Abstract
Postoperative radiotherapy or concurrent chemoradiotherapy are routine clinical options for the treatment of head and neck squamous cell carcinoma (HNSCC). However, the benefit of adding chemotherapy to radiotherapy is contested. The present study aimed to develop a gene signature to predict the clinical benefit of postoperative chemoradiotherapy using public data from The Cancer Genome Atlas. A 22-gene signature was established, which demonstrated the best predictive value. Patients were separated into low-score and high-score subgroups based on the expression score of the 22-gene signature. In the high-score subgroup, patients who received chemoradiotherapy demonstrated improved overall survival, relapse-free survival and local regional control compared with those who received radiotherapy alone. However, in the low-score subgroup adding chemotherapy to radiotherapy was associated with worse patient outcomes. The predictive value of the 22-gene signature was independent of the conventional clinical variables. Gene set enrichment analysis revealed that the expression signatures of hypoxia phenotype and stem-like traits were significantly enriched in the low-score subgroup. In addition, the low-score subgroup was associated with the gene sets involved in resistance to anticancer drugs. In conclusion, hypoxia- or stem-like gene expression properties are associated with chemotherapy-resistance in HNSCC. The 22-gene signature may be useful as a predictive marker to help distinguish patients who will benefit from postoperative concurrent chemoradiotherapy.
Keywords: head and neck squamous cell carcinoma, 22-gene signature, outcome, adjuvant chemoradiotherapy, stem-like phenotype
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the tenth most common malignancy and the eleventh cause of cancer-related deaths in the United States (1). Patients with early stage HNSCC could usually get good curative results after surgery or radiotherapy. Unfortunately, many patients were diagnosed with advanced HNSCC and postoperative adjuvant therapy seems necessary for improving outcome (2). In patients with clinically high-risk HNSCC, postoperative concurrent chemoradiotherapy improved loco-regional control (LRC) compared with mono radiotherapy (3,4). However, the long-term relapse free survival (RFS) and overall survival (OS) showed no difference between patients who received chemoradiotherapy and those with radiotherapy alone (4). Meanwhile, adding chemotherapy to radiotherapy increases the risk of both acute and late toxicity, resulting in a decreased quality of life (5,6). For this reason, the proper decision of concurrent chemoradiotherapy or mono radiotherapy should be discussed for selecting patients who will get more clinical benefit and avoid unnecessary side effect. Thus, finding novel molecular signatures with superior predictive values will be helpful for better treatment decision in patients with HNSCC.
To date, gene expression profiles by microarrays or next-generation sequencing platforms have been widely used in developing biomarkers in various types of cancer including HNSCC. Multiple studies have reported molecular subtypes of HNSCC by gene expression profiles (7–9). These gene expression subtypes were correlated with biological features of tumors as well as the clinical outcome of patients. Linge et al reported that high expression levels of hypoxia-induced gene signature and cancer stem cell markers were correlated with poor prognosis after postoperative chemoradiotherapy in HNSCC patients (10). However, most of these gene signatures were reported as prognostic factors for HNSCC. Little biomarkers have been developed for predicting the clinical benefit of postoperative concurrent chemoradiotherapy compared with mono radiotherapy.
This study was designed to develop a gene signature for distinguishing HNSCC patients who would benefit from concurrent postoperative chemoradiotherapy compared with radiotherapy alone. RNA-sequencing data and corresponding therapeutic information of HNSCC patients were collected from The Cancer Genome Atlas (TCGA). Using a bioinformatic approach, we identified a 22-gene signature. This signature was helpful to stratify patients with different survival benefits of postoperative chemoradiotherapy compared with radiotherapy alone. However, there were no prognostic differences between concurrent chemoradiotherapy and mono radiotherapy groups when considering all patients. In addition, the predictive value of this signature was independent of traditional clinical parameters.
Materials and methods
Patients and clinical characteristics
The gene-expression profiles and clinical information from 528 patients with HNSCC were collected from TCGA. A subgroup of 281 patients from this cohort was involved in this analysis, with the following criteria: i) Without any neo-adjuvant therapy before surgery; ii) receiving postoperative mono radiotherapy or concurrent chemoradiotherapy before any recurrence or metastasis occurs; iii) with follow-up information after treatment; and iv) with gene-expression profiles of the primary tumors. A clinically high-risk factor was defined for 209 patients, by three parameters: With positive lymph nodes ≥2, extracapsular extension of nodal disease or positive surgical margins, as described previously (3).
OS was defined as the survival time from the surgery date to the time point of patient death, or the last follow-up time of living (censored data). RFS was defined as the survival time from the surgery date to the point of loco-regional or distant recurrence of cancer, or the last follow-up time without recurrence (censored data). LRC was defined as the survival time from the surgery date to the point of local or regional recurrence of cancer, or to the last follow-up time without loco-regional recurrence (censored data).
Postoperative radiation and chemoradiotherapy
Patients have received mono radiotherapy or concurrent chemoradiotherapy after primary surgical treatment. Both radiation and chemoradiotherapy were delivered without any evidence of loco-regional recurrence or distant metastasis. For irradiation, 66.9% (188/281) of patients received ≥60 Gy of total dosage. Conventional fractionation delivered 1.8–2.2 Gy/fraction, 1 fraction/day, 5 days/week. The other fraction types were characterized as none-conventional group. In the chemoradiotherapy group, patients received concomitant chemotherapy with irradiations. Chemotherapy agents were classified into three types: i) Mono therapy of platinum (cisplatin or carboplatin); ii) platinum plus paclitaxel; and iii) chemo-drug plus target drug. The clinical parameters between mono radiotherapy and concurrent chemoradiotherapy were showed in Table I.
Table I.
Association between treatment groups and other clinical parameters.
