Abstract
Previous studies have indicated that certain microRNAs (miRNAs/miRs) function as either tumor suppressors or oncogenes in human cancer. The present study identified the miR-23a/27a/24-2 cluster, containing miR-23, miR-27a and miR-24, as an oncogene in gastric cancer. The expression of the miR-23a/27a/24-2 cluster was upregulated in clinical gastric cancer tissues. Transfection with inhibitors of miR-23a, miR-27a, or miR-24, either independently or together, repressed in vitro colony formation and in vivo tumor formation. The miR23a/27a/24-2 cluster inhibitors repressed the growth of gastric cancer cells in a synergistic manner. In addition, treatment with lower doses of the miRNA inhibitor mixture induced the formation of apoptotic bodies. According to computational predictions using TargetScan, suppressor of cytokine-induced signaling 6 (SOCS6) was identified as one of the downstream target genes of the miR-23a/27a/24-2 cluster. The expression of SOCS6 was significantly lower in tumor tissues than in matched normal tissues (P<0.01) and was associated with poor survival (P<0.00001). Taken together, these results strongly suggested that the miR-23a/27a/24-2 cluster may mediate the progression of gastric cancer through the suppression of SOCS6 expression. The present study also provides a novel molecular target for the development of an anti-gastric cancer agent.
Keywords: microRNA-23a/27a/24-2 cluster, gastric cancer, suppressor of cytokine-induced signaling 6
Introduction
Gastric cancer was the second most common cause of cancer-associated mortality worldwide in 2014 (1). The long-term survival of patients with gastric cancer remains unsatisfactory because of increased incidences of recurrence and chemotherapy resistance (2). A substantial proportion of gastric cancer cases are either inherently resistant to chemotherapy or develop resistance during the course of therapy. Therefore, an improved understanding of the molecular mechanisms involved will ultimately result in more effective methods of overcoming chemotherapy resistance and developing novel antineoplastic treatment strategies.
MicroRNAs (miRNAs/miRs) are genomically encoded, small, non-coding RNAs (ncRNAs) that negatively regulate gene expression by controlling either translation or stability of mRNAs through an RNA interference-like pathway (3). miRNAs comprise 1–3% of the human genome (4) and regulate 30% of human gene expression (5). The majority of miRNAs are located within the introns of either the protein-coding or non-coding transcriptional units and are expressed with their host genes coordinately (6). A few miRNA genes are located in the exons of ncRNAs (6). miRNAs that cluster in the same genome region are transcribed as polycistronic transcripts (6). Studies have demonstrated that miRNAs are associated with multiple physiological processes, including aging, differentiation, hematopoiesis and endocrine functions, and also function as key regulators in the progression of a number of human diseases, including heart disease and cancer (7–11).
For the past three decades, changes to the expression of certain gene has been proposed to be the major factors of tumorigenesis as well as metastasis (12). The majority of cancer-associated genes are thought to be protein-coding genes. Previous studies have demonstrated that there are associations between cancer and miRNA (13–15). In addition, silencing the expression of miRNA-processing factors in a transgenic mouse model increases the susceptibility of patients to cancer (16). Evidence indicates that miRNAs serve significant functions in almost all aspects of cancer biology, including cell proliferation, apoptosis, angiogenesis, invasion, metastatic lesions and drug resistance (17). Recently, multiple dysregulated miRNAs were found to participate in numerous aspects of gastric cancer (18). miR-31, miR-106a and miR-21 are reported to possess clinical significance (9). Functions for miR-451, miR-141, miR-34a and miR-27a have been identified in the progression of gastric cancer (19–22). miR-15b and miR-16 modulate multidrug resistance of gastric cancer cells by negatively regulating B-cell lymphoma 2 (BCL2) expression (23). Although multiple miRNAs have been shown to function as bona fide oncogenes or tumor suppressors, the precise functions and the molecular mechanism underlying their dysregulation of the gastric cancer progression, as well as the development of chemotherapy resistance, remain largely unknown (15,24,25). In addition to individual miRNAs, the functions and regulation of the genomic miRNA cluster have also not been clearly elucidated in gastric cancer (26).
The present study analyzed the expression of microRNAs in gastric cancer tissues using microRNA arrays. In vitro and in vivo experiments were applied to investigate their oncogenic functions in gastric cancers.
Materials and methods
Patients and microRNA arrays
Gastric cancer and its corresponding adjacent normal tissues were obtained from 68 patients who received surgical resection in the Department of General Surgery, Taipei Veterans General Hospital (Taipei, Taiwan). A total of 5 patients were <65 years old, while the remainder were >65 years old. Overall, 5 patients were female and the remainder were male. Tumor and adjacent normal tissues were snap-frozen and stored in liquid nitrogen. The Institutional Review Board of Taipei Veterans General Hospital approved the use of these tissues. The total RNA from 68 pairs of gastric cancer tissues and their adjacent normal tissues were extracted by TRIzol® (Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA). MicroRNA analyses were commissioned by the High-throughput Genome Analysis Core Facility of the VYM Genome Research Center (National Yang-Ming University, Taipei, Taiwan) on NCode™ Multi-Species miRNA microarrays (Invitrogen; Thermo Fisher Scientific, Inc.). Only 21 pairs of total RNA passed the quality check of the core facility for microarray analysis and were subsequently used. Results were analyzed using the Partek Genomics suite V6.6 (Partek, Inc., St. Louis, MO, USA) for multi-dimensional scaling, clustering, and heat map drawing.
