Abstract
Upregulation of microRNA (miR)-10b has been confirmed in multiple types of cancer, however, the role of miR-10b in glycosylation remains unclear. Protein core-fucosylation is an important N-linked glycosylation modification and serves important roles in cancer progression. In a previous study, a glycogene array was applied to profile the alterations of glycogene expression in miR-10b-overexpressed MCF10A cells. Notably, fucosyltranferase 8 (FUT8), which is responsible for the addition of core-fucose to N-glycan, was significantly upregulated by miR-10b. In the present study, increased motility and proliferation were observed in miR-10b-overexpressed MCF10A cells. To assess the mechanism involved, the role of FUT8 in MCF10A cells was studied and it was confirmed that miR-10b promotes motility and proliferation by regulating FUT8 and activating the protein kinase B (AKT) signaling pathway. Consistent with the aforementioned result, decreased motility and proliferation were detected when miR-10b expression was inhibited in MDA-MB-231 cells, transforming growth factor-β-induced and Twist-overexpressed MCF10A cells. To conclude, the findings from the present study indicate that miR-10b promotes motility and proliferation by increasing FUT8 and activating AKT in breast cancer cells.
Keywords: breast cancer, microRNA-10b, fucosyltranferase 8, phosphorylated protein kinase B, motility, proliferation
Introduction
Breast cancer is the most common type of malignant cancer among women worldwide, and the second leading cause of cancer-associated mortality among women in Asia in 2012 (1). In 2012, figures published by the International Agency for Research on Cancer (IARC), the specialized cancer agency of the World Health Organization, indicate ~1.7 million women globally are diagnosed with breast cancer annually, which attributes for an estimated 520,000 mortalities each year (accounting for 15% of all cancer associated-loss of life) (2). Metastasis, together with recurrence and resistance to chemotherapy, are the leading causes of breast cancer mortality (3); however, the underlying mechanism of breast cancer metastasis is still poorly understood and warrants extensive study.
MicroRNAs (miRNAs), single-stranded noncoding RNAs, play pivotal roles in numerous biological processes, ranging from development, organogenesis, proliferation, apoptosis, and migration to invasion (4). In general, miRNAs attenuate or repress the expression of target genes at the post-transcriptional level through sequence-specific binding to the 3′untranslated region (3′UTR) of target genes (5). Aberrant miRNA expression is associated with diverse pathophysiological diseases, including cancer. Accumulating evidence demonstrates that miRNAs are implicated in tumor metastasis and control central metastasis regulators (6). miR-10b, one of the metastasis miRs, is upregulated in breast cancer tissues and cells (7,8). It positively regulates invasion and metastasis by inhibition of HOXD10 and resulting in increased expression of ras homolog family member C (RHOC) (8), negatively regulates phosphatase and tensin homolog (PTEN) and ultimately results in protein kinase B (AKT) activation in breast cancer stem cells (9). Furthermore, miR-10b plays a critical role in transforming growth factor (TGF)-β1-induced epithelial-mesenchymal transition (EMT) in breast cancer (7).
Abundant core fucosylation was reported in numerous types of cancer, including breast cancer (10), hepatocellular carcinoma (11), non-small cell lung cancer (12) and colon carcinoma (13). Fucosyltranferase 8 (FUT8) specifically catalyzes the transfer of a fucose residue to the innermost GlcNAc residue of N-linked type complex glycopeptide via the α1,6-linkage. Upregulated levels of FUT8 were revealed in the EMT process (12) and in activation of the β-catenin pathway, the PI3K/AKT signaling pathway and hypoxia condition (14). miR-26a, miR-34a and miR-146a repress FUT8 expression by binding to the 3′UTR of FUT8 in hepatocellular carcinoma (11). However, thus far, the regulation of FUT8 in breast cancer was not previously documented.
In the present study, it was confirmed that miR-10b promoted the motility and proliferation of breast cancer cells, by enhancing FUT8 expression, resulting in the activation of AKT signaling.
Materials and methods
Cells and cell culture
The immortalized human mammary epithelial cell line MCF10A and human breast cancer cell line MDA-MB-231 were obtained from the American Type Culture Collection (Manassas, VA, USA). MCF10A cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 complete medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), containing epidermal growth factor (20 ng/ml), hydrocortisone (0.5 mg/ml), cholera toxin (100 ng/ml) purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany), insulin (10 µg/ml), penicillin (100 units/ml) and streptomycin (100 µg/ml; Gibco), at 37°C in 5% CO2. MDA-MB-231 cells were cultured in DMEM (Hyclone; GE Healthcare, Chicago, IL, USA) at 37°C in 5% CO2. All cell cultures were supplemented with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare), 100 IU/ml penicillin and 100 µg/ml streptomycin.