| Treatment | ||||
|---|---|---|---|---|
| Variable | Parameter | Radiation | Chemoradiation | P-value |
| Sex | 0.2160 | |||
| Female | 34 (24.8) | 26 (18.1) | ||
| Male | 103 (75.2) | 118 (81.9) | ||
| Age | 0.0004 | |||
| <60 years | 54 (39.4) | 88 (61.1) | ||
| ≥60 years | 83 (60.6) | 56 (38.9) | ||
| Smoking index | 0.8450 | |||
| <40 pys | 34 (24.8) | 39 (27.1) | ||
| ≥40 | 43 (31.4) | 55 (38.2) | ||
| N/A | 60 (43.8) | 50 (34.7) | ||
| Anatomic site | 9.65×10−5a | |||
| Oral cavity | 93 (67.9) | 78 (54.2) | ||
| Oropharynx | 2 (1.5) | 4 (2.8) | ||
| Hypopharynx | 36 (26.3) | 31 (21.5) | ||
| Larynx | 6 (4.4) | 31 (21.5) | ||
| Nodal positive | 5.82×10−7 | |||
| <2 | 81 (59.1) | 38 (26.4) | ||
| ≥2 | 35 (25.5) | 69 (47.9) | ||
| N/A | 21 (15.3) | 37 (25.7) | ||
| Nodal extracapsular | 0.0003 | |||
| NO | 74 (54) | 43 (29.9) | ||
| YES | 27 (19.7) | 49 (34) | ||
| N/A | 36 (26.3) | 52 (36.1) | ||
| Margin status | 0.0340 | |||
| Negative | 117 (85.4) | 90 (62.5) | ||
| Positive | 12 (8.8) | 2 (15.3) | ||
| N/A | 8 (5.8) | 32 (22.2) | ||
| Clinical high risk | 4.51×10−9 | |||
| No | 59 (43.1) | 15 (10.4) | ||
| Yes | 49 (35.8) | 86 (59.7) | ||
| N/A | 29 (21.2) | 43 (29.9) | ||
| Tumor grade | 0.0080a | |||
| G1 | 23 (16.8) | 8 (5.6) | ||
| G2 | 74 (54) | 89 (61.8) | ||
| G3 | 39 (28.5) | 33 (22.9) | ||
| G4 | 0 (0) | 2 (1.4) | ||
| N/A | 1 (0.7) | 12 (8.3) | ||
| TNM stage | 0.0010a | |||
| I | 8 (5.8) | 1 (0.7) | ||
| II | 12 (8.8) | 4 (2.8) | ||
| III | 26 (19) | 11 (7.6) | ||
| IV | 79 (57.7) | 93 (64.6) | ||
| N/A | 12 (8.8) | 35 (24.3) | ||
| pT stage | 0.2180 | |||
| T1-2 | 45 (32.8) | 31 (21.5) | ||
| T3-4 | 80 (58.4) | 81 (56.2) | ||
| N/A | 12 (8.8) | 32 (22.2) | ||
| pN stage | 3.47×10−8 | |||
| N0-1 | 80 (58.4) | 33 (22.9) | ||
| N2-3 | 38 (27.7) | 76 (52.8) | ||
| N/A | 19 (13.9) | 35 (24.3) | ||
Fisher's exact test. pys, package years; N/A, not available.
Gene-expression data processing
Gene-expression data of 281 primary HNSCCs was profiled by RNA sequencing method. Level 3 data of fragments per kilobase of gene per million fragments mapped with upper quartile normalization (FPKM-uq) were downloaded from TCGA Data Portal. FPKM-uq value was log2-transformed for subsequent analysis. For each gene, the expression value was scaled to z-value with mean of 0 and standard deviation of 1 across all samples.
Selection of gene-expression signature for survival benefit from chemoradiotherapy
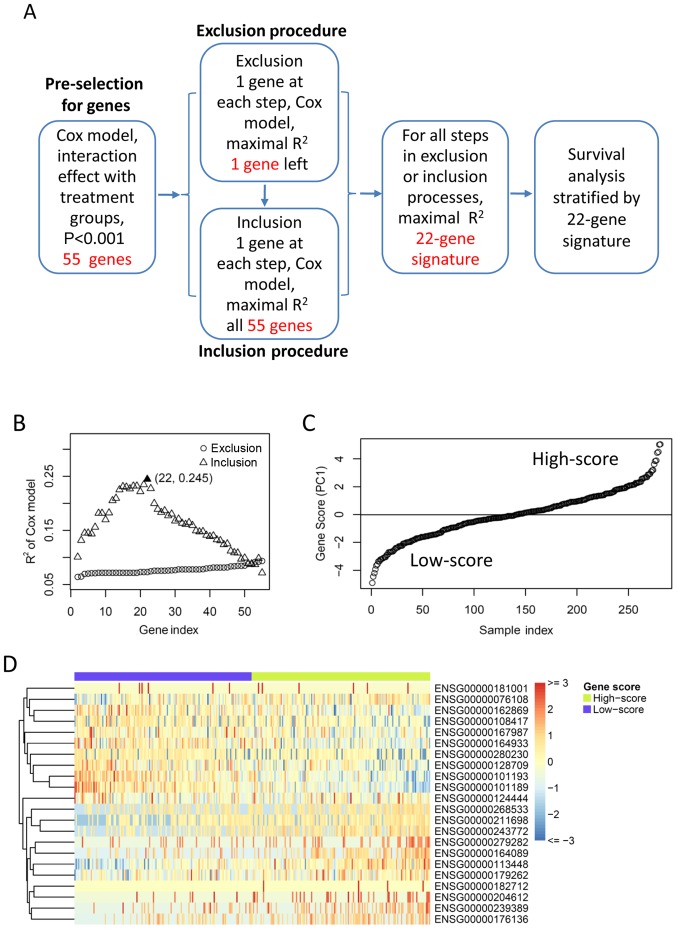
A workflow chart of signature development in this study is depicted in Fig. 1A. The genes were preselected by their interaction effect with treatment groups based on the OS. For each gene, all patients were grouped into two sub-groups according to the median of gene expression value. The interaction effect between gene groups and treatment groups were tested by Cox model based on OS. Genes with P<0.001 (test for interaction) were selected for subsequent exclusion and inclusion procedures for identifying a minimum set of genes that had the highest predictive ability. In the exclusion and inclusion processes, the Principal Component Analysis were performed for each gene set, and the first principal component (PC1) was used to group patients into two subgroups by median. Cox model was performed to test the interaction effect between PC1 groups and treatment groups. The exclusion procedure removes one gene at a time which yielded the maximal R Square (R2, goodness-of-fit) in the Cox model. The procedure was repeated until there was only one gene left. Starting with this gene, the inclusion procedure added one gene at a time which yielded the maximal resultant R2. The minimum number of genes having the largest R2 was chosen as the optimal predictive gene signature for survival benefit from chemoradiotherapy.
Figure 1.
Identifying optimal gene signature for predicting survival benefit of postoperative treatment. (A) A workflow of the method used for identifying the gene signature in this study. (B) Exclusion and inclusion procedure for gene selection. Scatter plotting shows the R square (y-axis) of the Cox model against gene index (x-axis), for each exclusion or inclusion step. The best selection of minimal gene number and maximal R square was highlighted. (C) Gene-expression score of the best 22-gene signature was ordered and plotted against the patient index. Horizontal line indicates the median gene-expression score, which was used as cut-off for patient grouping. (D) Gene-expression profile of the 22-gene signature across all patients. Relative gene-expression levels are showed by heat map, with red for high expression and blue for low expression. Patients/samples are grouped into low-score and high-score sub-groups as described in (B). PC1, the first principal component.
We also performed another method described by Zhu et al to identify the prognostic genes (11). This method were used to stratify patients for survival benefit analysis of chemoradiotherapy, and were compared with our method.
Bioinformatic and statistical methods
All tumor samples were separated into two subgroups by the median of gene-expression score (PC1) of the predictive gene signature identified above. Gene gene-expression profiles were visualized by heat map. Gene Set Enrichment Analysis (GSEA) of was performed for the two subgroups of samples using the ‘C2’ gene sets from the MSigDB database v6.0 (12). Gene sets with gene numbers <15 or >500 were not included in analysis. The enrichment P-value was obtained by using 1,000 iterations of sample-shuffling. The false discovery rate (FDR) <0.25 was considered to be significant cut-off as recommended previously (12).
