Abstract
Gastric cancer (GC) is the fifth most common type of malignant tumor worldwide and the most common cause of cancer-associated mortality in China. Recent studies revealed that microRNAs (miRNAs) function in the pathogenesis of GC, and that miR-155-5p expression is downregulated in GC tissues. However, the function of miR-155-5p has not been fully identified. In the present study, it was demonstrated that overexpression of miR-155-5p inhibited GC-cell proliferation and promoted apoptosis, while downregulation of miR-155-5p promoted GC-cell proliferation and decreased the cisplatin sensitivity of GC cells. Mitogen-activated protein kinase kinase kinase 10 was demonstrated to be a potential target gene of miR-155-5p. In conclusion, an antitumor role of miR-155-5p in gastric cancer was suggested.
Keywords: microRNA-155-5p, proliferation, apoptosis, gastric cancer
Introduction
Gastric cancer (GC) is the fifth most common type of malignant tumor worldwide and the third most common cause of cancer-associated mortality (1,2). Approximately 50% of cases occur in Eastern Asia (mainly in China) (3). Therefore, GC is a major public health problem, particularly in China. In the majority of cases, GC patients are diagnosed at advanced stages, at which effective therapeutic strategies are limited and the prognosis is relatively poor (4–6). Invasion and metastasis are hallmarks of advanced GC progression; therefore, investigation of the molecular pathogenesis of GC is critical to improve the survival of GC patients.
miRNAs are evolutionary conserved small non-coding RNAs and are involved in the regulation of gene expression and protein translation (7). miRNAs function in the pathogenesis of GC (8–14).
The role of miR-155-5p in various types of cancer has been revealed in recent studies. In colorectal carcinoma (CRC), miR-155-5p expression is upregulated and has been indicated to promote the proliferation, invasion and metastasis of CRC cells (15). In osteosarcoma, increased miR-155-5p and reduced miR-148a-3p expression were demonstrated to contribute to the suppression of tumor cell death (16). In GC, miR-155-5p was demonstrated to be downregulated in GC tissues (17), and miR-155-5p inhibition promoted the transformation of bone marrow mesenchymal stem cells into GC tissue-derived MSC-like cells via nuclear factor-κB p65 activation (18). However, the role of miR-155-5p in GC has not been fully elucidated.
In the present study, the effect of miR-155-5p on GC-cell proliferation and apoptosis was investigated. The data achieved may provide a novel therapeutic target for further investigation.
Materials and methods
Patients and tissues
A total of 14 GC tissues and corresponding adjacent normal tissues were collected from 14 patients with GC who underwent surgery at the Department of Gastroenterology, Center Hospital of Nanchong City. College (Nanchong, China) from January 2013 to December 2013. The histopathological diagnoses of all patients were confirmed by a senior pathologist at West China Hospital (Chengdu, China). The information of the 14 GC patients (average age: 60.2 years, age range: 36–63 years) are listed in Table I. The sex ratio was 50:50. The present study was approved by the ethics committee of Northern Sichuan Medical College (Nanchong, Sichuan), and all patients provided written informed consent.
Table I.
The characteristics of the patients with gastric cancer included in the present study.
| Patient | Age (years) | Sex | Stage | Differentiation |
|---|---|---|---|---|
| 1 | 45 | Female | III | Low |
| 2 | 63 | Female | II | Moderate |
| 3 | 56 | Female | II | Low |
| 4 | 45 | Female | III | Moderate |
| 5 | 59 | Male | III | Moderate |
| 6 | 50 | Male | IV | Moderate |
| 7 | 56 | Male | III | Moderate |
| 8 | 77 | Female | IV | Low |
| 9 | 66 | Male | II | Moderate |
| 10 | 73 | Female | II | Moderate |
| 11 | 48 | Male | III | Low |
| 12 | 67 | Male | III | Low |
| 13 | 53 | Female | IV | Moderate |
| 14 | 50 | Male | III | Moderate |
Cell lines and reagents
The GC cell lines, AGS and SGC-7901, were purchased from the Chinese Academy of Sciences (Shanghai, China). The human gastric epithelial mucosa cell line, GES-1, was gifted by the Department of Gastrology, West China Hospital, Sichuan University (Chengdu, China). Cells were cultured in RPMI-1640 medium, supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA) (19). Cisplatin was purchased from Hanson Pharma (Lianyungang, China; http://www.hansoh.cn/). Cisplatin was added to the cultures at a final concentration of 25 µM, as previously described (20).