Cell lines, cell culture and transfection
The gastric cancer SC-M1 cell line was obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.) containing 100 mg/ml penicillin-streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) and 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA). A total of 5×105 SC-M1 cells were seeded into 6-well plates for 12-h, with fresh medium replaced to a total volume of 500 µl 1 h prior to transfection. TransIT TKO reagent (6 µl; Mirus Bio, LLC, Madison, WI, USA) was mixed with 200 µl serum-free Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) for 20 min and then 2 µl microRNA inhibitors (Dharmacon Inc., Lafayette, CT, USA) for hsa-miR-23a (IH-300494-05-0010), hsa-miR-27a (cat. no. IH-300502-05-0020), hsa-miR-24-2 (IH-300497-05-0010) and the control oligo (cat. no. IN-001-005-01-20) were added for 20 min. The mixture was added into the well and incubated with the cells at room temperature for 30 min.
Nucleic acid (nuclear) staining/DAPI staining
Cells pre-treated with 30 nM miRNA inhibitor mix or scramble RNA for 24 h were washed 3 times in PBS. DAPI stain solution (300 nM; Thermo Fisher Scientific, Inc.) was added at a sufficient quantity to cover the cells, then protected from light and incubated at room temperature for 5 min. The cells were washed 3 times in PBS and visualized by fluorescence microscopy. Normal nuclear and apoptotic bodies were visualized and distinguished.
Clonogenic assays
For the clonogenic assay, 100 SC-M1 cells that were pre-treated with 30 or 90 nM of a miR-23a, miR-27a or miR-24 inhibitor, either independently or together at 24 h after transfection, were seeded in 35-mm dishes. The seeding density was 11 cells/cm2. All experiments were conducted in triplicate. The dishes were incubated at 37°C in a 5% CO2 incubator for 15 days and the medium was changed every 3 days. Colonies were fixed with methanol (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at room temperature for 10 min, stained with 1% crystal violet (Sigma-Aldrich; Merck KGaA) incubated at room temperature for 30 min, and then counted using ImageQuant TL (version 7.0; GE Healthcare, Chicago IL, USA).
Tumorigenesis in nude mice
All animal experiments were approved by and performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of National Yang-Ming University (Taipei, Taiwan). In total, 3×106 viable SC-M1 cells pre-treated with either the control oligo or miRNA inhibitor mixture were washed in PBS and injected subcutaneously into the 2 h indlimbs of 5-week-old male BALB/c nude (nu/nu) mice to assess tumor formation (initial bodyweight, 17.6±2.9 g). The mice were supplied by the National Laboratory Animal Center (Taipei, Taiwan). Each group contained 5 mice. The housing conditions followed the guidelines of the Animal Center, National Yang-Ming University (Taipei, Taiwan) (temperature, 20–22°C; humidity, 50–70%; normal diet, specific pathogen-free). Primary tumor volume was monitored with a ruler every 3 days. The tumor size was considered to be the humane endpoint, with a maximum tumor diameter of 20 mm. The mice were sacrificed after 27 days. The mice were sacrificed using 30 psi CO2 for <20 sec until cardiac arrest. Tumor volume was calculated by the formula: Tumor volume=length × width2 ×0.5.
microRNA target gene prediction by TargetScanHuman
TargetScanHuman (http://www.targetscan.org) predicts target genes of miRNAs by searching for the presence of conserved 6-8mer sites that match the seed region of each miRNA. It considers matches to human 3′-untranslated regions (UTRs) and their orthologs.
Dual luciferase reporter assay
Luciferase reporter constructs were generated by cloning a specific miRNA-binding sequence (wild-type/mutants), 3′UTR of SOCS6, into the NotI and XbaI site located at the psiCHECK2 dual luciferase expression vector (Promega Corporation, Madison, WI, USA). In total, 1.5×105 SC-M1 cells were seeded in 12-well plates for 16 h and co-transfected with 100 ng luciferase expression vector and 90 nM miRNA inhibitors, either independently or jointly, with TransIT TKO reagent (Mirus Bio, LLC). Subsequent to a 24-h incubation at 37°C, 5% CO2, cell lysates were collected and luciferase activity (Dual-Luciferase Reporter Assay System; Promega Corporation) was detected using a microplate reader. Activity was normalized by comparison with Renilla luciferase.