The correlations between various clinical variables and patient subgroups identified by the gene signature were tested by the Chi-square test or Fisher's exact test. Kaplan-Meier curves and log-rank test were used to compare the OS, RFS and LRC of different postoperative treatments, in each subgroup of patients. The interaction effect between clinical/genetic variables and treatment groups was estimated by multivariate Cox model. All statistical tests were two-sided, and P-value <0.05 was considered to be significant. All statistical analyses were performed by R software (version 2.15.2).
Results
Identifying gene-expression predictor for survival benefit from chemoradiotherapy
For the gene-expression profiles, each gene was tested by their interaction effect with treatment groups in Cox model based on OS. With a cut-off of P<0.001 (test for interaction), 55 genes showed significant interaction effect with treatment groups. In order to identify the minimum set of genes with the best predicting efficiency, the 55 candidate genes were used in the exclusion and inclusion procedure. As a result, a combination of 22 genes achieved the highest R2 in the Cox regression model (Fig. 1B). The details of the 22 genes were listed in Table II. The gene-expression score of the 22 genes (also referred to as 22-gene signature) was defined as the first principal component (PC1) by the PCA model. The patients were separated into low-score and high-score subgroups according to the median of gene-expression score (Fig. 1C). The two subgroups showed distinct gene-expression patterns, with 10 genes upregulated in the low-score subgroup and 12 genes upregulated in the high-score subgroup respectively (Fig. 1D).
Table II.
Gene information of the 22-gene signature.
| Subgroup | Ensemble ID | Entrez gene ID | Gene symbol | Description |
|---|---|---|---|---|
| A, Low-score | ||||
| ENSG00000128709 | 3235 | HOXD9 | Homeobox D9 | |
| ENSG00000181001 | 79473 | OR52N1 | Olfactory receptor family 52 subfamily N member 1 | |
| ENSG00000167987 | 55048 | VPS37C | VPS37C, ESCRT-I subunit | |
| ENSG00000101193 | 54994 | GID8 | GID complex subunit 8 homolog | |
| ENSG00000101189 | 55257 | MRGBP | MRG domain binding protein | |
| ENSG00000162869 | 129285 | PPP1R21 | Protein phosphatase 1 regulatory subunit 21 | |
| ENSG00000076108 | 11176 | BAZ2A | Bromodomain adjacent to zinc finger domain 2A | |
| ENSG00000164933 | 81034 | SLC25A32 | Solute carrier family 25 member 32 | |
| ENSG00000108417 | 8688 | KRT37 | Keratin 37 | |
| ENSG00000280230 | – | – | – | |
| B, High-score | ||||
| ENSG00000113448 | 5144 | PDE4D | Phosphodiesterase 4D | |
| ENSG00000179262 | 5886 | RAD23A | RAD23 homolog A, nucleotide excision repair protein | |
| ENSG00000164089 | 64850 | ETNPPL | Ethanolamine-phosphate phospho-lyase | |
| ENSG00000182712 | 1E+08 | CMC4 | C-X9-C motif containing 4 | |
| ENSG00000204612 | 442425 | FOXB2 | Forkhead box B2 | |
| ENSG00000239389 | 56136 | PCDHA13 | Protocadherin alpha 13 | |
| ENSG00000211698 | 6977 | TRGV4 | T cell receptor gamma variable 4 | |
| ENSG00000124444 | 79177 | ZNF576 | Zinc finger protein 576 | |
| ENSG00000176136 | 4161 | MC5R | Melanocortin 5 receptor | |
| ENSG00000243772 | 3804 | KIR2DL3 | Killer cell immunoglobulin like receptor, two immunoglobulin domains and long cytoplasmic tail 3 | |
| ENSG00000268533 | – | – | – | |
| ENSG00000279282 | – | – | – | |
Association of 22-gene signature with the clinical variables
The association between 22-gene signature subgroups and conventional clinical variables was analyzed (Table III). There was no significant correlation between the 22-gene subgroups and the postoperative treatment groups (P=0.592). The anatomic site of the primary tumor was associated with the gene signature sub-groups (P=0.033). Tumors in the low-score subgroup were associated with positive lymph nodes <2 (P=0.001), negative surgical margin (P=0.051), as well as the three-variable combined low-risk group (P=0.003). For pathological grade, the well differentiated tumors were enriched in the low-score subgroup while the poor differentiated tumors were enriched in the high-score subgroup (P=0.0004). The other clinical variables (such as gender, age, smoking index, TNM stage, irradiation dosage/fraction or chemo-reagents) showed no significant association with the 22-gene subgroups (P>0.1).
Table III.
Association between 22-gene subgroups and clinical parameters.
| 22-gene signature | ||||
|---|---|---|---|---|
| Variable | Parameter | Low-score | High-score | P-value |
| Sex | 0.640 | |||
| Female | 32 (22.9) | 28 (19.9) | ||
| Male | 108 (77.1) | 113 (80.1) | ||
| Age | 0.511 | |||
| <60 years | 74 (52.9) | 68 (48.2) | ||
| ≥60 years | 66 (47.1) | 73 (51.8) | ||
| Smoking index | 0.703 | |||
| <40 pys | 36 (25.7) | 37 (26.2) | ||
| ≥40 pys | 46 (32.9) | 52 (36.9) | ||
| N/A | 58 (41.4) | 52 (36.9) | ||
| Anatomic site | 0.033a | |||
| Oral cavity | 94 (67.1) | 77 (54.6) | ||
| Oropharynx | 11 (7.9) | 26 (18.4) | ||
| Hypopharynx | 2 (1.4) | 4 (2.8) | ||
| Larynx | 33 (23.6) | 34 (24.1) | ||
| Positive nodes | 0.001 | |||
| <2 | 73 (52.1) | 46 (32.6) | ||
| ≥2 | 48 (34.3) | 56 (39.7) | ||
| N/A | 19 (13.6) | 39 (27.7) | ||
| Extracapsular invasion | 0.127 | |||
| No | 59 (42.1) | 58 (41.1) | ||
| Yes | 44 (31.4) | 32 (22.7) | ||
| N/A | 37 (26.4) | 51 (36.2) | ||
| Margin status | 0.028 | |||
| Negative | 113 (80.7) | 94 (66.7) | ||
| Positive | 12 (8.6) | 22 (15.6) | ||
| N/A | 28 (20) | 37 (26.2) | ||
| Clinical high risk | 0.003 | |||
| No | 47 (33.6) | 27 (19.1) | ||
| Yes | 63 (45) | 72 (51.1) | ||
| N/A | 30 (21.4) | 42 (29.8) | ||
| Tumor grade | 0.0004a | |||
| G1 | 24 (17.1) | 7 (5) | ||
| G2 | 86 (61.4) | 77 (54.6) | ||
| G3 | 26 (18.6) | 46 (32.6) | ||
| G4 | 0 (0) | 2 (1.4) | ||
| N/A | 4 (2.9) | 9 (6.4) | ||
| TNM stage | 0.458a | |||
| I | 3 (2.1) | 6 (4.3) | ||
| II | 10 (7.1) | 6 (4.3) | ||
| III | 21 (15) | 16 (11.3) | ||
| IV | 86 (61.4) | 86 (61) | ||
| N/A | 20 (14.3) | 27 (19.1) | ||
| pT stage | 0.161 | |||
| T1-2 | 32 (22.9) | 44 (31.2) | ||
| T3-4 | 88 (62.9) | 73 (51.8) | ||
| N/A | 20 (14.3) | 24 (17) | ||
| pN stage | 0.100 | |||
| N0-1 | 65 (46.4) | 48 (34) | ||
| N2-3 | 52 (37.1) | 62 (44) | ||
| N/A | 23 (16.4) | 31 (22) | ||
| Radiation fraction | 0.945 | |||
| Conventional | 87 (62.1) | 85 (60.3) | ||
| None-conventional | 10 (7.1) | 11 (7.8) | ||
| N/A | 43 (30.7) | 45 (31.9) | ||
| Radiation dosage | 0.266 | |||
| <60 Gy | 17 (12.1) | 2 (17) | ||
| ≥60 Gy | 100 (71.4) | 88 (62.4) | ||
| N/A | 23 (16.4) | 29 (20.6) | ||
| Drug name | 0.878a | |||
| Platin | 21 (15) | 25 (17.7) | ||
| Platin + Taxel | 8 (5.7) | 9 (6.4) | ||
| Chemo + Target | 2 (1.4) | 3 (2.1) | ||
| N/A | 109 (77.9) | 104 (73.8) | ||
| Postoperative treatment group | 0.592 | |||
| Radiation | 71 (50.7) | 66 (46.8) | ||
| Radichem | 69 (49.3) | 75 (53.2) | ||
Fisher's exact test. pys, package years; N/A, not available; RadiChem, concurrent chemoradiotherapy; TNM, tumor, node metastasis.