Antibodies and reagents
Primary antibodies used were as follows: Mouse anti-epithelial (E)-cadherin immunoglobulin (Ig)G2a mAb (1:50,000; cat. no. 610181) (BD Biosciences; San Jose, CA, USA), mouse anti-Fut8 IgG1 mAb (1:1,000; cat. no. sc-271244), mouse anti-Twist (Twist2C1a; 1:1000; cat. no. sc-81417), mouse anti-N-cadherin IgG1 mAb (1:1,000; cat. no. sc-8424) (Santa Cruz Biotechnology, Inc., Dallas, TX, USA), rabbit anti-GAPDH (1:100,000; cat. no. G9545) and anti-Fibronectin polyclonal antibodies (FN; 1:10,000; cat. no. F3648) (Sigma-Aldrich; Merck KGaA), rabbit anti-AKT (1:1,000; cat. no. 9272) and anti-phosphorylated (p)-AKT (Ser473; D9E) XP® mAb (1:1,000; cat. no. 4060) (Cell Signaling Technology, Inc., Danvers, MA, USA). The secondary antibodies were horseradish peroxides (HRP)-labeled goat anti-mouse IgG (1:5,000; cat. no. A0216) and goat anti-rabbit IgG (1:5,000; cat. no. A0208) (Beyotime Institute of Biotechnology, Haimen, China).
The reagents used in the present study were as follows: MK2206 (Sigma-Aldrich; Merck KGaA) and TGF-β1 (BD Biosciences). MK2206, a PI3K/AKT signaling inhibitor, can persistently reduce the expression of p-AKT. MK2206 (working concentration 10 nM) was added to miR-10b-overexpressing MCF10A cells, using DMSO as a negative control, to confirm whether miR-10b promotes cell motility and proliferation via activating AKT. For induction of EMT, MCF10A cells (~30% confluence) were incubated with 5 ng/ml TGF-β1 at 37°C for 48 h.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA, including miRNA, was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. The concentration was determined using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.) and the RNA sample (A260/A280>1.8) was reversed transcribed using ReverTra Ace-α-® kit (Toyobo; Shanghai, China), according to the manufacturer's protocols. Specific primers used for multiple genes were as follows: FUT8 forward, 5′-TCCATGACCCTAATGGTCTTTT-3′; and reverse, 5′-TGTCCTGTACTTCATGCGCT-3′; β-actin forward, 5′-GCACAGAGCCTCGCCTT-3′; and reverse, 5′-GTTGTCGACGACGAGCG-3′; RNU6B forward, 5′-CTCGCTTCGGCAGCACA-3′; and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The specific hairpin-itTM miRNA primers for miR-10b were designed and synthesized by GenePharma (Suzhou, China; cat. no. F02001). RT-qPCR was performed using UltraSYBR Mixture (Beijing CoWin Biotech Co., Ltd., Beijing) and run on the CFX96 RT-PCR detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The relative expression levels of the target genes were quantified using 2−ΔΔCq method from triplicate experiments (15).
Plasmid construction
The plasmid pBABE-Puro-Twist was purchased from Addgene, Inc. (Cambridge, MA, USA) and for the plasmid pLVX-AcGFP1-FUT8, human gene FUT8 was amplified with the following primer: Sense, 5′-CCGCTCGAGCGGGCCACCATGCGGCCATGGACTG, and antisense, 5′-CGCGGATCCGCGGATCAGAGCCCTCTTCATCTACAG, using the same PCR conditions as aforementioned. The PCR product was digested with restriction enzyme XhoI and BamHI, and ligated into the vector pLVX-AcGFP1-N1 (Addgene Inc.) at the corresponding sites, according to the manufacturer's instructions. All the plasmids were confirmed by DNA sequencing.
Transfection and RNA interference
MCF10A cells were stably transfected with plasmids pBABE-Puro-Twist and the negative control vector, pBABE-puro, using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), selected with puro (0.5 µg/ml) and designated as MCF10A/Twist and MCF10A/Mock. Lentiviral vector pLVX-AcGFP1-FUT8 or pLVX-AcGFP1-N1 (negative control) and packaging plasmids were co-transfected into 293T cells using Lipofectamine® 2000. After 48 h post-transfection, supernatants containing lentiviruses were harvested and purified with 0.22 µM filters (Merck KGaA). Then, the lentiviruses titer were determined and infected into MCF10A cells. The stable clones were selected with puro and designated as MCF10A/FUT8 and MCF10A/Mock2 (negative control). All the stably transfected cells were confirmed by western blot analysis. For RNA interference assay, MDA-MB-231 cells were transfected with small interfering (si)RNAs for FUT8 and negative control (NC), and expression of FUT8 was detected by RT-qPCR and western blot. The sequences of validated siRNA for FUT8 were: Forward, 5′-GUGUCUCAGUUUGUCAAAUTT-3′; and reverse, 5′-AUUUGACAAACUGAGACACTT-3′; for si-1; forward, 5′-GGUGUGUAAUAUCAACAAATT-3′; and reverse, 5′-UUUGUUGAUAUUACACACCTT-3′ for si-2.
