However, little is known about the potential functions of FGFRL1 in osteosaroma. In current study, we investigated the relationship between miR-210 and FGFRL1 in osteosaroma.
In conclusion, miR-210 was upregulated not only in osteosaroma tissues but also in osteosaroma cells and its expression was correlated with clinicopathological features. In addition, we found that the expressions of miR-210 were negatively correlated with the expression of FGFRL1 in osteosarcoma tissues. miR-210 exerted tumor carcinogenic functions in osteosarcoma by targeting FGFRL1 directly. In summary, all the results in this study suggested that miR-210-FGFRL1 axis may be the novel biomarker for the diagnosis and treatment of osteosarcoma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XL and CZ perform the analysis with constructive discussions. CW contributed to analysis and manuscript preparation. JS performed the data analyses and wrote the manuscript. DW and YZ contributed to the conception of the study. XX contributed to manuscript preparation and the conception of the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study obtained approval from the Ethics Committee of the People's Hospital of Qingdao West Coast New Area (Shandong, China). Signed written informed consents were obtained from the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Angulo P, Kaushik G, Subramaniam D, Dandawate P, Neville K, Chastain K, Anant S. Natural compounds targeting major cell signaling pathways: A novel paradigm for osteosarcoma therapy. J Hematol Oncol. 2017;10:10. doi: 10.1186/s13045-016-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari S, Serra M. An update on chemotherapy for osteosarcoma. Expert Opin Pharmacother. 2015;16:2727–2736. doi: 10.1517/14656566.2015.1102226. [DOI] [PubMed] [Google Scholar]
- 3.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: Current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Xu SF, Xu M, Yu XC. Intentional marginal resection of periosteal osteosarcoma in combination with neoadjuvant chemotherapy: A report of two cases and a review of the literature. Oncol Lett. 2017;13:1343–1347. doi: 10.3892/ol.2017.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirotsu M, Setoguchi T, Sasaki H, Matsunoshita Y, Gao H, Nagao H, Kunigou O, Komiya S. Smoothened as a new therapeutic target for human osteosarcoma. Mol Cancer. 2010;9:5. doi: 10.1186/1476-4598-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margue C, Reinsbach S, Philippidou D, Beaume N, Walters C, Schneider JG, Nashan D, Behrmann I, Kreis S. Comparison of a healthy miRNome with melanoma patient miRNomes: Are microRNAs suitable serum biomarkers for cancer? Oncotarget. 2015;6:12110–12127. doi: 10.18632/oncotarget.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo K, Liang Z, Li F, Wang H. Comparison of miRNA and gene expression profiles between metastatic and primary prostate cancer. Oncol Lett. 2017;14:6085–6090. doi: 10.3892/ol.2017.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun B, Liu X, Gao Y, Li L, Dong Z. Downregulation of miR-124 predicts poor prognosis in pancreatic ductal adenocarcinoma patients. Br J Biomed Sci. 2016;73:152–307. doi: 10.1080/09674845.2016.1220706. [DOI] [PubMed] [Google Scholar]
- 9.Ohzawa H, Miki A, Teratani T, Shiba S, Sakuma Y, Nishimura W, Noda Y, Fukushima N, Fujii H, Hozumi Y, et al. Usefulness of miRNA profiles for predicting pathological responses to neoadjuvant chemotherapy in patients with human epidermal growth factor receptor 2-positive breast cancer. Oncol Lett. 2017;13:1731–1740. doi: 10.3892/ol.2017.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gee HE, Ivan C, Calin GA, Ivan M. HypoxamiRs and cancer: From biology to targeted therapy. Antioxid Redox Signal. 2014;21:1220–1238. doi: 10.1089/ars.2013.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manikandan J, Aarthi JJ, Kumar SD, Pushparaj PN. Oncomirs: The potential role of non-coding microRNAs in understanding cancer. Bioinformation. 2008;2:330–334. doi: 10.6026/97320630002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paudel D, Zhou W, Ouyang Y, Dong S, Huang Q, Giri R, Wang J, Tong X. MicroRNA-130b functions as a tumor suppressor by regulating RUNX3 in epithelial ovarian cancer. Gene. 2016;586:48–55. doi: 10.1016/j.gene.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Zhuang LK, Yang YT, Ma X, Han B, Wang ZS, Zhao QY, Wu LQ, Qu ZQ. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu D, Huang P, Wang L, Zhou Y, Pan H, Qu P. MicroRNA-143 inhibits cell migration and invasion by targeting matrix metalloproteinase 13 in prostate cancer. Mol Med Rep. 2013;8:626–630. doi: 10.3892/mmr.2013.1501. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S, Lai N, Liao K, Sun J, Lin Y. MicroRNA-210 regulates cell proliferation and apoptosis by targeting regulator of differentiation 1 in glioblastoma cells. Folia Neuropathol. 2015;53:236–244. doi: 10.5114/fn.2015.54424. [DOI] [PubMed] [Google Scholar]