miR-155-5p mimics and oligonucleotide transfection
The miR-155-5p mimics, miR-155-5 oligonucleotides, miR-155-5p antisense oligonucleotides (ASO) and negative controls were purchased from Dharmacon, Inc. (Chicago, IL, USA). Cells were seeded at 2×105 per well in 6-well plates, and transfected with mimics (50 nM), oligonucleotides (50 nM) or controls (50 nM) using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.), according to the standard protocol. The sequence of these molecules were listed as following: miR-155-5p mimics, 5′-UUAAUGCUAAUCGUCAUAGGGGU-3′; miR-155-5p-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′; miR-155-5p ASO, 5′-ACCCCUAUCACGAUUAGCAUUAA-3′; miR-155-5p ASO-NC, 5′-CAGUACUUUUGUGUAGUACAA-3′. A total of 24 h following transfection, the miR-155-5p levels were examined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Western blot analysis
The lysates were prepared with lysis buffer (Abcam, Cambridge, UK) containing protease inhibitors and then centrifuged (12,000 × g for 5 min in 4°C). The protein levels were determined by BCA protein quantification kit (cat. no. ab102536; Abcam). A total of 20 µg protein was separated by SDS-PAGE (10%) and transferred onto PVDF membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked in 5% skimmed milk for 15 min at room temperature. Following washing with PBS for 15 min, the membranes were incubated with MA3PK10 (cat. no. orb127031; 1:1,000; BioPike, LCC, Shanghai, China), or b-actin (1:4,000; cat. no. ab8227; Abcam) primary antibodies overnight at 4°C. The membranes were then washed by PBS three times. Next the membranes were incubated with a rabbit anti-mouse secondary antibody conjugated with horseradish peroxidase (1:5,000; cat. no. ab6728; Abcam) for 2 h at room temperature. The proteins were visualized using an enhanced chemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.).
Cell proliferation assay
Cell proliferation was assessed by MTT assay. Cells were seeded into 96-well plates at a density of 5×105/well. MTT was added to the medium at a final concentration of 0.1 mg/ml. A total of 100 µl dimethyl sulfoxide was used to dissolve the formazan crystals in each well. The OD was measured using a microplate reader at a wavelength of 570 nm.
RT-qPCR
GC tissues and cell lines were incubated with TRIzol® reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Total RNA was isolated from the tissues and cell lines using a mirVana miRNA Isolation kit (Thermo Fisher Scientific, Inc.). Reverse transcription was performed using an All-in-one™ miRNA First-Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.). The primers were synthesized by the Sangon Biotech Company (Shanghai, China, http://www.sangon.com/). PCR assay was performed as previously described, with miR-155-5p expression normalized to U6 snRNA expression (14,21–24). The primers were: miR-155-5p, forward, 5′-UAAUACCGUCUUAAAACCGU-3′, and reverse, 5′-UUCUGGGAACGUGAAACCT-3′; and U6 snRNA, forward, 5′-CGCTTCGGCAGCACATATACTAAAATTGGAAC-3′, and reverse, 5′-GCTTCACGAATTTGCGTGTCATCCTTGC-3′. All the reaction mixtures were incubated in a 96-well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 40 sec.
Cell apoptosis analysis
Cell apoptosis were analyzed by Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Staining Detection kit (cat. no. ab14085; Abcam) according to the manufacturer's protocol. In detail, 1×105 cells were suspended in 1X Annexin V Binding Buffer, then 5 µl Annexin V-FITC and 5 µl Propidium Iodide was added for incubation at room temperature for 5 min in the dark. Samples were analyzed using a flow assisted cell sorting analyzer instrument (BD LSR II, FACSDiva software v.1.1.0; BD Biosciences) using the 488 nm excitation line and emission was detected at 530 nm (green, FITC) and 575–610 nm (orange, PI) (22).