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay buffer on ice for 30 min, supplemented with protease inhibitor and phosphorylase inhibitor (Sigma-Aldrich; Merck KGaA), and centrifuged at 14,000 × g at 4°C for 15 min. Total protein samples (30 µg/lane) were separated on a 10% gel using SDS-PAGE and transferred onto polyvinylidene fluoride membranes by electrophoresis. After 2 h of blocking with 5% bovine serum albumin (BSA; cat. no. B2064; Sigma-Aldrich) at 37°C, membranes were incubated with E-cadherin, N-cadherin, Vimentin, FUT8, AKT, p-AKT, Fibronectin, Twist and GADPH primary antibodies overnight at 4°C, and blotted with appropriate HRP-conjugated secondary antibody at room temperature for 30 min. Protein bands were analyzed by ChemiDoc XRS+ system (Bio-Rad).
Wound healing
For the wound healing assay, cells were seeded into 6-well plates at a density of 5×105 cells/well for 24 h. Linear wounds were scratched on the 100% confluent monolayer using a pipette tip. Cells were cultured in medium without FBS; images were captured under an inverted microscope (×40, magnification) at 0 and 24 h and analyzed using the Scion image software (version 4.0.3.2; Scion Corporation, Frederick, MD, USA).
Transwell assays
Transwell assays were performed in Transwell inserts with an 8.0 µm-pore polycarbonate membrane (Costar; Corning Incorporated, Corning, NY, USA). Briefly, pretreated cells were suspended in serum-free media and seeded into the upper chamber (1.5×105 cells/chamber). The complete growth medium supplemented with 10% FBS was deposited in the lower chamber. After incubation for 16 h, cells on the inside of the membrane were removed with a cotton swab and the migrated cells on the outside of the membrane were fixed and stained with 0.1% crystal violet in methanol for 15 min, and imaged under a light microscope. The stained cells were lysed with 2% SDS in PBS and subjected to spectrophotometric analysis at 570 nm.
MTT assay
Cells at a density of 2×103 cells were plated in 96-well plates and at various hours of culturing (0, 24, 48, 72 and 92 h), 10 µl MTT solution (Cers, Yantai, China) was added to form formazan. The reaction was stopped by the addition of 100 µl DMSO. The absorbance was measured at 490 nm and assessed as previously described (16).
Statistical analysis
Data were presented as the mean ± standard deviation from 3 repeated experiments using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). Data between groups were analyzed by paired or unpaired Student's t-test and multiple testing was performed using one-way ANOVA and a t-test with Bonferroni's correction as a post-hoc test. Comparisons between means with P<0.05 were considered to indicate a statistically significant difference.
Results
miR-10b promotes cell motility and proliferation via activating AKT
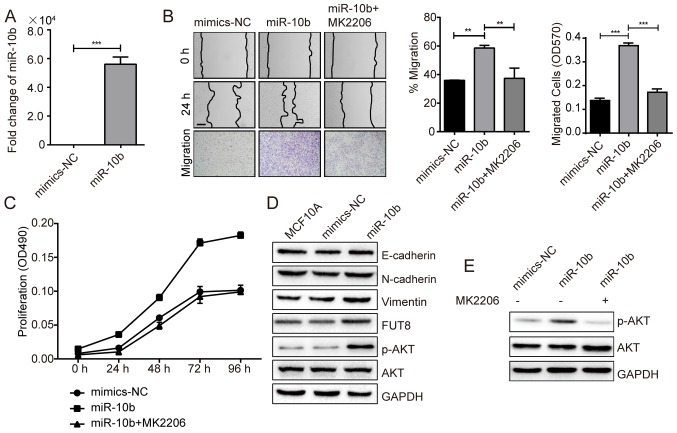
It has been revealed that miR-10b exerts positive effect on cell metastasis and invasion, in vitro and in vivo (8). Critical role of miR-10b is also demonstrated in TGF-β1-induced EMT process in breast cancer (7). In the present study, miR-10b was transfected into MCF10A cells, which demonstrated a 5.6×104-fold increase, compared with NC (Fig. 1A), and the impact of overexpressed miR-10b on cell motility and proliferation were examined. Compared with NC-mimic, cell motility evaluated by wound healing and transwell assays, and cell proliferation evaluated by MTT assay were significantly promoted for ~1.7-fold (P<0.05) and ~2-fold (P<0.01) with miR-10b overexpression, respectively, (Fig. 1B and C). The expression of the epithelial marker E-cadherin, mesenchymal markers N-cadherin and vimentin were detected to confirm the effects of miR-10b on EMT. miR-10b induced increased vimentin expression, however, no effects on E-cadherin and N-cadherin expression were observed (Fig. 1D). In addition, an increase in p-AKT expression was observed in miR-10b-overexpressed MCF10A cells (Fig. 1D). Conversely, the motility and proliferation of MCF10A cells were significantly reduced when p-AKT was inhibited (Fig. 1B and C) with an AKT antagonist MK2206, compared with miR-10b-transfected cells (Fig. 1E). Following the addition of the MK2206 inhibitor, the data indicated that miR-10b may promote the cell motility and proliferation via activating AKT.
