Abstract
Period 2 (per2) is a core circadian clock gene. Dysregulation of the per2 gene has been identified in a number of types of human cancer and may be associated with a poor prognosis. To confirm the influence of per2 gene on MNNG/HOS human osteosarcoma cells, small interfering (si)RNA against per2 or plasmids containing per2 were transfected into MNNG/HOS cells, and the proliferation, apoptosis and migration were observed. The present study demonstrated that per2 knockdown significantly enhanced MNNG/HOS cell proliferation and migration and protected MNNG/HOS cells from apoptosis. Per2 overexpression inhibited MNNG/HOS cell proliferation and migration and promoted apoptosis. Furthermore, the protein expression of phosphorylated (p)-protein kinase B (Akt) and Bcl-2 were inhibited in per2-overexpressing cells, while the expression of p27, p21 and cleaved caspase-3 was promoted. In contrast, the expression of p-Akt and Bcl-2 was promoted in per2-knockdown cells, and p27, p21 and cleaved caspase-3 were decreased. This initial study may provide an alternative therapeutic strategy for the treatment of osteosarcoma.
Keywords: period 2 gene, human osteosarcoma cells, proliferation, migration, apoptosis
Introduction
Osteosarcoma is the most common malignant bone tumor, with characteristics that include hematogenous metastasis and local invasion (1–3). Osteosarcoma primarily occurs in children aged 10–14 years old (2); the metaphysis of long bones is the most susceptible area, particularly the distal femur and proximal tibia. Patients without lung metastasis have a five-year survival rate range from 50 to 70% following radical tumor resection, whereas those with lung metastasis have a <20% five-year survival rate (4). The lung is the most common site of metastasis, followed by the bones (5).
At present, the standard treatment strategies include preoperative chemotherapy, surgical resection and postoperative chemotherapy. Although various chemotherapeutics have been developed, a large proportion of patients continue to succumb to disease recurrence and metastasis. The main reasons for treatment failure are early metastasis and the development of drug resistance. Therefore, it is necessary to explore novel alternative treatment options, particularly gene therapy (6,7), to prevent early metastasis, slow osteosarcoma progression and improve the quality of life and survival rate for patients with osteosarcoma.
Circadian rhythm is also known as the ‘biological rhythm’ and is a cycle of ~24 h. Nearly all living beings have a biological rhythm, which resembles the Earth's day and night cycle. This phenomenon is part of the biological evolution and the natural selection process. Biological rhythm affects not only macro-organisms, but also in various organs and even single cells, so biological rhythm is one of the most important basic characteristics of biological activity. A number of biochemical and physiological indexes, including body temperature, heart rate, blood pressure (8) and sleep-wake cycle (9), are significantly affected by circadian rhythm. The molecular biological foundation of these processes are the circadian rhythm genes, which serve important roles in the maintenance of circadian rhythm, as well as affecting other gene expression and biochemical processes (10). The disruption of circadian rhythm leads to the occurrence and development of certain diseases (11,12). A circadian variation in the onset of stroke (13) and myocardial infarction (14) has been demonstrated, and workers who work night shifts have a greater chance of developing diabetes (15). Circadian clock genes, such as Period 2 (per2) and Bmal1, have been demonstrated to serve important roles in angiogenesis (16).
Per2 is a core circadian clock gene. Per2 protein, which affects various signaling pathways, including the protein kinase B (Akt) cascade, (17) to regulate the biological activities of the body, is expressed variably in different tissues (18). Dysregulation of the per2 gene has been identified in various types of human cancer (12,19), including breast cancer (20), hepatoma (21), colorectal cancer (22) and pancreatic ductal adenocarcinoma (23), and may be associated with a poor prognosis. However, there is limited data regarding whether and how the circadian gene per2 affects the functions of osteosarcoma cells. As further research is required on this topic, the present study aimed to investigate the effect of per2 in osteosarcoma cells. To achieve this, a eukaryotic expression vector containing per2 or per2 small interfering (si)-RNA was transfected into MNNG/HOS osteosarcoma cells. Following transfection, the effect on cell proliferation, migration and apoptosis were analyzed. Furthermore, in order to investigate the mechanisms of per2 in MNNG/HOS cells, the effects of per2 overexpression and knockdown on proliferation and apoptosis-associated proteins and the Akt signal pathway were examined.