Western blot analysis
Cell lysates were prepared in RIPA buffer [(150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris-HCl, and 5 mM EDTA (pH 7.5)] containing cocktails of protease inhibitors (Roche, Mannheim, Germany). The relative concentration of protein in the lysates was determined by BCA protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). For each lane of 10% SDS-PAGE, 20 µg of protein lysates was separated in gel and subsequently transferred onto Hybond ECL membranes (Amersham; GE Healthcare, Chicago, IL, USA). The membranes were gently agitated in the blocking solution (5% skimmed mik in 1X Tris-buffered saline with Tween-20 at 4°C overnight) and then were probed with anti-SOCS6 antibody (cat. no. sc-5608; 1:500 dilution; Santa Cruz Biotechnology, Inc., Callas, TX, USA) and with anti-β-actin antibody (cat. no. A5316; 1:20,000 dilution; Sigma-Aldrich; Merck KGaA), as an internal control for protein loading at 4°C 6 h. The secondary antibodies were anti-mouse IgG and anti-rabbit IgG (cat. nos. NA931 and NA934; 1:10,000 dilution, Amersham; GE Healthcare, Chicago, IL, USA). The western blot analysis was detected with SuperSignal enhanced chemiluminescence reagents (Pierce; Thermo Fisher Scientific, Inc.) and film autoradiography.
Statistical analysis
Based on SPSS v20 (IBM Corp, Armonk, NY, USA) differences between groups were analyzed using three-way analysis of variance followed by Tukey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference. Kaplan-Meier survival analysis was also conducted, with P-values calculated by log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression profile of miRNA in gastric cancer
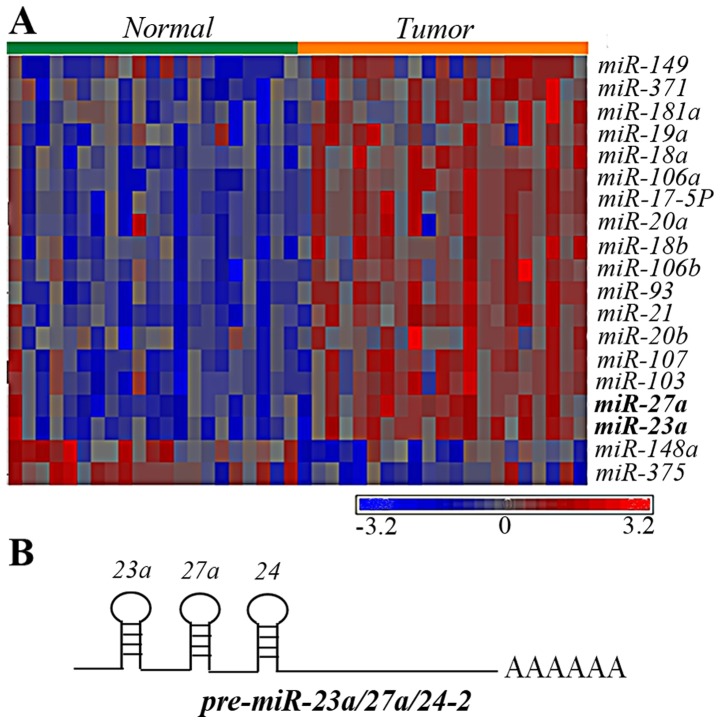
To identify dysregulated miRNAs that may participate in the tumorigenesis of gastric cancer, miRNA microarray analyses were performed on 21 pairs of gastric cancer tissues and adjacent non-neoplastic stomach tissues. Using a three-way analysis of variance, dysregulated miRNAs were defined as having ≥1.5 folds of change in their expression levels. The P-value was set at 0.01. A total of 19 miRNAs were significantly differentially expressed compared with the paired tissues (Fig. 1A). Of the upregulated miRNAs, miR-23a and miR27a, attracted attention, since they belong to the miR-23a/27a/24-2 cluster (Fig. 1B). In addition, even though miR-24 barely reached a significant P-value (P=0.0319), it was upregulated in 15 out of 21 patients. Therefore, in the present study it was hypothesized that the miR-23a/27a/24-2 cluster was upregulated in gastric cancer and possessed oncogenic activities.
Figure 1.
miR expression pattern in gastric cancer. (A) Heat map showing expression levels of the indicated miRNA in relation to tissue type. The color represents the expression level of the microRNA, according to the color key shown below the heat map. Red represents high expression; blue represents low expression. (B) The structure of the pre-miRNA-transcript of miR-23a/27a/24-2. mi, micro; miR, microRNA.