Dual luciferase reporter assays
Cells were seeded at 2×105 cells/well and were serum-starved for 6 h pre-transfection. A mutated version of the mitogen-activated protein kinase kinase kinase 10 (MAP3K10) 3′untranslated region (3′UTR) was generated using a Site-Directed Mutagenesis kit (Promega Corporation), as previously described (25). The intact 3′UTR of MAP3K10 and the mutated version were cloned into a luciferase reporter plasmid (500 ng; cat. no. K801-200; NeoBioscience, Shanghai, China; http://www.nbs-bio.com/). The plasmid, and a pGL3-control (100 ng; Thermo Fisher Scientific, Inc.) were co-transfected using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. A total of 24 h later, cells were harvested and the luciferase activities were analyzed using a dual-luciferase reporter assay system (Promega Corporation, Madison, WI, USA). The luciferase activity was normalized to Renilla luciferase activity.
Prediction of the possible targets of miR-155-5p
Targetscan software (http://www.targetscan.org/) (26–31) was used to predict possible targets of miR-155-5p.
Statistical analysis
Statistical analysis was performed using SPSS 10.0 software (SPSS, Inc., Chicago, IL, USA). The difference between two groups was analyzed using a two-tailed Student's t-test. Analysis of variance was used to analyze differences among ≥3 groups, with post hoc contrasts performed using Student-Newman-Keuls test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-155-5p expression is low in GC tissues
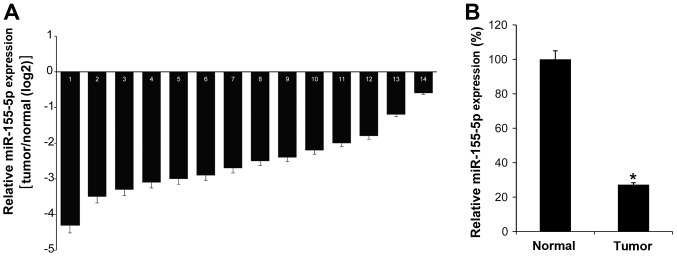
To investigate the function of miR-155-5p in the pathogenesis of GC, the miR-155-5p expression levels in 14 GC tissues were investigated by RT-qPCR. It was demonstrated that all GC tissues exhibited low levels of miR-155-5p compared with their matched adjacent normal tissues (Fig. 1A). As expected, the mean expression of miR-155-5p in GC tumor tissues was low compared with that in normal control tissues (Fig. 1B). Thus, we hypothesized that miR-155-5p may serve an antitumor role in GC.
Figure 1.
The expression level of miR-155-5p is low in GC tissues. (A) miR-155-5p expression was analyzed in 14 GC tissues and their matched adjacent normal tissues by reverse transcription-quantitative polymerase chain reaction. (B) The mean expression value of miR-155-5p in GC tissues was calculated, using U6 snRNA as an internal control. The data are represented as the mean ± standard deviation, and the experiment was repeated 3 times. *P<0.05 vs. normal control.
Overexpression of miR-155-5p inhibits GC cell proliferation and promotes apoptosis
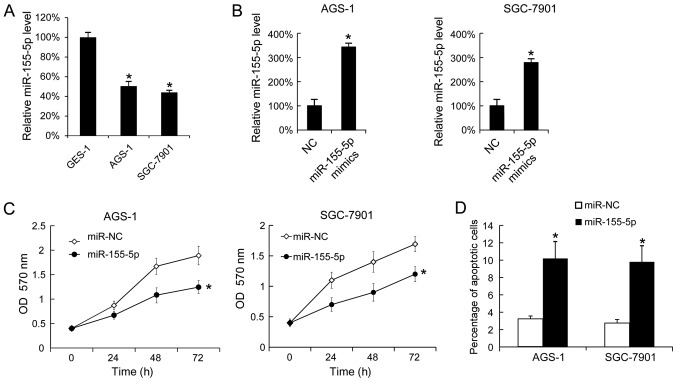
Subsequently, miR-155-5p expression levels were analyzed in the GC cell lines, AGS-1 and SGC-7901, using the gastric epithelial mucosa cell line, GES-1, as a negative control. Using RT-qPCR, it was demonstrated that AGS-1 and SGC-7901 cells expressed increased levels of miR-155-5p compared with GES-1 cells (Fig. 2A). Overexpression of miR-155-5p in AGS-1 and SGC-7901 was confirmed by RT-qPCR (Fig. 2B). Following transfection with miR-155-5p mimics, proliferation was analyzed by MTT assay. This revealed that miR-155-5p overexpression inhibited AGS-1 and SGC-7910 cell proliferation (Fig. 2C). A total of 48 h post-transfection, apoptosis was analyzed by Annexin V-FITC and PI staining. This indicated that miR-155-5p overexpression increased the apoptotic rates in AGS-1 and SGC-7910 cells (Fig. 2D).