Materials and methods
Cell culture and transfection
The MNNG/HOS Cl #5 human osteosarcoma cell line (cat. no. R-1059-D) was purchased from the Cell Resource Center of the Shanghai Institute of Life Sciences, Chinese Academy of Sciences (Shanghai, China). MNNG/HOS cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS; both Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 IU/ml penicillin and 100 IU/ml streptomycin. Cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
The cells were allocated into five groups, i.e., blank control (no treatment), pcDNA3.1-per2, pcDNA3.1-only, per2 siRNA and control siRNA groups. Cells were transfected with the plasmid and siRNA with Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.). Briefly, according to the manufacturer's protocol, the transfection complex was prepared based on an optimized proportion of plasmid or siRNA and Lipofectamine® 2000 reagent, which was transfected into MNNG/HOS cells at 70–80% confluence. After 48 h, cells were washed with PBS and collected for subsequent assays.
Immunofluorescence
Cells were plated on coverslips in six-well plates and transfected as described, cells were fixed with 4% paraformaldehyde at room temperature for 10 min and followed by 0.1% Triton X-100 for 10 min, blocked with 1% bovine serum albumin (cat no. 10270106; Gibco; Thermo Fisher Scientific, Inc.) for 20 min at room temperature, and incubated with rabbit anti-per2 (dilution, 1:250; cat no. ab180655; Abcam, Cambridge, UK) overnight at 4°C. Subsequent to incubation with an fluorescein isocyanide-conjugated AffiniPure goat anti-rabbit secondary antibody (dilution, 1:1,000; cat no. 1095047; Cell Signaling Technology, Danvers, MA, USA) for 1 h at room temperature, cells were stained with DAPI (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 15 min at room temperature. Photomicrographs were then captured from laser confocal microscopy (LCSM, Zeiss KS 400; Zeiss AG, Oberkochen, Germany).
Cell proliferation assay
Cell proliferation was detected using cell counting kit-8 (CCK8, cat no. C0038; Beyotime Institute of Biotechnology, Haimen, China). Following transfection as described, cells were seeded at a density of 5,000 cells per well in 96-well plates. Following starvation (growth without serum) for 24 h, an aliquot of 10 µl CCK-8 solution was added to the wells and incubated for 2 h at 37°C. Absorbance was then measured at 450 nm to determine the optical density (OD) value.
Cell migration assay
The migration assay was performed using an 8-µm pore size Transwell invasion chamber (24-well; Corning Incorporated, Corning, NY, USA) according to the manufacturer's protocol. Briefly, 1×105 cells were transferred to the top chamber in 200 µl serum-free DMEM, and 500 µl DMEM supplemented with 10% FBS was added to the lower chambers. After a 24-h incubation, cells that remained in the top chamber were removed with a cotton swab, and the migrated cells in the bottom chamber were fixed with 4% formaldehyde for 10 min at room temperature and stained with hematoxylin for 10 min at room temperature. Cells were photographed using an inverted microscope and cell numbers were quantified by ImageJ software (1.51; National Institutes of Health, Bethesda, MD, USA).
TUNEL staining
Preconditioned cells were collected for terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) apoptosis assay using a In Situ Cell Death Detection kit (Merck KGaA) according to the manufacturer's protocol. The rate of apoptosis was calculated as the ratio of TUNEL-positive cells to total cells as determined under a light microscope (Leica DM 3000; Leica Microsystems GmbH, Wetzlar, Germany) in five randomly determined fields.