Figure 1.
miR-10b promotes the motility and proliferation of BC cells via activation of AKT. (A) Fold-changes of miR-10b were assessed in miR-10b transiently overexpressed MCF10A cells. (B) MCF10A cells transiently transfected with miR-10b and treated with or without MK2206 were subjected to wound healing assay (Scale bar, 200 µm) and transwell assay (×100 magnification). (C) Analysis of proliferation of miR-10b transiently overexpressed MCF10A cells cultured ± MK2206 inhibitor. (D) Western blot analysis of E-cadherin, N-cadherin, vimentin, p-AKT and AKT in miR-10b transiently overexpressed MCF10A cells. (E) Western blot analysis of p-AKT and AKT in miR-10b transiently overexpressed MCF10A cells ± MK2206 inhibitor. **P<0.01, ***P<0.001 vs. NC. BC, breast cancer; AKT, protein kinase B; p-AKT, phosphorylated protein kinase B; miR, microRNA; NC, negative control; E, epithelial; N, neural.
Roles of FUT8 on cell motility and proliferation
Accumulated evidence has highlighted the important role of glycosylation in cancer progression (17). In a previous study, we performed a gene array to analyze the changes of glycogenes in miR-10b-overexpressed MCF10A cells. Specifically, it was notable that FUT8 was significantly upregulated by miR-10b (Fig. 1D). It was thus proposed that miR-10b may function through the regulation of the target gene, FUT8.
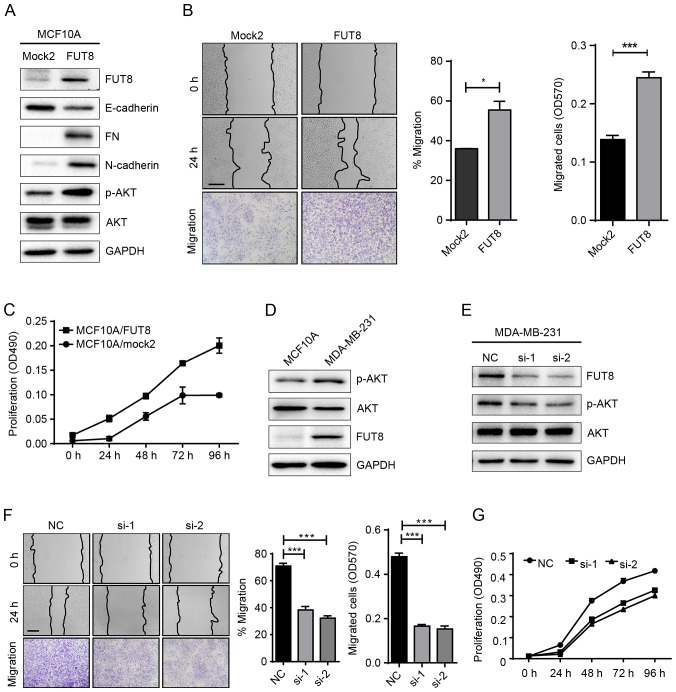
To validate our hypothesis, we established stable FUT8-overexpressed cells MCF10A/FUT8 (Fig. 2A). There was a 2.5-fold increase (P<0.05) of p-AKT expression in MCF10A/FUT8 cells, compared with Mock2 cells. In addition, overexpressed FUT8 resulted in decreased E-cadherin and enhanced Fibronectin (FN) and N-cadherin expression (Fig. 2A), though no obvious change on cell morphology was observed (data not shown). The effects of FUT8 overexpression significantly promoted the motility and proliferation of MCF10A cells at 24 h, compared with Mock2-transfected cells (Fig. 2B and C). Metastatic MDA-MB-231 cells, in comparison with MCF10A cells, displayed increased p-AKT and FUT8 expression (Fig. 2D). Transient silencing of FUT8 in MDA-MB-231 cells attenuated FUT8 and p-AKT expression (Fig. 2E), and decreased cell motility and proliferation, compared with NC cells (Fig. 2F and G).
Figure 2.
Effects of FUT8 on cell motility and proliferation. (A) Western blot analysis of EMT markers, p-AKT and AKT, detected in FUT8-overexpressed MCF10A cells. (B) FUT8-overexpressed MCF10A cells were subjected to wound healing and Transwell assay. *P<0.05, ***P<0.001 vs. Mock. (C) Proliferation analysis of FUT8-overexpressed MCF10A cells. (D) Western blot analysis of p-AKT, AKT and FUT8 expression in MCF10A and MDA-MB-231 cells. (E) Western blot analysis of p-AKT, AKT and FUT8 in FUT8-knockdown MDA-MB-231 cells. (F) Wound healing and Transwell assay (Scale bar, 200 µm; ×100 magnification) in FUT8-knockdown MDA-MB-231 cells. (G) MTT was performed in FUT8-knockdown MDA-MB-231 cells. ***P<0.001 vs. NC. FUT8, fucosyltranferase 8; p-AKT, phosphorylated protein kinase B; AKT, protein kinase B; NC, negative control.