Figure 2.
miR-155-5p mimics-transfection inhibits AGS-1 and SGC-7901 cell proliferation, and promotes apoptosis. The miR-155-5p levels in GES-1, AGS-1 and SGC-7901 cells were tested by reverse transcription-quantitative polymerase chain reaction. (A) The expression level of miR-155-5p in GES-1 cells was arbitrarily defined as 100% for comparison among cell lines. (B) The expression level of miR-155-5p in miR-NC-transfected cells was arbitrarily defined as 100% for comparison with transfected cells. (C) A total of 24, 48 and 72 h after transfection, proliferation was analyzed by MTT assay. (D) A total of 48 h following miR-155-5p mimics-transfection, apoptosis was measured by Annexin V-propidium iodide staining. The data are presented as the mean ± standard deviation, and the experiments were repeated 3 times. *P<0.05 vs. NC. NC, negative control; ASO, antisense oligonucleotide.
Downregulation of miR-155-5p promotes GC cell growth and decreases the sensitivity of GC cells to cisplatin
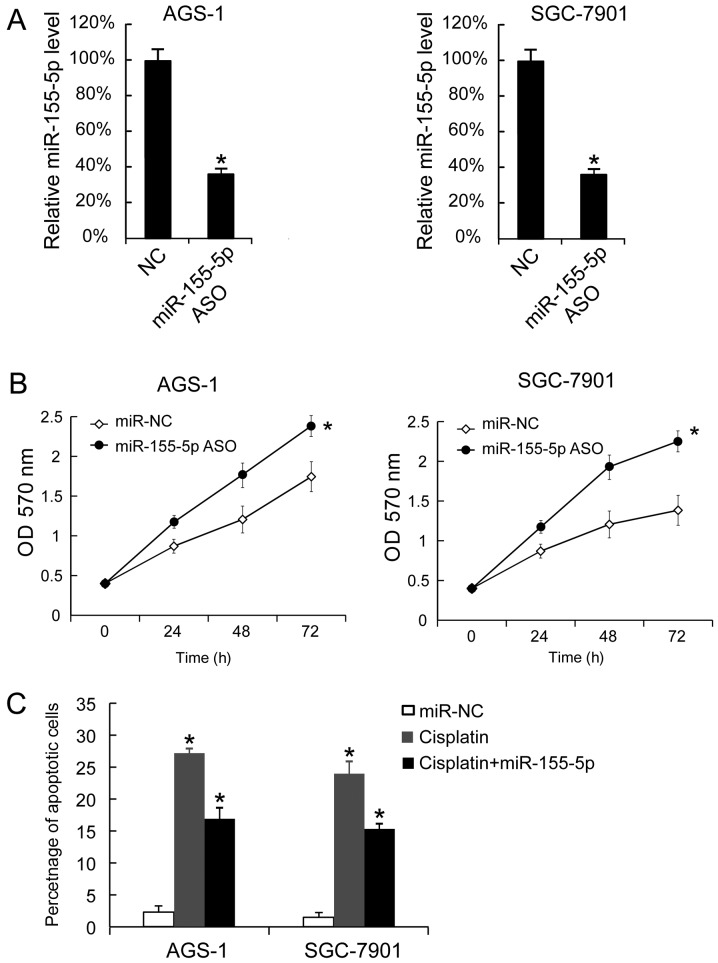
It was demonstrated that miR-155-5p ASO transfection downregulated miR-155-5p expression levels in AGS-1 and SGC-7901 cells (Fig. 3A). miR-155-5p ASO transfection also promoted the proliferation of AGS-1 and SGC-7901 cells (Fig. 3B). miR-155-5p ASO transfection also decreased the apoptotic rates of AGS-1 and SGC-7901 cells following treatment with cisplatin. Untransfected cells treated with cisplatin exhibited an increased apoptotic rate compared with untreated untransfected cells. Thus, miR-155-5p reduced the effect of cisplatin-induced apoptosis (Fig. 3C).