Western blot analysis
Cells were solubilized in cold radioimmunoprecipitation lysis buffer (Beyotime Institute of Biotechnology) to extract protein. The protein concentration was detected using a BCA kit (Beyotime Institute of Biotechnology). Cell lysates containing 30 µg of protein were separated with 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (GE Healthcare, Chicago, IL, USA). Subsequent to blocking with 5% skim milk at room temperature for 1 h, membranes were incubated overnight at 4°C with the following primary antibodies: Rabbit anti-per2 (dilution, 1:1,000; cat no. ab180655; Abcam), rabbit anti-Akt (dilution, 1:3,000; cat no. 4691s), rabbit anti-phosphorylated (p)-Akt (dilution, 1:3,000; cat no. 4060s), rabbit anti-Bcl-2 (dilution, 1:500; cat no. 3498s), rabbit anti-p21 (dilution 1:5,000; cat no. 2947s), rabbit anti-p27 (dilution, 1:500; cat no. 3686s; all Cell Signaling Technology), rabbit anti-caspase-3 (dilution, 1:250; cat no. ab44976; Abcam), rabbit anti-cleaved caspase-3 (dilution, 1:500; cat no. 9664s; Cell Signaling Technology), rabbit anti-CDK4 (dilution, 1:500; cat no. ab68266; Abcam) and mouse anti-β-actin (dilution, 1:1,000; cat no. 4970s; Cell Signaling Technology). The membranes were washed 3 times in Tris-buffered saline with Tween-20 (TBST) for 10 min, then incubated with anti-rabbit or anti-mouse horseradish peroxidase-conjugated IgG (dilution, 1:5,000; cat nos. TA130023 and TA130003, respectively; OriGene Technologies, Inc., Beijing, China) at room temperature for 1 h. The membranes were washed 3 times in TBST for 15 min. Finally, membranes were treated with an enhanced chemiluminescence reagent (Merck KGaA). Imaging apparatus (AI 600 RGB; GE Healthcare) was used to detect chemiluminescence according to the manufacturer's protocol.
Statistical analysis
Data were expressed as the mean ± standard deviation. SPSS 17.0 (SPSS Inc., Chicago, IL, USA) was used to conduct statistical analysis. A t test was used to compare differences between two groups, and one-way analysis of variance followed by a least significant difference post-hoc test was used for the comparison of multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Per2 was successfully overexpressed and downregulated
Following transfection for 24 h, per2 expression was detected by the immunofluorescence staining of the five groups of cells (i.e., Blank control, per2 overexpression, plasmid-only, per2 siRNA and control siRNA groups). The per2 overexpression group exhibited an increase in per2 fluorescence intensity, while there was reduced intensity in the per2 siRNA group. There were no differences between the blank control, plasmid-only and siRNA control groups (Fig. 1A). Western blot analysis was also performed to investigate per2 protein expression. The results of western blotting corresponded with the results of immunofluorescence.
Figure 1.