The low-score subgroup was associated with gene signatures of stemness and chemo-resistance
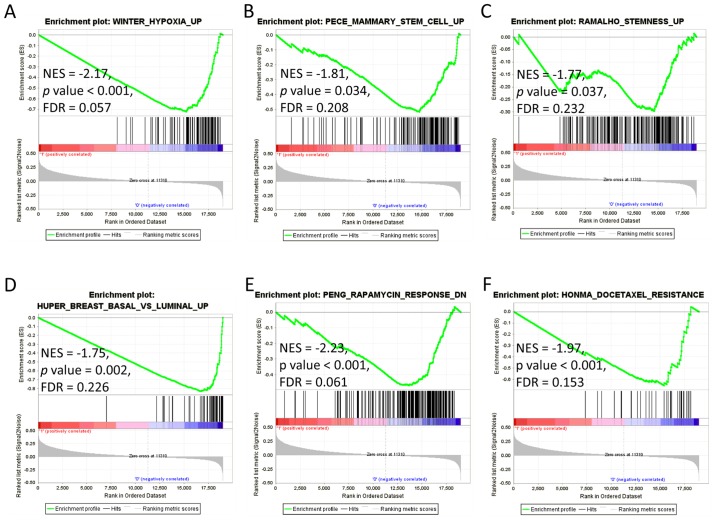
We next examined the biological meanings of the 22-gene signature subgroups. GSEA was performed using the 4,731 curated gene sets (C2), which contain canonical pathways and expression signatures from various experiments. When using FDR <25% as the significant cut-off (12), 69 gene sets were enriched in the low-score subgroup while there was no gene sets significantly enriched in the high-score subgroup. The hypoxia upregulated gene signature identified in HNSCC was significantly enriched in the low-score subgroup (Fig. 2A, NES=−2.17, P-value <0.001, FDR=0.057). Two gene sets of human mammary stem cells (Fig. 2B, NES=−1.81, P-value=0.034, FDR=0.208) and the mouse embryonic, neural, and hematopoietic stem cells (Fig. 2C, NES=−1.77, P-value=0.037, FDR=0.232) were enriched in the low-score subgroup. Meanwhile, the low-score subtype was also associated with genes upregulated in the basal subtype of breast cancer (Fig. 2D, NES=−1.75, P-value=0.002, FDR=0.226). These results suggest the low-score subgroup may carry characteristics of stemness and basal phenotype of cancer.
Figure 2.
GSEA for the two subgroups of HNSCC. GSEA was performed for the two subgroups identified by 22-gene signature. Label ‘1’ shows the high-score subgroup, label ‘0’ for the low-score subgroup. Green lines indicate the Enrichment score of each gene sets. The P-value was obtained by 1,000 iterations of sample-shuffling. (A) Gene set upregulated in hypoxia in HNSCC. (B) Gene set upregulated in human mammary stem cells. (C) Gene set upregulated in mouse embryonic, neural, and hematopoietic stem cells. (D) Gene set upregulated in basal subtype compared with luminal subtype. (E) Gene set downregulated in cells in response to rapamycin. (F) Gene set upregulated in cells resistant to docetaxel. GSEA, Gene Set Enrichment Analysis; HNSCC, head and neck squamous cell carcinoma; ES, enrichment score; NES, new enrichment score; FDR, false discovery rate.
Particularly, the low-score subtype was significantly associated with gene sets involved in resistance of cancer cells to antitumor drugs. Genes downregulated in B-lymphoma cells in response to rapamycin were significantly enriched in the low-score subgroup (Fig. 2E, NES=−2.23, P-value <0.001, FDR=0.061). Another gene set which was upregulated in breast cancer cells resistant to docetaxel was also significantly enriched in the low-score subgroup (Fig. 2F, NES=−1.97, P-value <0.001, FDR=0.153).