Figure 3.
Transfection of miR-155-5p ASO promoted AGS-1 and SGC-7901 cellular proliferation and decreased the cisplatin-induced apoptosis. (A) miR-155-5p expression in AGS-1 and SGC-7901 cells was analyzed 48 h after transfection with miR-155-5p ASO. (B) The miR-155-5p level in miR-NC ASO-transfected cells was arbitrarily defined as 100%. A total of 24, 48 and 72 h after transfection, proliferation was analyzed by MTT assay. (C) Cisplatin (final concentration: 25 µM) was added 24 h after miR-155-5p ASO transffection, and apoptosis was measured by Annexin V-propidium iodide staining. The data are presented as the mean ± standard deviation, and the experiments were repeated 3 times. *P<0.05 vs. NC. miR, microRNA; NC, negative control; ASO, antisense oligonucleotide.
MAP3K10 is targeted by miR-155-5p
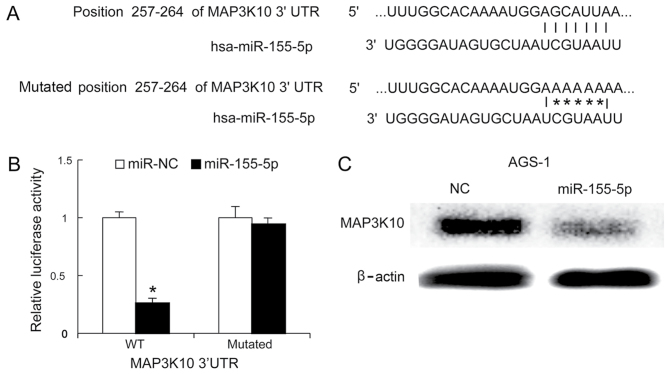
A previous study demonstrated that MAP3K10 was overexpressed in pancreatic ductal adenocarcinoma (PDAC), and that it promoted proliferation and decreased therapeutic impact in pancreatic cancer cells (32). Bioinformatics analysis suggested that MAP3K10 is a targeted gene of miR-155-5p (Fig. 4A). The luciferase activity of the reporter carrying the mutated 3′UTR of MAP3K10 was not significantly different in miR-NC and miR-155-5p-transfected cells, whereas miR-155-5p was demonstrated to reduce the luciferase activity of the wild type 3′UTR reporter (Fig. 4B). To investigate whether miR-155-5p reduced MAP3K10 protein levels, western blotting was performed 48 h following miR-155-5p mimics transfection. It was demonstrated that miR-155-5p mimics significantly decreased the protein expression level of MAP3K10 proteins in AGS-1 cells, compared with negative control (Fig. 4C).
Figure 4.
MAP3K10 is targeted by miR-155-5p in AGS-1 cells. (A) The intact and mutated versions of the MAP3K10 3′UTR and miR-155-5p are presented. (B) miR-155-5p mimics inhibited the activity of luciferase in the plasmid containing the 3′UTR of the WT MAP3K10 gene. (C) AGS-1 cells were transfected with miR-155-5p mimics, and 48 h later, the MAP3K10 protein expression level was analyzed by western blotting. Data are presented as the mean±standard deviation, and the experiments were repeated 3 times. *P<0.05 miR-NC vs.miR-155-5p MAP3K10, mitogen-activated protein kinase kinase kinase 10; miR, microRNA; 3′UTR, 3′untranslated region; hsa, homo sapiens; NC, negative control; WT, wild type.