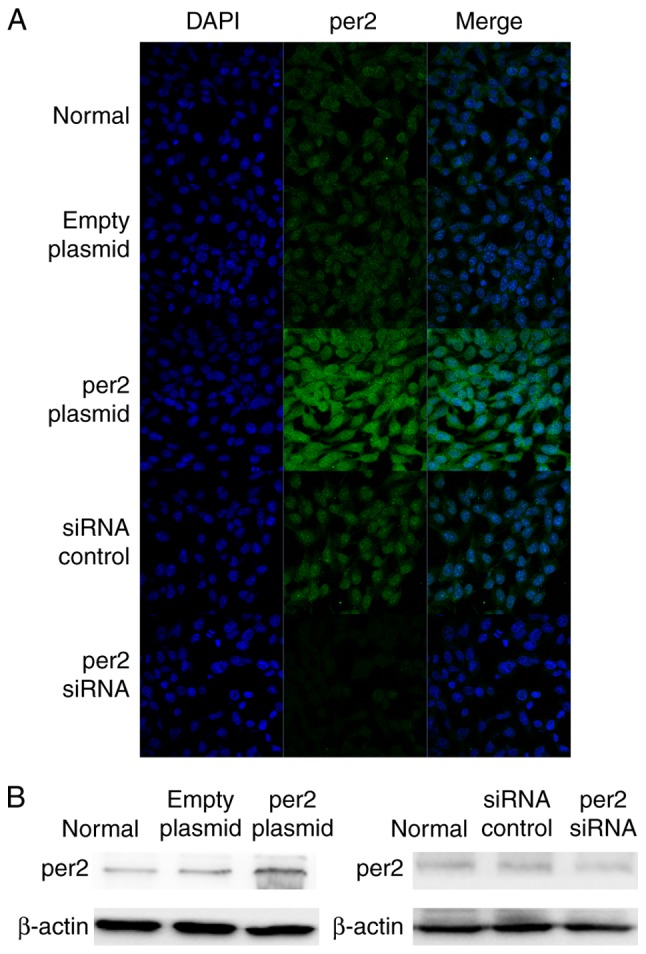
Overexpression and knockdown of per2 in MNNG/HOS cells in vitro. (A) Immunofluorescence indicated that the highest fluorescence intensity expression in per2 overexpression cells and the lowest fluorescence intensity expression in per2 knockdown cells (magnification, ×40). (B) Western blotting demonstrated similar results. Per2, Period 2; siRNA, small interfering RNA.
Per2 overexpression decreased osteosarcoma cell proliferation and prevented apoptosis, in contrast with per2 knockdown
The CCK8 assay was used to analyze MNNG/HOS cell proliferation. Per2 overexpression induced a significant decrease in proliferation (Fig. 2A). Conversely, per2 knockdown significantly upregulated proliferation (Fig. 2A). There were no statistically significant differences between the blank control, plasmid-only and siRNA control groups. A TUNEL assay was also used to analyze the rate of MNNG/HOS cell apoptosis. The results indicated that the per2 overexpression group demonstrated an increase in apoptosis whereas per2 siRNA prevented apoptosis. Plasmid-only and control siRNA groups did exhibited a significantly increased rate of apoptosis compared with blank control (P<0.05). The reason may be that damage was caused by Lipofectamine 2000. Per2 Plasmid was significantly increased compared with the normal control (P<0.01) and the empty plasmid (P<0.01). Per2 siRNA was significantly decreased compared with siRNA control (P<0.05) (Fig. 2B and C).
Figure 2.
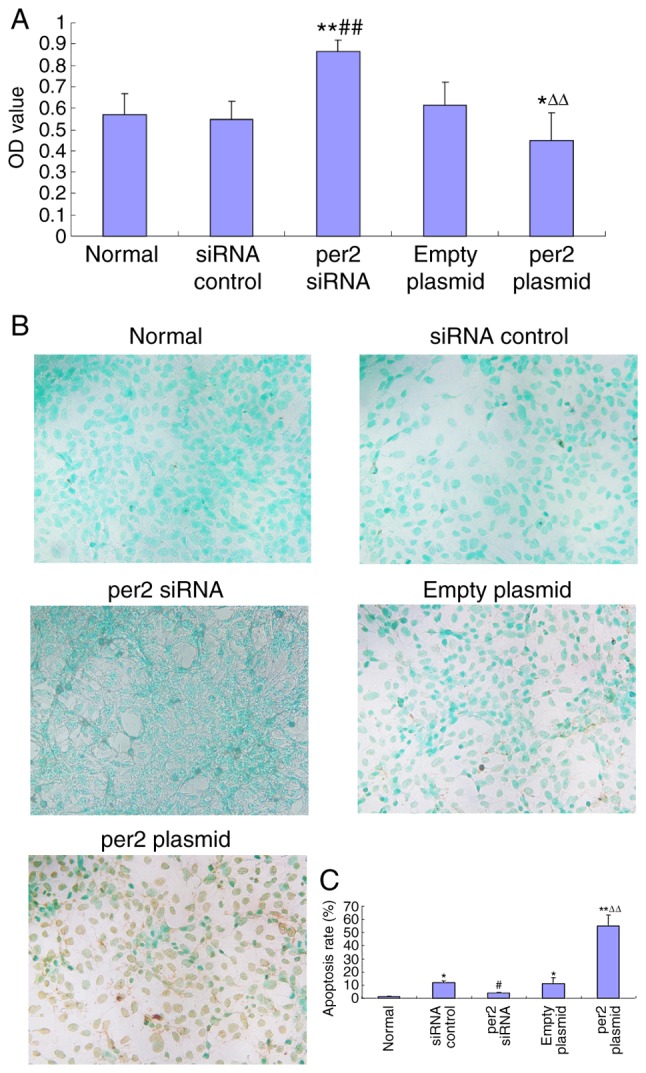