Survival benefit of chemoradiotherapy predicted by 22-gene signature
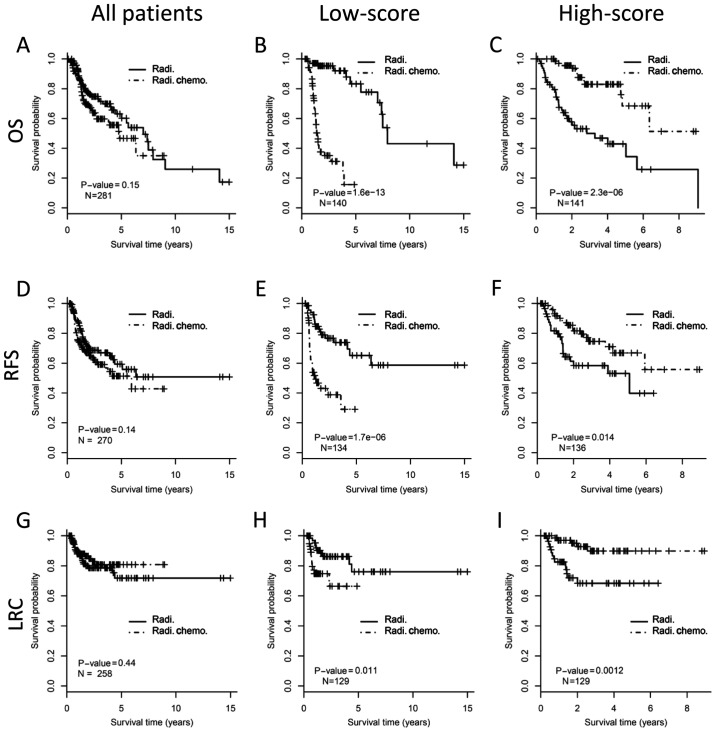
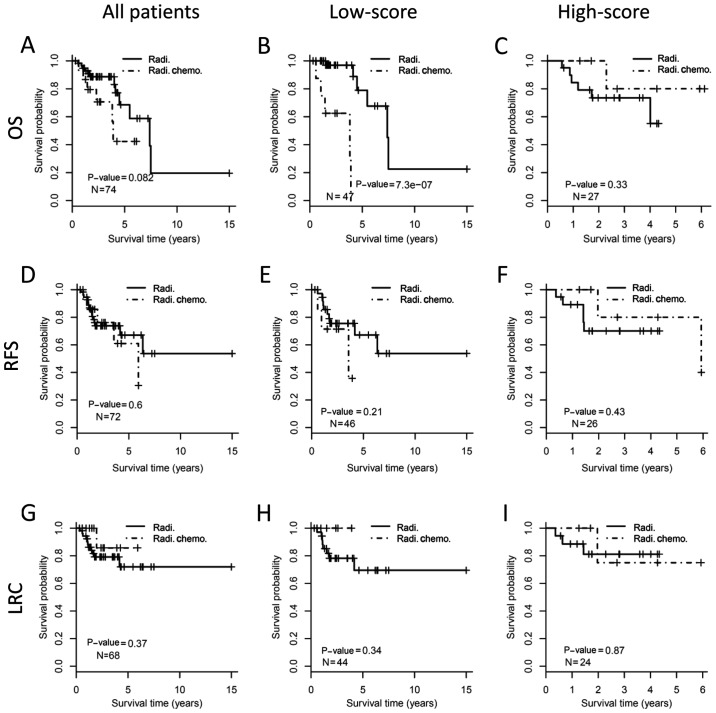
The survival benefit from concurrent chemoradiotherapy was assessed in different subgroups of patients. There was no OS improvement for adding chemotherapy to radiotherapy when considering the whole cohort (Fig. 3A, log-rank test, P=0.15). In the low-score subgroup, patients who received chemoradiotherapy showed even worse OS than those with radiotherapy (P<0.001; Fig. 3B). However, for the high-score subgroup, concurrent chemoradiotherapy demonstrated significantly improved OS than radiotherapy (P<0.001; Fig. 3C). For RFS, patients who received concurrent chemoradiotherapy showed no benefit for the whole cohort (P=0.14; Fig. 3D), poorer outcome in the low-score subgroup (P<0.001; Fig. 3E), and better outcome in the high-score subgroup (P=0.014; Fig. 3F). Similarly, patients who received chemoradiotherapy have no benefit on LRC than those who received radiotherapy alone (P=0.44; Fig. 3G) when considering all patients. Patients who received chemoradiotherapy in low-score subgroup demonstrated worse LRC (P=0.011; Fig. 3H), but patients in the high-score subgroup showed improved LRC (P=0.001; Fig. 3I), compared with those who received radiotherapy alone.
Figure 3.
Outcome benefit from concurrent chemoradiotherapy stratified by 22-gene signature. The upper, middle and the bottom panels show OS, RFS and LRC, respectively. The whole cohort of HNSC patients (the left column), low-score (the middle column) and high-score (the right column) stratified by 22-gene signature are analyzed respectively. Kaplan-Meier curves show the outcome of patients with different postoperative treatments. The P-value of each pair of Kaplan-Meier curves was calculated by log-rank test. (A) OS analysis for all patients. (B) OS analysis for patients with low-score by 22-gene signature. (C) OS analysis for patients with high-score by 22-gene signature. (D) RFS analysis for all patients. (E) RFS analysis for patients with low-score by 22-gene signature. (F) RFS analysis for patients with high-score by 22-gene signature. (G) LRC analysis for all patients. (H) LRC analysis for patients with low-score by 22-gene signature. (I) LRC analysis for patients with high-score by 22-gene signature. OS, overall survival; RFS, relapse free survival; LRC, local regional control; HNSCC, head and neck squamous cell carcinoma; Radi., radiation; Radi.Chemo., concurrent chemoradiotherapy.
Predictive value of 22-gene signature is better than routine clinical variables
The prognostic value and interaction effect with postoperative treatment of clinical/genetic variables were analyzed by Cox regression model (Table IV). The positive lymph nodes >2 (P=0.028), surgical margin (P=0.032), as well as the three-variable combined risk group (P=0.015) were significant prognostic factors for OS. The high-score subgroup of 22-gene signature was a high risk factor of patient death (P<0.001). A significant interaction effect was observed between 22-gene signature and postoperative treatment (test for interaction, P<0.001). Although the three-variable combined risk group was a prognostic factor for OS (P=0.015), but it was not a predictive factor for treatment benefit (test for interaction, P=0.121). The other clinical parameters showed no statistical significance in the Cox analysis.
Table IV.
Cox regression of clinical/genetic variables based on OS.
| Main effect of variables | Interaction effects with therapy | |||
|---|---|---|---|---|
| Variables | Hazard ratio | P-value | Hazard ratio | P-value |
| Gender (male vs. female) | 0.86 (0.2–3.75) | 0.839 | 0.89 (0.35–2.25) | 0.807 |
| Age (≥60 vs. <60), years | 1.942 (0.49–7.74) | 0.347 | 0.77 (0.33–1.79) | 0.547 |
| Smoking index (≥40 vs. <40), py | 1.17 (0.19–7.24) | 0.866 | 0.85 (0.29–2.53) | 0.776 |
| Anatomic site (others vs. oral cavity) | 1.09 (0.27–4.36) | 0.902 | 0.84 (0.36–1.94) | 0.679 |
| Positive node (≥2 vs. <2) | 5.06 (1.19–21.47) | 0.028 | 0.53 (0.21–1.35) | 0.185 |
| Extracapsular invasion (yes vs. no) | 3.49 (0.75–16.23) | 0.111 | 0.92 (0.35–2.44) | 0.869 |
| Margin status (positive vs. negative) | 6.43 (1.17–35.33) | 0.032 | 0.48 (0.17–1.37) | 0.171 |
| High riska (yes vs. no) | 7.48 (1.48–37.83) | 0.015 | 0.42 (0.14–1.26) | 0.121 |
| High riskb (yes vs. no) | 6.19 (1.41–27.17) | 0.016 | 0.58 (0.22–1.48) | 0.251 |
| Tumor grade (G3-4 vs. G1-2) | 1.68 (0.401–7.01) | 0.480 | 0.64 (0.26–1.59) | 0.331 |
| TNM stage (IV vs. I–III) | 4.66 (0.76–28.68) | 0.097 | 0.56 (0.16–2.02) | 0.379 |
| pT stage (T3-4 vs. T1-2) | 4.43 (0.79–24.75) | 0.090 | 0.59 (0.2–1.71) | 0.327 |
| pN stage (N2-3 vs. N0-1) | 3.56 (0.8–15.93) | 0.096 | 0.72 (0.27–1.94) | 0.518 |
| 22-gene signature (high-score vs. low-score) | 300.32 (62.07–1453.13) | 1.32×10−12 | 0.02 (0.01–0.05) | 7.99×10−15 |
Clinical high-risk by Positive node ≥2, extracapsular invasion and positive margin
Clinical high-risk by extracapsular invasion and positive margin. y, years; py, package years.