Clonogenic, apoptosis and proliferation assays for miR inhibitor treatment
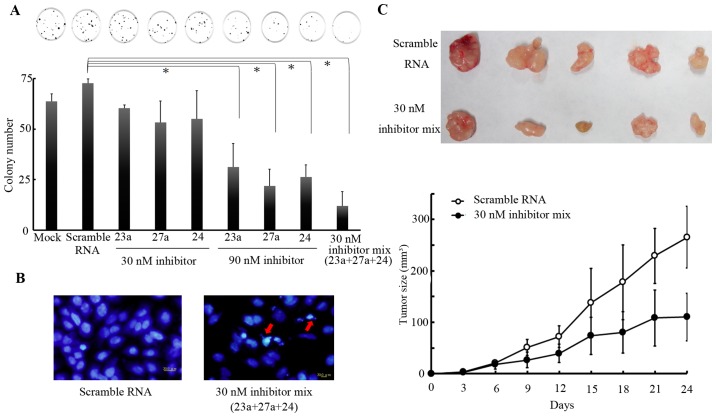
To confirm the aforementioned hypothesis, SC-M1 cells were selected, owing to their increased expression levels of miR-23a/27a/24-2 cluster. Clonogenic assays were performed in SC-M1 cells that were treated with 30 or 90 nM of a miR-23a, miR-27a or miR-24 inhibitor, either independently or together at 24 h after transfection. Results revealed that treatments with either or both miRNA inhibitors repressed cell proliferation (Fig. 2A). The miR23a/27a/24-2 cluster inhibitors caused repression of the proliferation of gastric cancer cells in a synergetic manner. In addition, the treatment with the decreased dose, 30nM, of the 3 miRNA inhibitors induced the formation of apoptotic body (Fig. 2B). These results indicated that the miR-23a/27a/24-2 cluster promoted the proliferation of gastric cancer cells. To validate these observations in vivo, SC-M1 cells pre-treated with either the control oligo or miRNA inhibitor mixture were injected into limbs of nude mice. Tumor size was measured once every 3 days for 24 days. Tumor growth in the group treated with miRNA inhibitor mixture was significantly decreased compared with that in the control group following an incubation period of 21 days (P<0.05; Fig. 2C). The tumor masses isolated from mice in the group treated with the 3 miRNA inhibitors were smaller compared with those in the control group (Fig. 2C). Taken together, these results indicated that miR-23a/27a/24-2 cluster may possess oncogenic activities.
Figure 2.
Functional assay for miR inhibitor treatment. (A) Clonogenic assays for SC-M1 cells were treated with miR-23a, miR-27a and miR-24 inhibitor. Colony numbers were counted using Image J. *P<0.05 vs. scramble RNA pre-treated SC-M1 cells. (B) SC-M1 cells were treated with miR-23a, miR-27a and miR-24 inhibitors mixture for 24 h and stained with DAPI. Red arrows indicate apoptotic bodies. (C) miR-23a, miR-27a and miR-24 inhibitor mixture pre-treated SC-M1 cells were implanted subcutaneously into nude mice, resulting in the observed tumors. P<0.05 vs. scramble RNA pre-treated SC-M1 cells. miR, microRNA.
Search for the downstream target genes of miR-23a, miR-27a and miR-24
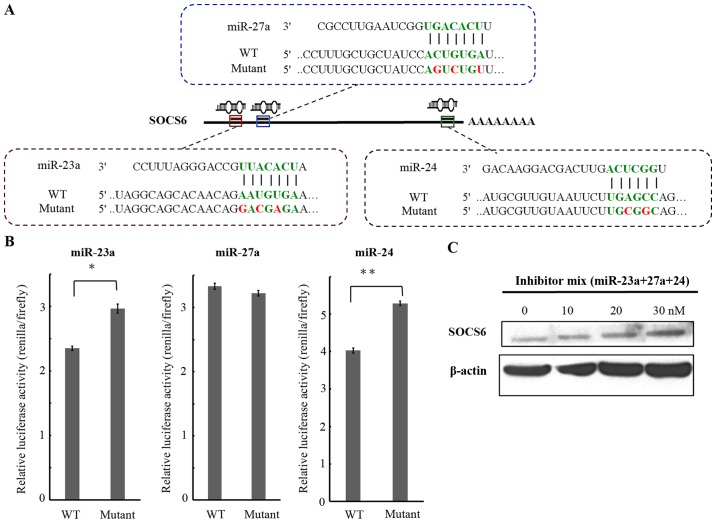
The downstream targets of miRNAs are critical in determining the roles of miRNA. Since expression consistency and growth repression by the mRNA inhibitor mixture was presented in a synergistic manner, it was presumed that miR-23a, miR-27a, and miR-24 may act in combination to perform their functions by targeting either a common downstream gene or various genes in the same signaling pathway. TargetScanHuman was used to identify the putative downstream target genes of the miR-23a/27a/24-2 cluster that may control either cell growth or cell death. Accordingly, the suppressor of cytokine singaling-6 (SOCS6), which belongs to the SOCS family of negative regulators of the cytokine signaling pathway (27), was predicted to be a common potential target for miR-23a, miR-27a and miR-24. There were 3 putative binding sites, located at 240–282, 298–319, and 3099–3120 nt after the stop codon of the SOCS6 mRNA for miR-23a, miR-27a and miR-24, respectively (Fig. 3A). To demonstrate that the miR-23a/27a/24-2 cluster is able to target directly to the 3′-UTR of the SOCS6 mRNA, a luciferase reporter assay was conducted. To perform this assay, 3 reporter plasmids containing dual luciferase genes followed by DNA fragments containing the putative target sites for miR-23a, miR-27a, and miR-24 were constructed. There were also three mutant constructs with a 2/3-nucleotide alteration within the seed regions of binding sites generated (Fig. 3A). SC-M1 cells were transfected with the reporter plasmid and three microRNA inhibitors, either independently or jointly and incubated. Luciferase activity was measured at 24 h after transfection. In the three mutant type constructs, when treated with the miRNA inhibitors individually, the relative luciferase activity increased 31% in the miR-23 mutant and 25% in the miR-24a mutant, compared with the wild-type-treated group (Fig. 3B). Analysis of the protein expression of SOCS6 revealed that treatment with the miR-23a, miR27a and miR24 inhibitors were able to restore the SOCS6 protein expression level in a dose-dependent manner (Fig. 3C).