Alteration of MNNG/HOS cell proliferation and apoptosis by per2 overexpression or knockdown. (A) Overexpression of per2 reduced the rate of proliferation; per2 inhibition increased proliferation. (B) Representative figures of the apoptosis assay. The overexpression of per2 increased the rate of apoptosis; per2 interference protected the cells from apoptosis (magnification, ×20). (C) Quantification and statistical analysis of the rate of apoptosis. **P<0.01 vs. untreated group; #P<0.05, ##P<0.01 vs. control siRNA group; ∆∆P<0.01 vs. plasmid-only group. Per2, Period 2; OD, optical density; siRNA, small interfering RNA.
Per2 overexpression decreases MNNG/HOS cell migration, in contrast with per2 knockdown
The migration ability of MNNG/HOS cells was evaluated with a Transwell assay. It was demonstrated that per2 overexpression decreased MNNG/HOS cell migration, whereas per2 siRNA increased MNNG/HOS cell migration. There were no statistically significant differences between the blank control, plasmid-only and siRNA control groups (Fig. 3A and B).
Figure 3.
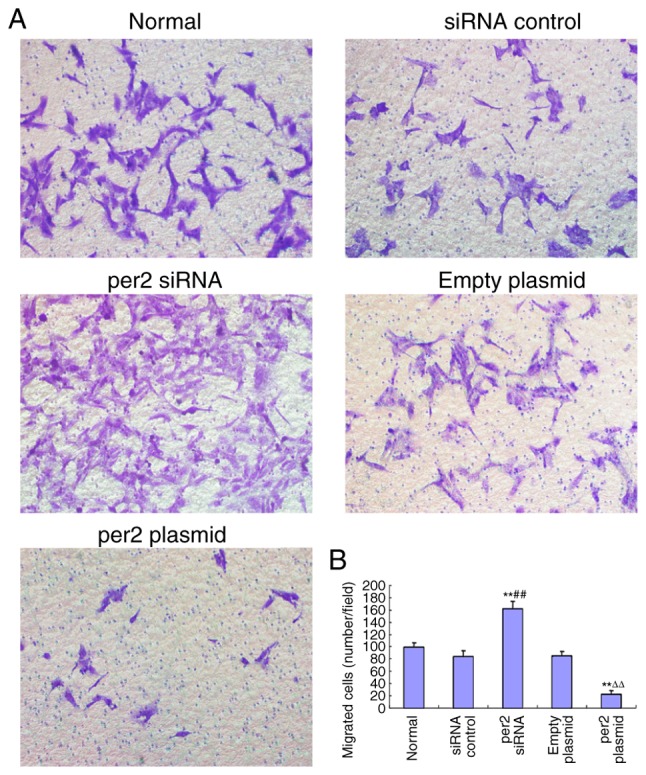
Alteration of MNNG/HOS cell migration following the overexpression or knockdown of per2. (A) Representative images of MNNG/HOS cell migration (magnification, ×20). The overexpression of per2 inhibited migration, whereas per2 interference increased migration. (B) Quantification and statistical analysis of MNNG/HOS cell migration in the five examined groups **P<0.01 vs. normal group; ##P<0.01 vs. siRNA control group; ∆∆P<0.01 vs. empty plasmid group. Per2, Period 2; siRNA, small interfering RNA.