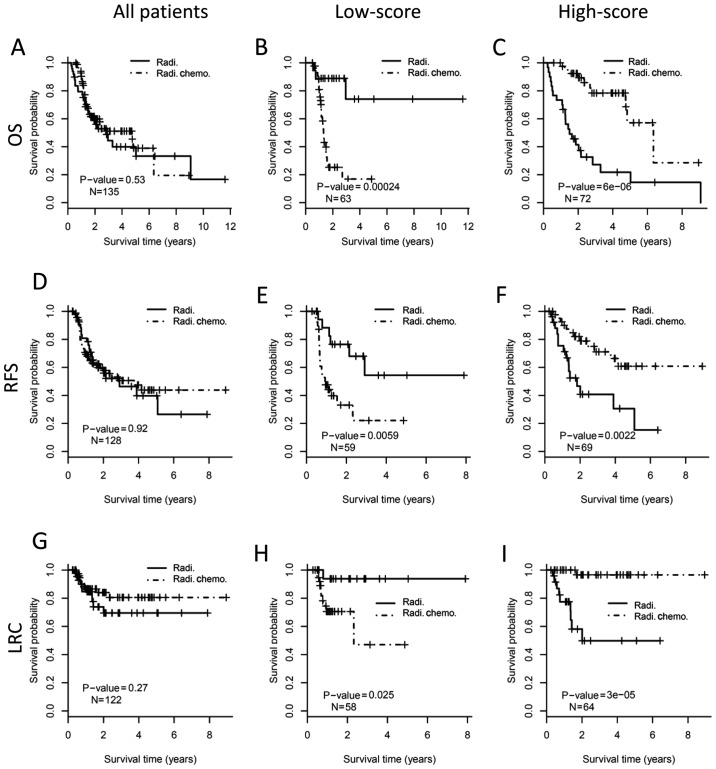
We next investigate the relationship between the 22-gene signature and the conventional clinical risk factors in predicting the outcome benefit of adjuvant chemoradiotherapy (Fig. 4). When considering all patients at clinical high-risk, concurrent chemoradiotherapy after surgical resection did not demonstrate better outcome than radiotherapy alone (P>0.05; Fig. 4A, D and G). For the patients in low-score subgroup by the 22-gene signature, concurrent chemoradiotherapy was correlated with worse OS (log-rank test, P<0.001; Fig. 4B), poorer RFS (P=0.006; Fig. 4E) and worse LRC (P=0.025; Fig. 4H), compare with mono radiotherapy. In the high-score sub-group, however, chemoradiotherapy demonstrated better OS (P<0.001; Fig. 4C), better RFS (P=0.002; Fig. 4F), and improved LRC (P<0.001; Fig. 3I) than mono radiotherapy. However, the predictive value of the 22-gene signature was not significant anymore in the sub-cohort of patients at clinical low-risk (Fig. 5). These results suggested that a combination of the 22-gene signature and conventional clinical risk factors may be more precise in predicting treatment benefit.
Figure 4.
Predictive value of 22-gene signature in patients at clinical high-risk. The entire cohort of high-risk patients (the left column), low-score subgroup (the middle column) and high-score subgroup (the right column) stratified by 22-gene signature are analyzed respectively. The upper, middle and the bottom panels show OS, RFS and LRC, respectively. Kaplan-Meier curves show the outcome of patients with different postoperative treatments. The P-value of each pair of Kaplan-Meier curves is calculated by log-rank test. (A) OS analysis for all patients at clinical high-risk. (B) OS analysis for patients with low-score by 22-gene signature. (C) OS analysis for patients with high-score by 22-gene signature. (D) RFS analysis for all patients at clinical high-risk. (E) RFS analysis for patients with low-score by 22-gene signature. (F) RFS analysis for patients with high-score by 22-gene signature. (G) LRC analysis for all patients at clinical high-risk. (H) LRC analysis for patients with low-score by 22-gene signature. (I) LRC analysis for patients with high-score by 22-gene signature. OS, overall survival; RFS, relapse free survival; LRC, local regional control; Radi., radiotherapy; Radi.Chemo., concurrent chemoradiotherapy.
Figure 5.
Predictive value of 22-gene signature in patients at clinical low-risk. The entire cohort of low-risk patients (the left column), low-score subgroup (the middle column) and high-score subgroup (the right column) stratified by 22-gene signature are analyzed respectively. The upper, middle and the bottom panels show OS, RFS and LRC, respectively. Kaplan-Meier curves show the outcome of patients with different postoperative treatments. The P-value of each pair of Kaplan-Meier curves is calculated by log-rank test. (A) OS analysis for all patients at clinical low-risk. (B) OS analysis for patients with low-score by 22-gene signature. (C) OS analysis for patients with high-score by 22-gene signature. (D) RFS analysis for all patients at clinical low-risk. (E) RFS analysis for patients with low-score by 22-gene signature. (F) RFS analysis for patients with high-score by 22-gene signature. (G) LRC analysis for all patients at clinical low-risk. (H) LRC analysis for patients with low-score by 22-gene signature. (I) LRC analysis for patients with high-score by 22-gene signature. OS, overall survival; RFS, relapse free survival; LRC, local regional control; Radi., radiotherapy; Radi.Chemo., concurrent chemoradiotherapy.
We also compared our bioinformatic approach with a previously described method which identified the prognostic genes (11). Using the previous method, we developed a gene-expression profile (Fig. 6A) which could predict poor prognosis for patients who received mono radiotherapy (Fig. 6B, left column). The patients who received concurrent chemoradiotherapy were also classified as low-risk or high-risk groups by this gene-expression profile. In the low-risk subgroup, concurrent chemoradiotherapy could not improve patient outcome, but an even worse prognosis was observed (Fig. 6B, middle column). In the high-risk group, there was no significant improvement of survival by concurrent chemoradiotherapy (Fig. 6B, right column).
Figure 6.
Identification of a prognostic signature for HNSCC patients. (A) Gene-expression profile for the patients who received mono radiotherapy. Genes were pre-selected by univariate Cox regression based on OS (P<0.005, n=66). Patients/samples are grouped into low-risk and high-risk sub-groups by unsupervised hierarchical clustering. Relative gene-expression levels are showed by heat map, with red for high expression and blue for low expression. The patients who received concurrent chemoradiotherapy were also classified as low-risk or high-risk sub-groups by the centroids of those who received mono radiotherapy. (B) Kaplan-Meier analysis of HNSCC patients in different groups. The left column indicates the survival difference of patients in low/high-risk groups in A. The middle and right column indicate survival benefit from concurrent chemoradiotherapy in the low-risk and high-risk subgroups respectively. The upper, middle and the bottom panels show OS, RFS and LRC, respectively. The P-value of each pair of Kaplan-Meier curves is calculated by log-rank test. HNSCC, head and neck squamous cell carcinoma; OS, overall survival; RFS, relapse free survival; LRC, local regional control; Radi., radiotherapy; Radi.Chemo., concurrent chemoradiotherapy.