The effects of miR-10b on p-AKT expression in TGF-β-treated and Twist-overexpressed MCF10A cells
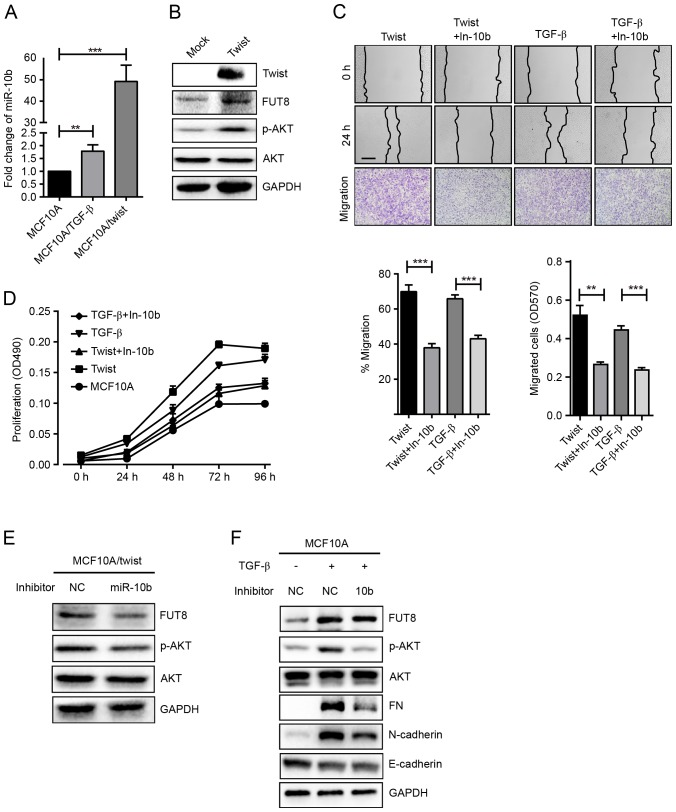
Transcriptional factor Twist has been revealed to induce the EMT process in multiple cell models (18). Twist directly binds to an E-box proximal to the putative promoter of miR-10b and regulates its transcription (8). Herein, a stable Twist-overexpressed cell MCF10A/Twist was established, and the expression of miR-10b, FUT8 and p-AKT were detected. It was demonstrated that overexpressed Twist significantly upregulated miR-10b and FUT8 and activated the phosphorylation of AKT (Fig. 3A and B). As expected, increased cell motility and proliferation were also observed, compared with MCF10A cells (Fig. 3C and D). Conversely, repression of miR-10b by its inhibitor, resulted in significantly decreased FUT8 and p-AKT expression in Twist-overexpressed MCF10A cells (Fig. 3E). Consistently, cell motility and proliferation were decreased (Fig. 3C and D).
Figure 3.
Effects of miR-10b on p-AKT expression in TGF-β-treated and Twist-overexpressed MCF10A cells. (A) miR-10b expression was detected in TGF-β-treated and Twist-overexpressed MCF10A cells and presented as fold-changes. **P<0.01, ***P<0.001 vs. MCF10A. (B) Western blot analysis of FUT8, p-AKT and AKT, detected in Twist-overexpressed MCF10A cells. (C) TGF-β-treated and Twist-overexpressed MCF10A cells were subjected to wound healing and transwell assay ± inhibitor of miR-10b. **P<0.01; ***P<0.001 vs. TGF-β-treated or Twist-overexpressed MCF10A cells. (D) Proliferation analysis in TGF-β-treated and Twist-overexpressed MCF10A cells ± inhibitor of miR-10b. (E) Western blot analysis of FUT8, p-AKT and AKT protein in Twist-overexpressed MCF10A cells, ± inhibitor of miR-10b. (F) Western blot analysis of EMT markers, FUT8, p-AKT and AKT, detected in TGF-β-treated MCF10A cells, ± inhibitor of miR-10b. (Scale bar, 200 µm; ×100 magnification). p-AKT, phospho- protein kinase B; TGF-β, transforming growth factor-β; FUT8, fucosyltranferase; p-AKT, phosphorylated protein kinase B; AKT, protein kinase B; EMT, epithelial-mesenchymal transition; miR, microRNA.
The same phenomenon was observed in TGF-β-induced cells. TGF-β repressed the expression of E-cadherin, enhanced the expression of Fibronectin, N-cadherin and FUT8, and activated the AKT pathway (Fig. 3A). When TGF-β-induced MCF10A cells were treated with miR-10b inhibitor, cell motility and proliferation were decreased (Fig. 3C and D). In addition, expression of Fibronectin, N-cadherin and p-AKT were decreased, while the expression of FUT8 was slightly attenuated (Fig. 3F).