Affects of the alteration of per2 on the expression of proliferation and apoptosis-associated proteins
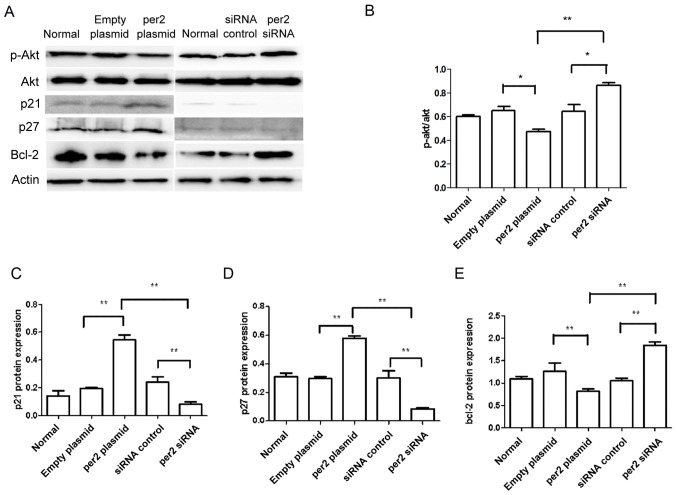
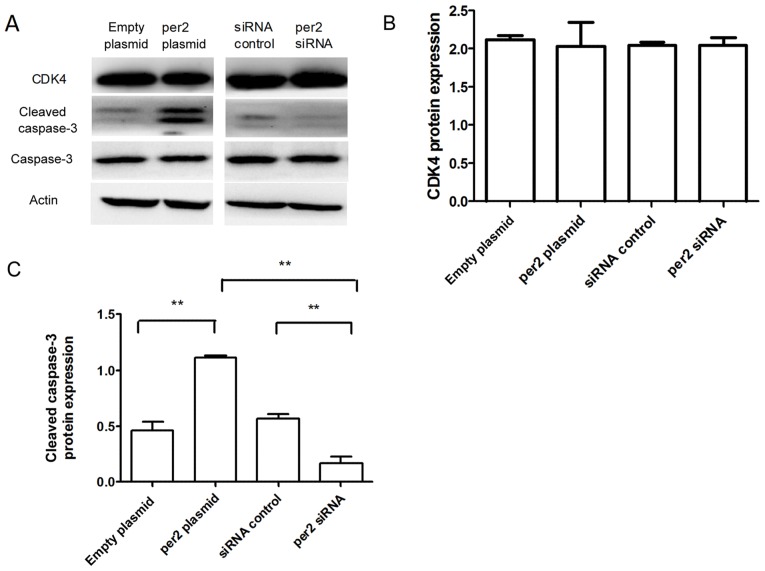
The activation of Akt in tumor cells enhances cell migration and growth (24). Western blot analysis was used to detect proteins from the Akt signaling pathway, and other proteins associated with proliferation and apoptosis. As demonstrated in Fig. 4, decreased protein levels of p-Ser473 Akt were observed in MNNG/HOS cells with per2 overexpression, whereas per2 siRNA increased the p-Ser473 Akt level (Fig. 4A and B). Per2 overexpression also elevated the levels of p27 and p21 protein expression. Conversely, per2 knockdown decreased the p27 and p21 levels (Fig. 4A, C and D). In addition, the overexpression of per2 reduced the protein expression of Bcl-2, whereas the suppression of per2 increased the expression of Bcl-2 (Fig. 4A and E). Per2 overexpression increased cleaved caspase-3 protein expression, whereas per2 siRNA decreased cleaved caspase-3 protein expression (Fig. 5A and C). There was no alteration of CDK4 expression in per2 overexpression and siRNA cells (Fig. 5A and B). There were no evident differences in expression among the control groups.