Discussion
Postoperative radiotherapy or concurrent chemoradiotherapy are common for improving patient outcome in advanced HNSCC. However, the proper decision of using mono radiotherapy or concurrent chemoradiotherapy is controversial. In this study, we developed a 22-gene expression signature which distinguished HNSCC patients with different outcome after receiving chemoradiotherapy. Patients classified as high-score subgroup might get improved outcome from concurrent chemoradiotherapy compared with mono radiotherapy. In contrast, in the low-score subgroup, patient receiving chemoradiotherapy would get even worse prognosis. The high-score subtype was associated with some proliferation promoting genes. Genes upregulated in the low-score subtype were related to stemness traits and drug resistance features in cancer cells.
For HNSCC patients with clinically high-risk (at least one of the three factors: With two or more positive lymph nodes, extracapsular extension of nodal disease or positive surgical margins), postoperative chemoradiotherapy improved local control and RFS compared with mono radiotherapy initially (3). However, the long-term RFS and OS showed no significant differences (4). Further analysis showed that only patients with extracapsular extension and/or positive surgical margins could benefit from adjuvant concurrent chemoradiotherapy (4,13). For the TCGA cohort analyzed in this study, we found that neither the three-variable nor the two-variable combined risk factor could predict the survival benefit of adjuvant chemoradiotherapy (Fig. 4; Table IV). These results suggest a novel signature is needed to improve the predictive efficiency of the conventional clinical factors. Here, the 22-gene signature showed a significant interaction effect with postoperative concurrent chemoradiotherapy on the patient outcome. Adding chemotherapy to radiotherapy improved postoperative outcome only in patients from the high-score subgroup but not in those from the low-score subgroup stratified by the 22-gene signature. Moreover, we also found that the 22-gene signature exhibited predictive value only in the clinically high-risk sub-cohort, but not in patients with low-risk. These results suggested that a combination of the 22-gene signature and the clinical risk factors may serve as a robust predictive marker for identifying patients who would benefit from postoperative chemoradiotherapy.
A previous study identified a gene expression signature which could predict survival benefit from adjuvant chemotherapy for non-small cell lung cancer (11). In the study, colleagues focused on a prognostic gene signature which could group patients into high/low-risk subgroups, and the individuals at high-risk group were more likely to benefit from chemotherapy (11). Using the same approach, we developed a gene-expression profile which could predict poor prognosis for patients who received mono radiotherapy (Fig. 6A and B, left column). The patients who received concurrent chemoradiotherapy were also classified by this gene-expression profile. For the low-risk subgroup, patients who received concurrent chemoradiotherapy had even worse outcome than those who received radiotherapy alone (Fig. 6B, middle column). However, the high-risk group could not serve as a predictor for benefit of chemoradiotherapy (Fig. 6B, right column). On contrary, our method focused on the interaction effect between genes and therapeutic groups on the patient outcome. The 22-gene signature identified by our approach could distinguished patients who would benefit from concurrent chemoradiotherapy (Figs. 3 and 4, right column). Collectively, the signature identified in the present study may be more likely to reflect the drug sensitivity/resistance traits of cancer cells. Furthermore, there are also some disadvantages of our method. For instance, the 22-gene signature was developed in a single HNSCC cohort, and another independent cohort is warranted to validate the signature in future work.
The high-score subgroup predicted poor prognosis of HNSCC when adjusted by the postoperative treatments (Table IV). The genes identified by the 22-gene signature may have some important biological insights in cancer cells. Some of the genes upregulated in the high-score subtype have been reported to serve as cancer-promoting genes. Phosphodiesterase 4D (PDE4D) is a subtype of metallohydrolases, and was characterized as a tumor-promoting factor in several types of cancers (14). Previous studies reported that high expression of PDE4D promotes cell proliferation and cancer growth in prostate cancer (15–17). A recent study found that PDE4D was over-expressed in nasopharyngeal carcinoma (NPC) tissues and cells. Knockdown of PDE4D resulted in cell cycle arrest and growth inhibition in NPC cells, which indicated that PDE4D saves as a promoting factor in NPC (18). However, the potential role of PDE4D in non-NPC HNSCC have been unknown. Our results showed that upregulation of PDE4D was correlated with poor prognosis in HNSCC patients. The mechanisms underlying the potential role of PDE4D on HNSCC progression are with great interest in the next study.
The low-score subgroup identified by the 22-gene signature was correlated to unfavorable outcome after receiving chemoradiotherapy, suggesting some possible chemo-resistance mechanisms existing in this subtype. Indeed, we found that two gene sets involved in drug resistance were significantly enriched in the low-score phenotype. Hypoxia is commonly observed in many solid tumors including HNSCC, and is linked to chemo-resistance (19,20). Our results revealed that the low-score subtype was significantly enriched by a hypoxia-induced gene set in HNSCC (21). The cancer stem cell-like phenotype was associated with chemo-resistance in head and neck cancer cells (22–24). We found that two stemness expression signatures were enriched in the low-score phenotype. The genes upregulated in basal-like breast cancer (25), which showed stem-like features (26), were associated with the low-score subgroup of HNSCC. In our 22-gene signature, HOXD9 was upregulated in the low-score subtype. A previous study found that HOXD9 was upregulated in a side population with cancer stem-like cells of glioma (27). However, the role of HOXD9 in HNSCC is poorly understood and further studies are warranted. Taken together, the chemo-resistance trait of the low-score subtype was supported by its significant association with biological features of hypoxia, basal phenotype as well as stemness.
In summary, this retrospective study established a 22-gene signature which has the potential to predict outcome benefit of postoperative concurrent chemoradiotherapy in HNSCC. The hypoxia- or stem-like gene expression properties of HNSCC may contribute to chemo-resistance. The 22-gene signature as a predictor for patient selection may be helpful in the personalized medicine field of HNSCC. A large prospective cohort is proposed to validate this gene signature.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- HNSCC
head-and-neck squamous cell carcinoma
- TCGA
The Cancer Genome Atlas
- OS
overall survival
- RFS
relapse free survival
- LRC
locoregional control
- PCA
principal component analysis
- GSEA
gene set enrichment analysis
- FDR
false discovery rate
Funding
No funding was received.
Availability of data and materials
The dataset analyzed during the current study is publicly available in the TCGA database portal (portal.gdc.cancer.gov/).