- 16.Liu D, Xia H, Wang F, Chen C, Long J. MicroRNA-210 interacts with FBXO31 to regulate cancer proliferation cell cycle and migration in human breast cancer. Onco Targets Ther. 2016;9:5245–5255. doi: 10.2147/OTT.S110969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amponsah PS, Fan P, Bauer N, Zhao Z, Gladkich J, Fellenberg J, Herr I. microRNA-210 overexpression inhibits tumor growth and potentially reverses gemcitabine resistance in pancreatic cancer. Cancer Lett. 2017;388:107–117. doi: 10.1016/j.canlet.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 18.Trueb B. Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cell Mol Life Sci. 2011;68:951–964. doi: 10.1007/s00018-010-0576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang L, Karotki AV, Bruecker P, Trueb B. Comparison of the receptor FGFRL1 from sea urchins and humans illustrates evolution of a zinc binding motif in the intracellular domain. BMC Biochem. 2009;10:33. doi: 10.1186/1471-2091-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kähkönen TE, Ivaska KK, Jian M, Büki KG, Väänänen HK, Härkönen PL. Role of fibroblast growth factor receptors (FGFR) and FGFR like-1 (FGFRL1) in mesenchymal stromal cell differentiation to osteoblasts and adipocytes. Mol Cell Endocrinol. 2018;461:194–204. doi: 10.1016/j.mce.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Sun Z, Liu Q, Hong H, Zhang H, Zhang T. miR-19 promotes osteosarcoma progression by targeting SOCS6. Biochem Biophys Res Commun. 2018;495:1363–1369. doi: 10.1016/j.bbrc.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Urciuoli E, Coletta I, Rizzuto E, De Vito R, Petrini S, D'Oria V, Pezzullo M, Milano GM, Cozza R, Locatelli F, Peruzzi B. Src nuclear localization and its prognostic relevance in human osteosarcoma. J Cell Physiol. 2018;233:1658–1670. doi: 10.1002/jcp.26079. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Song Z, Wang Z. Common mechanism of pathogenesis in various types of metastatic osteosarcoma. Oncol Lett. 2017;14:6307–6313. doi: 10.3892/ol.2017.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo T, Yi X, Si W. Identification of miRNA and genes involving in osteosarcoma by comprehensive analysis of microRNA and copy number variation data. Oncol Lett. 2017;14:5427–5433. doi: 10.3892/ol.2017.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diao H, Liu B, Shi Y, Song C, Guo Z, Liu N, Song X, Lu Y, Lin X, Li Z. MicroRNA-210 alleviates oxidative stress-associated cardiomyocyte apoptosis by regulating BNIP3. Biosci Biotechnol Biochem. 2017;81:1712–1720. doi: 10.1080/09168451.2017.1343118. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Deng M, Liu Z, Wu S. Hypoxia-induced miR-210 promoter demethylation enhances proliferation, autophagy and angiogenesis of schwannoma cells. Oncol Rep. 2017;37:3010–3018. doi: 10.3892/or.2017.5511. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Yang C, Zhang L, Yang P. MicroRNA-210 induces endothelial cell apoptosis by directly targeting PDK1 in the setting of atherosclerosis. Cell Mol Biol Lett. 2017;22:3. doi: 10.1186/s11658-017-0033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Shi L, Yi C, Yang Y, Chang L, Song D. MiR-210-3p inhibits the tumor growth and metastasis of bladder cancer via targeting fibroblast growth factor receptor-like 1. Am J Cancer Res. 2017;7:1738–1753. [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan K, Song G, Ouyang G. Role of hypoxia in the hallmarks of human cancer. J Cell Biochem. 2009;107:1053–1062. doi: 10.1002/jcb.22214. [DOI] [PubMed] [Google Scholar]
- 31.Arvelo F, Cotte C. Hypoxia in cancer malignity. Review. Invest Clin. 2009;50:529–546. (In Spanish) [PubMed] [Google Scholar]
- 32.Qin Q, Furong W, Baosheng L. Multiple functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer Res. 2014;33:50. doi: 10.1186/1756-9966-33-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beenken A, Mohammadi M. The FGF family: Biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou WY, Zheng H, Du XL, Yang JL. Characterization of FGFR signaling pathway as therapeutic targets for sarcoma patients. Cancer Biol Med. 2016;13:260–278. doi: 10.20892/j.issn.2095-3941.2015.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhuang L, Steinberg F, Trueb B. Receptor FGFRL1 acts as a tumor suppressor in nude mice when overexpressed in HEK 293 Tet-On cells. Oncol Lett. 2016;12:4524–4530. doi: 10.3892/ol.2016.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, Steinberg F, Zhuang L, Bessey R, Trueb B. Receptor FGFRL1 does not promote cell proliferation but induces cell adhesion. Int J Mol Med. 2016;38:30–38. doi: 10.3892/ijmm.2016.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.