Inhibitor of miR-10b attenuated p-AKT expression and reduced the migration and proliferation capacity in MDA-MB-231 cells
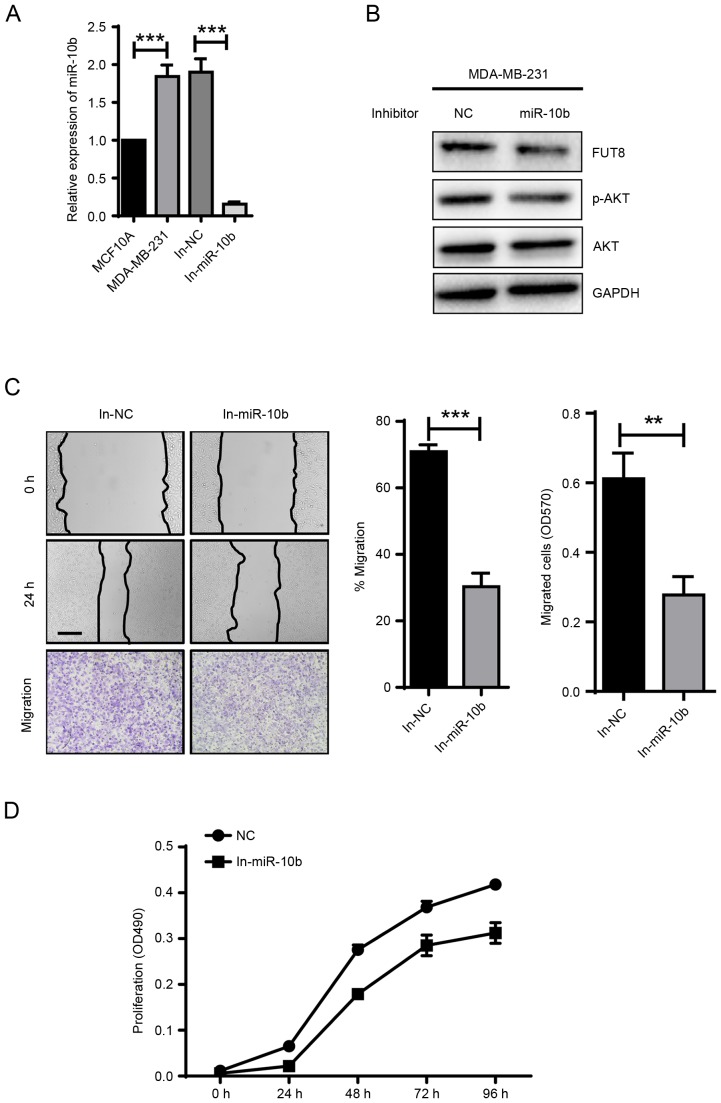
miR-10b is reported to be abundant in breast cancer cells. Notably, higher miR-10b expression was observed in triple-negative breast cancer cell lines compared with 6 human breast cancer-derived cell lines (19). Consistent with the previous report, miR-10b expression in MDA-MB-231 cells was significantly higher than in MCF10A cells (Fig. 4A). In MDA-MB-231 cells treated with miR-10b inhibitor for 48 h, miR-10b expression was significantly repressed (Fig. 4A). In response to the downregulation of miR-10b, FUT8 and p-AKT expression were also decreased (Fig. 4B). These results were consistent with the observation in both Twist-overexpressed and TGF-β-induced MCF10A cells. Furthermore, downregulated cell motility and proliferation were detected with the addition of miR-10b inhibitor in MDA-MB-231 cells, compared with NC cells (Fig. 4C and D).
Figure 4.
Inhibitor of miR-10b significantly attenuated the expression of p-AKT and reduced migration and proliferation in MDA-MB-231 cells. (A) miR-10b expression were detected in MDA-MB-231 cells with addition of miR-10b inhibitor. ***P<0.001 vs. MCF10A. (B) Western blot analysis. Expression of FUT8, p-AKT and AKT in MDA-MB-231cells with miR-10b inhibitor. (C) MDA-MB-231 cells were subjected to wound healing and transwell assay (Scale bar, 200 µm; ×100 magnification) ± inhibitor of miR-10b. **P<0.01; ***P<0.001 vs. NC. (D) Analysis of proliferation in MDA-MB-231 cells with addition of miR-10b inhibitor. FUT8, fucosyltranferase; p-AKT, phosphorylated protein kinase B; miR, microRNA; NC, negative control.