Figure 3.
SOCS6 is a target gene of miR-27a and miR-24. (A) The putative binding sites of miR-23a, miR-27a and miR-24 on SOCS6 were predicted using Targetscan. The interspecies sequence alignment revealed the mature miRNA sequences and the putative binding sites. Luciferase reporter constructs containing WT or mutant target sites of the 3′-untraslated region of SOCS6 mRNA for miR-23a, miR-27a and miR-24, respectively. (B) SC-M1 cells were co-transfected with dual luciferase reporter plasmid, containing a specific miRNA-binding sequence (wild-type or mutant), and 90 nM miR inhibitor for 24 h. Dual-luciferase activities were measured using a microplate reader. *P<0.05 and **P<0.01 for mutant vs. wild-type. (C) SC-M1 cells were treated with miR-23a, miR-27a and miR-24 inhibitors mixture for 48 h. The expression of SOCS6 was examined by western blotting. SOCS6, suppressor of cytokine-induced signaling 6; miR, microRNA; WT, wild-type.
Analysis of the expression of SOCS6 in gastric cancer patients
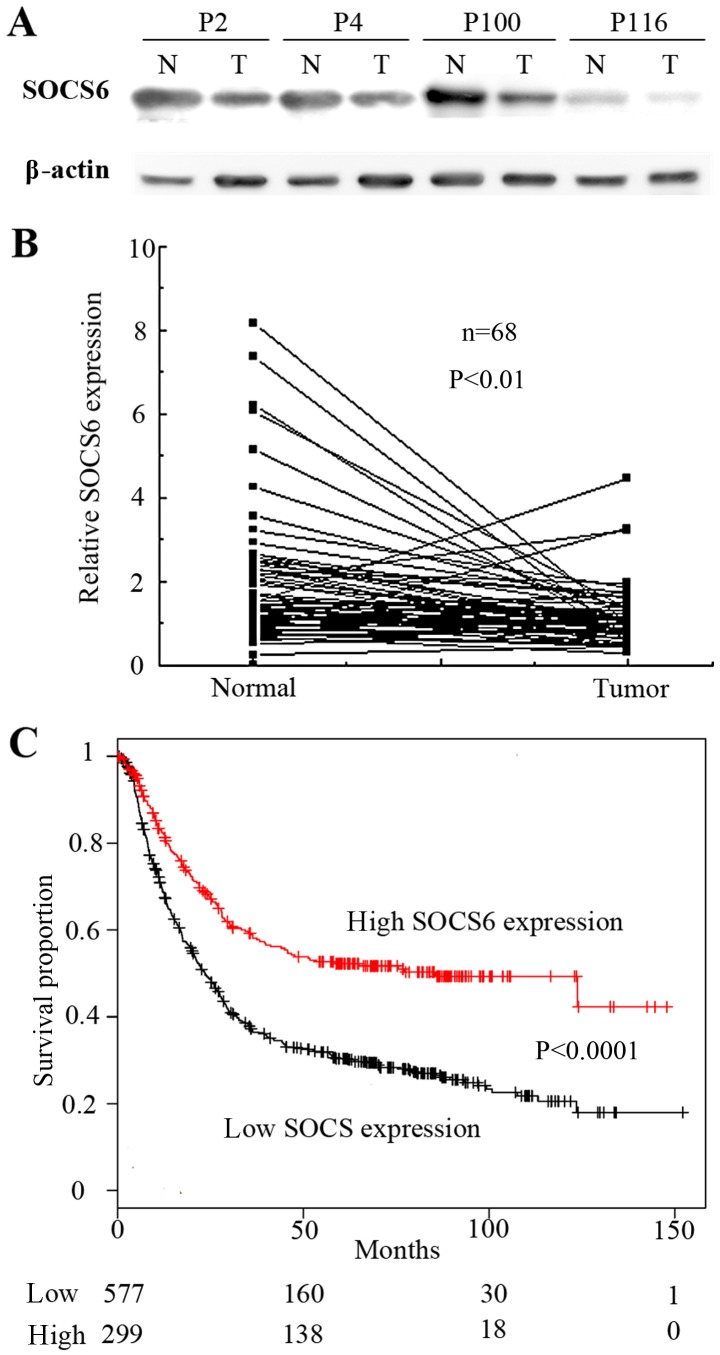
To confirm that SOCS6 was downregulated by the miR-23a/27a/24-2 cluster in gastric cancer, 68 pairs of clinical tissue samples were examined by western blot analysis (Fig. 4A). Quantification of western blot analysis revealed that the relative expression of SOCS6 in the tumor tissue was significantly lower than that in the normal tissue (P<0.01; Fig. 4B). To validate these observations, data from a microarray of 876 gastric cancer tissues obtained from the Kaplan-Meier plotter (http://kmplot.com/analysis/) (28). Kaplan-Meier analyses showed downregulation of SOCS6 was significantly associated with poor patient survival rates (P<0.00001; Fig. 4C). Taken together, these results indicate that SOCS6 is a downstream target gene of the miR-23a/27a/24-2 cluster in tumorigenesis of gastric cancer.