Authors' contributions
Conception and design, ZW and GZ; development of methodology, JC, GF and YC; acquisition of data, JC, GF and YC; analysis and interpretation of data, JC and GF; writing, review and revision of the manuscript, JC, GZ and ZW; administrative, technical or material support, GZ and ZW; study supervision, JC and YC. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Cao B, Wang Q, Zhang H, Zhu G, Lang J. Two immune-enhanced molecular subtypes differ in inflammation, checkpoint signaling and outcome of advanced head and neck squamous cell carcinoma. Oncoimmunology. 2017;7:e1392427. doi: 10.1080/2162402X.2017.1392427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper JS, Pajak TF, Forastiere AA, Jacobs J, Campbell BH, Saxman SB, Kish JA, Kim HE, Cmelak AJ, Rotman M, et al. Postoperative concurrent radiotherapy and chemotherapy for high-risk squamous-cell carcinoma of the head and neck. N Engl J Med. 2004;350:1937–1944. doi: 10.1056/NEJMoa032646. [DOI] [PubMed] [Google Scholar]
- 4.Cooper JS, Zhang Q, Pajak TF, Forastiere AA, Jacobs J, Saxman SB, Kish JA, Kim HE, Cmelak AJ, Rotman M, et al. Long-term follow-up of the RTOG 9501/intergroup phase III trial: Postoperative concurrent radiation therapy and chemotherapy in high-risk squamous cell carcinoma of the head and neck. Int J Radiat Oncol Biol Phys. 2012;84:1198–1205. doi: 10.1016/j.ijrobp.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janot F, de Raucourt D, Benhamou E, Ferron C, Dolivet G, Bensadoun RJ, Hamoir M, Géry B, Julieron M, Castaing M, et al. Randomized trial of postoperative reirradiation combined with chemotherapy after salvage surgery compared with salvage surgery alone in head and neck carcinoma. J Clin Oncol. 2008;26:5518–5523. doi: 10.1200/JCO.2007.15.0102. [DOI] [PubMed] [Google Scholar]
- 6.Wolff HA, Overbeck T, Roedel RM, Hermann RM, Herrmann MK, Kertesz T, Vorwerk H, Hille A, Matthias C, Hess CF, Christiansen H. Toxicity of daily low dose cisplatin in radiochemotherapy for locally advanced head and neck cancer. J Cancer Res Clin Oncol. 2009;135:961–967. doi: 10.1007/s00432-008-0532-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/S1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 8.Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, Ang MK, Hayward MC, Salazar AH, Hoadley KA, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;8:e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wichmann G, Rosolowski M, Krohn K, Kreuz M, Boehm A, Reiche A, Scharrer U, Halama D, Bertolini J, Bauer U, et al. The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int J Cancer. 2015;137:2846–2857. doi: 10.1002/ijc.29649. [DOI] [PubMed] [Google Scholar]
- 10.Linge A, Lock S, Gudziol V, Nowak A, Lohaus F, von Neubeck C, Jütz M, Abdollahi A, Debus J, Tinhofer I, et al. Low Cancer Stem Cell Marker Expression and Low Hypoxia Identify Good Prognosis Subgroups in HPV(−) HNSCC after Postoperative Radiochemotherapy: A Multicenter Study of the DKTK-ROG. Clin Cancer Res. 22:2639–2649. doi: 10.1158/1078-0432.CCR-15-1990. [DOI] [PubMed] [Google Scholar]
- 11.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernier J, Cooper JS, Pajak TF, van Glabbeke M, Bourhis J, Forastiere A, Ozsahin EM, Jacobs JR, Jassem J, Ang KK, Lefèbvre JL. Defining risk levels in locally advanced head and neck cancers: A comparative analysis of concurrent postoperative radiation plus chemotherapy trials of the EORTC (#22931) and RTOG (# 9501) Head Neck. 2005;27:843–850. doi: 10.1002/hed.20279. [DOI] [PubMed] [Google Scholar]
- 14.Lin DC, Xu L, Ding LW, Sharma A, Liu LZ, Yang H, Tan P, Vadgama J, Karlan BY, Lester J, et al. Genomic and functional characterizations of phosphodiesterase subtype 4D in human cancers. Proc Natl Acad Sci USA. 2013;110:6109–6114. doi: 10.1073/pnas.1218206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Böttcher R, Dulla K, van Strijp D, Dits N, Verhoef EI, Baillie GS, van Leenders GJ, Houslay MD, Jenster G, Hoffmann R. Human PDE4D isoform composition is deregulated in primary prostate cancer and indicative for disease progression and development of distant metastases. Oncotarget. 2016;7:70669–70684. doi: 10.18632/oncotarget.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powers GL, Hammer KD, Domenech M, Frantskevich K, Malinowski RL, Bushman W, Beebe DJ, Marker PC. Phosphodiesterase 4D inhibitors limit prostate cancer growth potential. Mol Cancer Res. 2015;13:149–160. doi: 10.1158/1541-7786.MCR-14-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahrmann EP, Collier LS, Knutson TP, Doyal ME, Kuslak SL, Green LE, Malinowski RL, Roethe L, Akagi K, Waknitz M, et al. Identification of PDE4D as a proliferation promoting factor in prostate cancer using a Sleeping Beauty transposon-based somatic mutagenesis screen. Cancer Res. 2009;69:4388–4397. doi: 10.1158/0008-5472.CAN-08-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu T, Wu S, Yuan Y, Yan G, Xiao D. Knockdown of phosphodiesterase 4D inhibits nasopharyngeal carcinoma proliferation via the epidermal growth factor receptor signaling pathway. Oncol Lett. 2014;8:2110–2116. doi: 10.3892/ol.2014.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doktorova H, Hrabeta J, Khalil MA, Eckschlager T. Hypoxia-induced chemoresistance in cancer cells: The role of not only HIF-1. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2015;159:166–177. doi: 10.5507/bp.2015.025. [DOI] [PubMed] [Google Scholar]
- 20.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 21.Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, Shah KA, Cox GJ, Corbridge RJ, Homer JJ, et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67:3441–3449. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- 22.Masui T, Ota I, Yook JI, Mikami S, Yane K, Yamanaka T, Hosoi H. Snail-induced epithelial-mesenchymal transition promotes cancer stem cell-like phenotype in head and neck cancer cells. Int J Oncol. 2014;44:693–699. doi: 10.3892/ijo.2013.2225. [DOI] [PubMed] [Google Scholar]
- 23.Ota I, Masui T, Kurihara M, Yook JI, Mikami S, Kimura T, Shimada K, Konishi N, Yane K, Yamanaka T, Kitahara T. Snail-induced EMT promotes cancer stem cell-like properties in head and neck cancer cells. Oncol Rep. 2016;35:261–266. doi: 10.3892/or.2015.4348. [DOI] [PubMed] [Google Scholar]
- 24.Hsu DS, Lan HY, Huang CH, Tai SK, Chang SY, Tsai TL, Chang CC, Tzeng CH, Wu KJ, Kao JY, Yang MH. Regulation of excision repair cross-complementation group 1 by Snail contributes to cisplatin resistance in head and neck cancer. Clin Cancer Res. 2010;16:4561–4571. doi: 10.1158/1078-0432.CCR-10-0593. [DOI] [PubMed] [Google Scholar]
- 25.Huper G, Marks JR. Isogenic normal basal and luminal mammary epithelial isolated by a novel method show a differential response to ionizing radiation. Cancer Res. 2007;67:2990–3001. doi: 10.1158/0008-5472.CAN-06-4065. [DOI] [PubMed] [Google Scholar]
- 26.Ricardo S, Vieira AF, Gerhard R, Leitão D, Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F, Paredes J. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J Clin Pathol. 2011;64:937–946. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- 27.Tabuse M, Ohta S, Ohashi Y, Fukaya R, Misawa A, Yoshida K, Kawase T, Saya H, Thirant C, Chneiweiss H, et al. Functional analysis of HOXD9 in human gliomas and glioma cancer stem cells. Mol Cancer. 2011;10:60. doi: 10.1186/1476-4598-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset analyzed during the current study is publicly available in the TCGA database portal (portal.gdc.cancer.gov/).