Discussion
Altered glycosylation is frequently associated with tumor development and progression and the therapeutics and diagnostics based on glycans have been investigated (20–22). In addition, miRNAs are potential therapeutic targets for cancer due to the key regulatory roles, identified as oncogenes and tumor suppressors in previous years (23). miRNA dysregulation has been identified as a frequent phenomenon in breast cancer and certain dysregulated miRNAs have been validated (24,25). However, the roles of miRNAs in glycosylation during tumor progression were rarely reported.
miR-10b, which is upregulated in breast cancer tissues and breast cancer cells, may contribute to lymph node metastases in breast cancer (26). In the present study, a glycan-related gene array was performed to analyze the variation of glycosylation in miR-10b-overexpressed MCF10A cells. It was observed that FUT8 expression was significantly increased at the mRNA and protein levels, and the AKT pathway was significantly activated. However, there were no significant changes in E-cadherin and N-cadherin detected, although a previous report demonstrated that miR-10b exerted a critical function in TGF-β-induced EMT process in breast cancer (7).
Upregulated core fucosylation has been reported in multiple types of cancer, including, prostate (27), non-small cell lung (28) and breast cancer (10). Increased core fucosylation was also detected in EMT (12), consistent with the result in the present study in TGF-β-induced MCF10A cells. In the present study, a decline in the expression of epithelial molecule E-cadherin and an increase in that of mesenchymal markers (Fibronectin and N-cadherin) were observed in FUT8-overexpressed MCF10A cells. Furthermore, upregulated p-AKT expression, increased cell migration and proliferation were also demonstrated, consistent with the role of FUT8 in regulating the activity of the PI3K/AKT signaling pathway (29). Similar results were discovered when miR-10b or FUT8 were knocked-down in MDA-MB-231 cells, revealing a potential association between miR-10b, FUT8 and the AKT pathway. In addition, the analysis of inhibited miR-10b in TGF-β1-induced and Twist-overexpressed MCF10A cells, led to the following observations: (i) EMT process was partially reversed and the level of p-AKT was significantly attenuated, although FUT8 was slightly altered in TGF-β1-induced MCF10A cells. (ii) FUT8 and p-AKT were significantly attenuated with inhibition of miR-10b in Twist-overexpressed MCF10A cells. (iii) Cell migration and proliferation were decreased in response to the inhibition of miR-10b.
miRNAs regulate the expression of target genes at the post-transcriptional level through sequence-specific binding to the 3′UTR of target genes (5). miR-26a, miR-34a and miR-146a repress FUT8 expression by binding to the 3′UTR of FUT8 in hepatocellular carcinoma (11). However, the exact mechanism by which miR-10b manipulates FUT8 expression remains unclear. A potential miR-10b/TFAP2C/STAT3 pathway is proposed by querying the Pathway Commons (http://www.pathwaycommons.org).
Taken together, the results of the present study demonstrated that miR-10b upregulates FUT8 expression, which further activates the AKT pathway, enhancing the migration and proliferation of breast cancer cells. Further follow-up studies will be focused on the mechanism of miR-10b on the regulation of FUT8 in breast cancer cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81672537), the Natural Science Foundation of Jiangsu Province, China (grant nos. BK20160173 and BK20161132) and the Fundamental Research Funds for the Central Universities (grant nos. JUSRP51619B and JUSRP116032).
Availability of data and materials
All data generated or analyzed during current study are available from the corresponding author on reasonable request.
Authors' contributions
FG and DG designed the experiments. DG, JG and XL performed the experiments and analyzed the data. DG and FG wrote the paper.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fan L, Goss PE, Strasser-Weippl K. Current status and future projections of breast cancer in asia. Breast Care (Basel) 2015;10:372–378. doi: 10.1159/000441818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Peart O. Metastatic breast cancer. Radiol Technol. 2017;88:519M–539M. [PubMed] [Google Scholar]
- 4.Bushati N, Cohen SM. microRNA functions. Ann Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 5.Samantarrai D, Dash S, Chhetri B, Mallick B. Genomic and epigenomic cross-talks in the regulatory landscape of miRNAs in breast cancer. Mol Cancer Res. 2013;11:315–328. doi: 10.1158/1541-7786.MCR-12-0649. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han X, Yan S, Weijie Z, Feng W, Liuxing W, Mengquan L, Qingxia F. Critical role of miR-10b in transforming growth factor-β1-induced epithelial-mesenchymal transition in breast cancer. Cancer Gene Ther. 2014;21:60–67. doi: 10.1038/cgt.2013.82. [DOI] [PubMed] [Google Scholar]