Figure 4.
Expression of SOCS6 in gastric cancer. (A) Western blot analysis of the expression of SOCS6 in patients with gastric cancer. (B) The relative expression of SOCS6 in gastric tumor and adjacent tissues was analyzed by western blot analysis of 68 samples. (C) Kaplan-Meier plot for overall survival of patients with gastric cancer categorized according to the expression of SOCS6. SOCS6, suppressor of cytokine-induced signaling 6; N, normal tissue; T, tumor tissue.
Discussion
The present study revealed that the miR-23a/27a/24-2 cluster was highly expressed in gastric cancer. In vivo and in vitro experiments indicated that miR-23a, miR-27a and miR-24 promoted tumor formation by enhancing cell growth. Target site prediction and luciferase reporter assays indicated that SOCS6 was regulated by miR-23a and miR-24. Finally, western blotting and survival analysis revealed that SOCS6 was downregulated in gastric cancers and positively associated with patient survival rates.
The miR-23a/27a/24-2 cluster is associated with multiple diseases (29–33). Even though the 3 miRNAs in this cluster are derived from a single primary transcript, their expression patterns vary depending on different biological conditions (34). For example, studies on colorectal cancer demonstrated that miR23a and miR27a were downregulated, but that miR-24 was upregulated in tumor samples (35). Such complex expression patterns may be due to post-transcriptional regulation during precursor processing; however, this mechanism requires further investigation (36). In the analysis of miRNA expression profiles in gastric cancer tissues, the miR-23a/27a/24-2 cluster was considered to be an oncogene owing to its increased fold-change of upregulation compared with that in adjacent normal regions. Although the degree of alteration to this upregulated expression of miR-23a, miR-27a and miR-24 exhibited variance, this may be due to individual differences between the miRNAs (data not shown). However, the trend of miR-23a, miR-27a and miR-24 upregulation was consistent.
Functionally, the miR-23a/27a/24-2 cluster has been proposed to control the cell cycle, cell proliferation, cell death and cell differentiation (24–26,28). In gastric cancer, miR-23a has been reported to promote cell proliferation by targeting interferon regulator factor 1 and interleukin-6 receptor (37,38). miR-27a also inhibits the expression of B-cell translocation gene 2 (BTG2) and prohibitin to facilitate cell proliferation in gastric cancer cell lines (39,40). BCL2L11 was targeted by miR-24 to regulate cell proliferation and apoptosis (41). Yuan et al (42) demonstrated that miRNAs of a miRNA cluster may work in combination to accomplish their function. In this study, miR-23a, miR-27a, and miR-24 were consistently upregulated in gastric cancer and may function cooperatively to serve the same function. The results of the present study, which indicated that the mixture of 3 microRNA inhibitors repressed cell proliferation in a synergistic manner, strongly supports this possibility.
SOCS6, located at chromosome 18q22, belongs to a member of the SOCS family of E3 ubiquitin ligases, a number of which have been implicated in the negative regulation of cytokine receptor signaling (27). Unlike other members of the SOCS family, SOCS6 neither binds to intermediate components of cytokine signaling pathways nor represses cytokine receptor signaling (43). SOCS6 is mainly associated with the negative regulation of receptor signaling by increasing the degradation, mediated by ubiquitination, of receptors or substrate proteins and induces apoptosis by targeting mitochondrial proteins (27). In gastric cancer cells, SOCS6 was reported to inhibit cell proliferation and colony formation ability, with SOCS6 expression inactivated by either loss of heterozygosity or epigenetic modification (44). These results also indicated that SOCS6 is commonly downregulated in patients with gastric cancer and is associated with poor survival rates. However, these results cannot fully explain those lower-expressed SOCS6 cases, the SOCS6 promoter regions of which were not hypermethylated. By contrast, the present study revealed that the expression of SOCS6 was negatively regulated by miR-23a and miR-24 in gastric cancer, providing an additional novel mechanism of SOCS6 regulation.