Discussion
In a previous study, patients with GC exhibiting low expression of miR-124-3pm miR-146a-5p, miR-335-5p and miR-155-5p were associated with increased lymph node metastasis, lymphatic invasion, venous invasion, high Bormann stage, lymphatic invasion and poor differentiation compared with those exhibiting high expression (17). The present may contribute to the elucidation of the underlying molecular mechanism of the clinical significance of miR-155-5p. Low expression of miR-155-5p was demonstrated to promote cellular proliferation and decrease the cisplatin-sensitivity of GC cells. MAP3K10 was also indicated to be a target gene of miR-155-5p. MAP3K10 is a member of the mitogen-activated protein kinase kinase kinase (MAP3K) family, it activates the C-Jun N-terminal kinase (JNK) signaling pathway and the p38 mitogen-activated-protein kinase (MAPK) pathway, and regulates apoptosis in numerous neurodegenerative diseases (32–34). It was demonstrated in the present study that MAP3K10 may be targeted by miR-155-5p in GC cell lines.
MAP3K10 has been demonstrated to promote the proliferation of pancreatic cancer cells and decrease the sensitivity to gemcitabine by upregulating the expression of GLI family zinc finger (Gli)-1 and Gli-2 (32). Whether Gli-1 and Gli-2 promote or inhibit GC tumor growth requires further investigation. In conclusion, the present study suggests that miR-155-5p serves an antitumor role in GC.
Acknowledgements
The authors would like to thank Ms. Miao Chen (Clinical Laboratory Department, The Affiliated Hospital of North Sichuan Medical College, Nanchong, China) for technical her assistance.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
SL, TZ and XZ collected patient data and performed cell experiments. ZD and FC performed RT-qPCR, western blot analysis and other molecular experiment. JL and QL contributed to study design and manuscript writing.
Ethics approval and consent to participate
The present study was approved by the ethics committee of Northern Sichuan Medical College (Nanchong, China), and all patients provided written informed consent.
Consent for publication
All patients gave informed consent for the use of their tissues and publication of the data and images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cance incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 4.Mocellin S, Verdi D, Pooley KA, Nitti D. Genetic variation and gastric cancer risk: A field synopsis and meta-analysis. Gut. 2015;64:1209–1219. doi: 10.1136/gutjnl-2015-309168. [DOI] [PubMed] [Google Scholar]
- 5.Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–655. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riquelme I, Saavedra K, Espinoza JA, Weber H, García P, Nervi B, Garrido M, Corvalán AH, Roa JC, Bizama C. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget. 2015;6:24750–24779. doi: 10.18632/oncotarget.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giordano S, Columbano A. MicroRNAs: New tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 8.Chen DL, Zhang DS, Lu YX, Chen LZ, Zeng ZL, He MM, Wang FH, Li YH, Zhang HZ, Pelicano H, et al. microRNA-217 inhibits tumor progression and metastasis by downregulating EZH2 and predicts favorable prognosis in gastric cancer. Oncotarget. 2015;6:10868–10879. doi: 10.18632/oncotarget.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, Liu L, Jia D, Tian Q, Wu J, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–7583. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Dong G, Wang B, Gao W, Yang Q. miR-543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem Biophys Res Commun. 2016;469:15–21. doi: 10.1016/j.bbrc.2016.03.112. [DOI] [PubMed] [Google Scholar]