- 8.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 9.Bahena-Ocampo I, Espinosa M, Ceballos-Cancino G, Lizarraga F, Campos-Arroyo D, Schwarz A, Maldonado V, Melendez-Zajgla J, Garcia-Lopez P. miR-10b expression in breast cancer stem cells supports self-renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep. 2016;17:648–658. doi: 10.15252/embr.201540678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue L, Han C, Li Z, Li X, Liu D, Liu S, Yu H. Fucosyltransferase 8 expression in breast cancer patients: A high throughput tissue microarray analysis. Histol Histopathol. 2016;31:547–555. doi: 10.14670/HH-11-693. [DOI] [PubMed] [Google Scholar]
- 11.Cheng L, Gao S, Song X, Dong W, Zhou H, Zhao L, Jia L. Comprehensive N-glycan profiles of hepatocellular carcinoma reveal association of fucosylation with tumor progression and regulation of FUT8 by microRNAs. Oncotarget. 2016;7:61199–61214. doi: 10.18632/oncotarget.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CY, Jan YH, Juan YH, Yang CJ, Huang MS, Yu CJ, Yang PC, Hsiao M, Hsu TL, Wong CH. Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc Natl Acad Sci USA. 2013;110:630–635. doi: 10.1073/pnas.1220425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osumi D, Takahashi M, Miyoshi E, Yokoe S, Lee SH, Noda K, Nakamori S, Gu J, Ikeda Y, Kuroki Y, et al. Core fucosylation of E-cadherin enhances cell-cell adhesion in human colon carcinoma WiDr cells. Cancer Sci. 2009;100:888–895. doi: 10.1111/j.1349-7006.2009.01125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta A, Comunale MA, Rawat S, Casciano JC, Lamontagne J, Herrera H, Ramanathan A, Betesh L, Wang M, Norton P, et al. Intrinsic hepatocyte dedifferentiation is accompanied by upregulation of mesenchymal markers, protein sialylation and core alpha 1,6 linked fucosylation. Sci Rep. 2016;6:27965. doi: 10.1038/srep27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Guo D, Guo J, Li X, Guan F. Differential effects of Pax3 on expression of polysialyltransferases STX and PST in TGF-β-treated normal murine mammary gland cells. Exp Biol Med (Maywood) 2017;242:177–183. doi: 10.1177/1535370216669838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munkley J, Elliott DJ. Hallmarks of glycosylation in cancer. Oncotarget. 2016;7:35478–35489. doi: 10.18632/oncotarget.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng Y, Li X. The roles of HLH transcription factors in epithelial mesenchymal transition and multiple molecular mechanisms. Clin Exp Metastasis. 2014;31:367–377. doi: 10.1007/s10585-013-9621-6. [DOI] [PubMed] [Google Scholar]
- 19.Fkih M'hamed I, Privat M, Ponelle F, Penault-Llorca F, Kenani A, Bignon YJ. Identification of miR-10b, miR-26a, miR-146a and miR-153 as potential triple-negative breast cancer biomarkers. Cell Oncol (Dordr) 2015;38:433–442. doi: 10.1007/s13402-015-0239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuchlova Horynova M, Raska M, Clausen H, Novak J. Aberrant O-glycosylation and anti-glycan antibodies in an autoimmune disease IgA nephropathy and breast adenocarcinoma. Cell Mol Life Sci. 2013;70:829–839. doi: 10.1007/s00018-012-1082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho WL, Hsu WM, Huang MC, Kadomatsu K, Nakagawara A. Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma. J Hematol Oncol. 2016;9:100. doi: 10.1186/s13045-016-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Leoz ML, Young LJ, An HJ, Kronewitter SR, Kim J, Miyamoto S, Borowsky AD, Chew HK, Lebrilla CB. High-mannose glycans are elevated during breast cancer progression. Mol Cell Proteomics. 2011;10:M110.002717. doi: 10.1074/mcp.M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwan JY, Psarianos P, Bruce JP, Yip KW, Liu FF. The complexity of microRNAs in human cancer. J Radiat Res. 2016;57(Suppl 1):i106–i111. doi: 10.1093/jrr/rrw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piva R, Spandidos DA, Gambari R. From microRNA functions to microRNA therapeutics: Novel targets and novel drugs in breast cancer research and treatment (Review) Int J Oncol. 2013;43:985–994. doi: 10.3892/ijo.2013.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christodoulatos GS, Dalamaga M. Micro-RNAs as clinical biomarkers and therapeutic targets in breast cancer: Quo vadis? World J Clin Oncol. 2014;5:71–81. doi: 10.5306/wjco.v5.i2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min W, Wang B, Li J, Han J, Zhao Y, Su W, Dai Z, Wang X, Ma Q. The expression and significance of five types of miRNAs in breast cancer. Med Sci Monit Basic Res. 2014;20:97–104. doi: 10.12659/MSMBR.891246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Chen J, Li QK Peskoe SB, Zhang B, Choi C, Platz EA, Zhang H. Overexpression of α (1,6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology. 2014;24:935–944. doi: 10.1093/glycob/cwu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honma R, Kinoshita I, Miyoshi E, Tomaru U, Matsuno Y, Shimizu Y, Takeuchi S, Kobayashi Y, Kaga K, Taniguchi N, Dosaka-Akita H. Expression of fucosyltransferase 8 is associated with an unfavorable clinical outcome in non-small cell lung cancers. Oncology. 2015;88:298–308. doi: 10.1159/000369495. [DOI] [PubMed] [Google Scholar]
- 29.Cheng L, Luo S, Jin C, Ma H, Zhou H, Jia L. FUT family mediates the multidrug resistance of human hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell Death Dis. 2013;4:e923. doi: 10.1038/cddis.2013.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during current study are available from the corresponding author on reasonable request.