Taken together, the data presented in the present study indicated the miR-23a/27a/24-2 cluster functions as oncogenic miRNAs in gastric cancer. Downregulation of SOCS6 expression by miR-23a and miR-24 may cause the activation of those cell proliferation signaling pathways and the suppression of the apoptosis signals, resulting in poorer survival of patients with gastric cancer. The combined inhibition of miR-23a, miR-27a and miR-24 may represent an efficient gastric cancer therapy.
Acknowledgements
The authors acknowledge the technical services provided by High-throughput Genome & Big Data Analysis Core Facility of VYM Genome Research Center (National Yang-Ming University). The National Core Facility Program supports the Core Facility for Biotechnology (NCFPB), Ministry of Science and Technology.
Funding
The present study was supported by the Yen Tjing Ling Medical Foundation (grant nos. CI-100-16 and CI-103-14), the Center of Excellence for Cancer Research at Taipei Veterans General Hospital (grant nos. DOH101-TD-C-111-007, DOH102-TD-C-111-007 and MOHW103-TD-B-111-02), the Ministry of Education, Aim for the Top University Plan (grant nos. 101AC-T406, 103AC-T607 and 104AC-T506) and the Ministry of Science and Technology, Taiwan (grant no. MOST105-2319-B-010-001).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
KH designed the study, performed experiments and bioinformatics analysis, analyzed data, and wrote the manuscript. YTC performed experiments and analyzed the data. CFC conducted bioinformatics analysis and assisted interpretation of results. Y-ST performed experiments and analyzed data. TTH performed animal experiments and analyzed data. YCL performed experiments and analyzed data. TSY provided technical and material support and assisted with analysis and interpretation of results. KHY provided clinical samples. HCL provided technical support and assisted with analysis and interpretation of results. MTH assisted with analysis and interpretation of results. CWC provided clinical samples and assisted with analysis and interpretation of results. CWW provided clinical samples. CHL assisted with analysis and interpretation of results. YHP designed the study, assisted with analysis and interpretation of results, and wrote the manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Taipei Veterans General Hospital and written informed consent was obtained from all patients. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of National Yang Ming University.
Consent for publication
The study was approved by the Ethics Committee of Taipei Veterans General Hospital and written informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621–628. doi: 10.11622/smedj.2014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu CW, Lo SS, Shen KH, Hsieh MC, Chen JH, Chiang JH, Lin HJ, Li AF, Lui WY. Incidence and factors associated with recurrence patterns after intended curative surgery for gastric cancer. World J Surg. 2003;27:153–158. doi: 10.1007/s00268-002-6279-7. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 4.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 5.Liang H, Li WH. Lowly expressed human microRNA genes evolve rapidly. Mol Biol Evol. 2009;26:1195–1198. doi: 10.1093/molbev/msp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu K, Liu D, Lai H, Li J, Wang C. Developing miRNA therapeutics for cardiac repair in ischemic heart disease. J Thorac Dis. 2016;8:E918–E927. doi: 10.21037/jtd.2016.08.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang W, Gao S, Liang L, Huang X, Hu N, Lu X, Zhao Y. miRNA-34b is directly involved in the aging of macrophages. Aging Clin Exp Res. 2017;29:599–607. doi: 10.1007/s40520-016-0611-9. [DOI] [PubMed] [Google Scholar]
- 10.Shi C, Huang F, Gu X, Zhang M, Wen J, Wang X, You L, Cui X, Ji C, Guo X. Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget. 2016;7:40830–40845. doi: 10.18632/oncotarget.8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petriv OI, Hansen CL, Humphries RK, Kuchenbauer F. Probing the complexity of miRNA expression across hematopoiesis. Cell Cycle. 2011;10:2–3. doi: 10.4161/cc.10.1.14289. [DOI] [PubMed] [Google Scholar]
- 12.Hunter T. Cooperation between oncogenes. Cell. 1991;64:249–270. doi: 10.1016/0092-8674(91)90637-E. [DOI] [PubMed] [Google Scholar]
- 13.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Q, Yang X, Duan W, Li C, Luo Y, Lu S. miRNA-346 promotes proliferation, migration and invasion in liver cancer. Oncol Lett. 2017;14:3255–3260. doi: 10.3892/ol.2017.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Wu YY, Jiang JN, Liu XS, Ji FJ, Fang XD. MiRNA-3978 regulates peritoneal gastric cancer metastasis by targeting legumain. Oncotarget. 2016;7:83223–83230. doi: 10.18632/oncotarget.12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 17.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143:35–47. doi: 10.1053/j.gastro.2012.05.003. e2. [DOI] [PubMed] [Google Scholar]