- 11.Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, Wu Y, Qi L, Fan Y, Chen Y, et al. Diagnostic value of a plasma microRNA signature in gastric cancer: A microRNA expression analysis. Sci Rep. 2015;5:11251. doi: 10.1038/srep11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibarrola-Villava M, Llorca-Cardeñosa MJ, Tarazona N, Mongort C, Fleitas T, Perez-Fidalgo JA, Roselló S, Navarro S, Ribas G, Cervantes A. Deregulation of ARID1A, CDH1, cMET and PIK3CA and target-related microRNA expression in gastric cancer. Oncotarget. 2015;6:26935–26945. doi: 10.18632/oncotarget.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge X, Liu X, Lin F, Li P, Liu K, Geng R, Dai C, Lin Y, Tang W, Wu Z, et al. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget. 2016;7:24466–24482. doi: 10.18632/oncotarget.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Y, Wang L, Wu H, Zhang Y, Wang K, Wu D. MicroRNA-141 inhibits migration of gastric cancer by targeting zinc finger E-box-binding homeobox 2. Mol Med Rep. 2015;12:3416–3422. doi: 10.3892/mmr.2015.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu YL, Wang HF, Sun ZQ, Tang Y, Han XN, Yu XB, Liu K. Up-regulated miR-155-5p promotes cell proliferation, invasion and metastasis in colorectal carcinoma. Int J Clin Exp Pathol. 2015;8:6988–6994. [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya S, Chalk AM, Ng AJ, Martin TJ, Zannettino AC, Purton LE, Lu J, Baker EK, Walkley CR. Increased miR-155-5p and reduced miR-148a-3p contribute to the suppression of osteosarcoma cell death. Oncogene. 2016;35:5282–5294. doi: 10.1038/onc.2016.68. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Xie S, Liu M, Chen Z, Liu X, Wang L, Li D, Zhou Y. The clinical significance of downregulation of mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric cancer tumorigenesis. Int J Oncol. 2014;45:197–208. doi: 10.3892/ijo.2014.2415. [DOI] [PubMed] [Google Scholar]
- 18.Zhu M, Wang M, Yang F, Tian Y, Cai J, Yang H, Fu H, Mao F, Zhu W, Qian H, Xu W. miR-155-5p inhibition promotes the transition of bone marrow mesenchymal stem cells to gastric cancer tissue derived MSC-like cells via NF-κB p65 activation. Oncotarget. 2016;7:16567–16580. doi: 10.18632/oncotarget.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y, Zhang C, Gu C, Li Q, Wu N. Function of deubiquitinating enzyme USP14 as oncogene in different types of cancer. Cell Physiol Biochem. 2016;38:993–1002. doi: 10.1159/000443051. [DOI] [PubMed] [Google Scholar]
- 20.Price PM, Yu F, Kaldis P, Aleem E, Nowak G, Safirstein RL, Megyesi J. Dependence of cisplatin-induced cell death in vitro and in vivo on cyclin-dependent kinase 2. J Am Soc Nephrol. 2006;17:2434–2442. doi: 10.1681/ASN.2006020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Liu X, Lin L, Hou J, Li N, Wang C, Wang P, Zhang Q, Zhang P, Zhou W, et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. 2011;286:36677–36685. doi: 10.1074/jbc.M111.270561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song B, Zhang C, Li G, Jin G, Liu C. MiR-940 inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell Physiol Biochem. 2015;35:1167–1177. doi: 10.1159/000373995. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q, Yu Y. Upregulated CDK16 expression in serous epithelial ovarian cancer cells. Med Sci Monit. 2015;21:3409–3414. doi: 10.12659/MSM.894990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Xu Y, Qiu W, Zhao D, Zhang Y. Tissue miR-193b as a novel biomarker for patients with ovarian cancer. Med Sci Monit. 2015;21:3929–3934. doi: 10.12659/MSM.895407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan B, Liang Y, Wang D, Luo F. MiR-940 inhibits hepatocellular carcinoma growth and correlates with prognosis of hepatocellular carcinoma patients. Cancer Sci. 2015;106:819–824. doi: 10.1111/cas.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John B, Sander C, Marks DS. Prediction of human microRNA targets. Methods Mol Biol. 2006;342:101–113. doi: 10.1385/1-59745-123-1:101. [DOI] [PubMed] [Google Scholar]
- 31.Lee S, Paulson KG, Murchison EP, Afanasiev OK, Alkan C, Leonard JH, Byrd DR, Hannon GJ, Nghiem P. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J Clin Virol. 2011;52:272–275. doi: 10.1016/j.jcv.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An Y, Cai B, Chen J, Lv N, Yao J, Xue X, Tu M, Tang D, Wei J, Jiang K, et al. MAP3K10 promotes the proliferation and decreases the sensitivity of pancreatic cancer cells to gemcitabine by upregulating Gli-1 and Gli-2. Cancer Lett. 2013;329:228–235. doi: 10.1016/j.canlet.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol Cell Biol. 2002;3:663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- 34.Xu Z, Maroney AC, Dobrzanski P, Kukekov NV, Greene LA. The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Mol Cell Biol. 2001;21:4713–4724. doi: 10.1128/MCB.21.14.4713-4724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.