- 19.Su Z, Zhao J, Rong Z, Geng W, Wang Z. MiR-451, a potential prognostic biomarker and tumor suppressor for gastric cancer. Int J Clin Exp Pathol. 2015;8:9154–9160. [PMC free article] [PubMed] [Google Scholar]
- 20.Du Y, Xu Y, Ding L, Yao H, Yu H, Zhou T, Si J. Down-regulation of miR-141 in gastric cancer and its involvement in cell growth. J Gastroenterol. 2009;44:556–561. doi: 10.1007/s00535-009-0037-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X, Yang L, Hu J. Down-regulation of miR-27a might inhibit proliferation and drug resistance of gastric cancer cells. J Exp Clin Cancer Res. 2011;30:55. doi: 10.1186/1756-9966-30-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao W, Fan R, Wang L, Cheng S, Li H, Jiang J, Geng M, Jin Y, Wu Y. Expression and regulatory function of miRNA-34a in targeting survivin in gastric cancer cells. Tumour Biol. 2013;34:963–971. doi: 10.1007/s13277-012-0632-8. [DOI] [PubMed] [Google Scholar]
- 23.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 24.Yang L, Liang H, Wang Y, Gao S, Yin K, Liu Z, Zheng X, Lv Y, Wang L, Zhang CY, et al. MiRNA-203 suppresses tumor cell proliferation, migration and invasion by targeting Slug in gastric cancer. Protein Cell. 2016;7:383–387. doi: 10.1007/s13238-016-0259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R, Li F, Wang W, Wang X, Li S, Liu J. The effect of antisense inhibitor of miRNA 106b~25 on the proliferation, invasion, migration, and apoptosis of gastric cancer cell. Tumour Biol. 2016;37:10507–10515. doi: 10.1007/s13277-016-4937-x. [DOI] [PubMed] [Google Scholar]
- 26.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 27.Kabir NN, Sun J, Ronnstrand L, Kazi JU. SOCS6 is a selective suppressor of receptor tyrosine kinase signaling. Tumour Biol. 2014;35:10581–10589. doi: 10.1007/s13277-014-2542-4. [DOI] [PubMed] [Google Scholar]
- 28.Szász AM, Lánczky A, Nagy A, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, Lao K, Kosik KS. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetic. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- 30.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 31.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 33.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Mol Cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res. 2006;12:2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- 36.Ma S, Liu M, Xu Z, Li Y, Guo H, Ge Y, Liu Y, Zheng D, Shi J. A double feedback loop mediated by microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage polarization and thus regulates cancer progression. Oncotarget. 2016;7:13502–13519. doi: 10.18632/oncotarget.6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Ru J, Zhang J, Zhu LH, Liu M, Li X, Tang H. miR-23a targets interferon regulatory factor 1 and modulates cellular proliferation and paclitaxel-induced apoptosis in gastric adenocarcinoma cells. PLoS One. 2013;8:e64707. doi: 10.1371/journal.pone.0064707. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu M, Li X. MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 2010;277:3726–3734. doi: 10.1111/j.1742-4658.2010.07773.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhou L, Liang X, Zhang L, Yang L, Nagao N, Wu H, Liu C, Lin S, Cai G, Liu J. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. Oncotarget. 2016;7:51943–51954. doi: 10.18632/oncotarget.10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T, Tang H, Lang Y, Liu M, Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 2009;273:233–242. doi: 10.1016/j.canlet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Duan J, Qu Y, Deng T, Liu R, Zhang L, Bai M, Li J, Ning T, Ge S, et al. Onco-miR-24 regulates cell growth and apoptosis by targeting BCL2L11 in gastric cancer. Protein Cell. 2016;7:141–151. doi: 10.1007/s13238-015-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan X, Liu C, Yang P, He S, Liao Q, Kang S, Zhao Y. Clustered microRNAs' coordination in regulating protein-protein interaction network. BMC Syst Biol. 2009;3:65. doi: 10.1186/1752-0509-3-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masuhara M, Sakamoto H, Matsumoto A, Suzuki R, Yasukawa H, Mitsui K, Wakioka T, Tanimura S, Sasaki A, Misawa H, et al. Cloning and characterization of novel CIS family genes. Biochem Biophys Res Commun. 1997;239:439–446. doi: 10.1006/bbrc.1997.7484. [DOI] [PubMed] [Google Scholar]
- 44.Lai RH, Hsiao YW, Wang MJ, Lin HY, Wu CW, Chi CW, Li AF, Jou YS, Chen JY. SOCS6, down-regulated in gastric cancer, inhibits cell proliferation and colony formation. Cancer Lett. 2010;288:75–85. doi: 10.1016/j.canlet.2009.06.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.