Figure 4.
Western blot analysis of the effect of per2 expression on Akt, p21, p27 and Bcl-2. (A) Per2 overexpression inhibited the phosphorylation of Akt, while per2 siRNA increased the phosphorylation of Akt. (B) Quantification of p-Akt/Akt ratio. *P<0.05 and **P<0.01. Overexpression of per2 elevated the protein levels of (C) p21 and (D) p27 compared with the control groups; suppression of per2 decreased the protein levels of p27 and p21. *P<0.05 and **P<0.01. (E) Overexpression of per2 decreased the protein levels of Bcl-2 compared with the control groups; knockdown of per2 significantly increased the protein levels of Bcl-2 compared with the control groups. *P<0.05 and **P<0.01. Per2, Period 2; siRNA, small interfering RNA; Akt, protein kinase B; Bcl-2, B-cell lymphoma 2 -associated X protein.
Figure 5.
Western blot analysis of the effect of per2 expression on CDK4 and cleaved caspase-3. (A) Per2 overexpression increased the protein expression of cleaved caspase-3. Per2 siRNA decreased cleaved caspase-3 protein expression. (B) Quantification of CDK4 protein expression. (C) Quantification of cleaved caspase 3 protein expression. **P<0.01. Per2, Period 2; CDK4, cyclin dependent kinase 4; siRNA, small interfering RNA.
Discussion
Osteosarcoma continues to present a threat to human health and survival. Proliferation and metastasis are critical events in the pathogenesis of cancer (25). Oncogenes and tumor suppressor genes are associated with cancer cell proliferation and differentiation; therefore, it is necessary to identify genes which promote osteosarcoma cell growth, survival and metastasis.
Previous studies have demonstrated that the aberrant expression or rhythm of circadian clock genes is associated with carcinogenesis and cancer progression (26–28). The core clock genes include per1, 2 and 3, CLOCK, cryptochrome1 (CRY1), cryptochrome2 (CRY2), Bmal1, casein kinase1 epsilon (CSNK1ε), timeless (TIM) and timeless-interacting protein (Tipin) (23). Among the clock genes, per2 has been demonstrated to serve an important role in cancer progression, and its altered expression has been identified in a number of types of cancer (20–23). The overexpression of per2 has been demonstrated to suppress tumor growth in vivo (29) and to induce apoptosis in cancer cells (30). Additionally, mice deficient in per2 exhibited a higher incidence of cancer development (31). It was previously reported that per2 expression level is associated with patient age, tumor histological grade, invasion depth, lymph node metastasis and tumor-node-metastasis stage (28). It was also demonstrated that a reduced expression of per2 accelerated tumor growth in vitro (32). Furthermore, Koyanagi et al (33) reported that per2 inhibited tumor angiogenesis through inhibiting vascular endothelial growth factor agonist activity.
In the present study, a per2 plasmid and anti-per2 siRNA were constructed, and transfected into MNNG/HOS cells. Per2 expression was detected by western blotting and immunofluorescence. MNNG/HOS cell proliferation, migration and apoptosis were detected by CCK-8, Transwell and TUNEL assays. Per2 overexpression inhibited MNNG/HOS cell proliferation and migration, while apoptosis was upregulated. In cells with per2 siRNA, the effect was the opposite.
The results of the present study suggest that the overexpression of per2 may alter MNNG/HOS cell function through the Akt cascade, as per2 overexpression inhibited Akt phosphorylation. The Akt signaling pathway is a downstream signal transduction pathway involving numerous growth factors and cytokines. The Akt signaling pathway inhibits cell apoptosis, promotes cell proliferation and is associated with tumor occurrence, while also affecting the migration and invasion of tumor cells and promoting tumor progression. The associated PI3K/PKB signaling pathway also affects glucose metabolism and promotes the growth of tumor cells (34). It was previously demonstrated that cells with downregulated per2 expression have sustained high levels of Akt phosphorylation after growth factor stimulation or DNA damage (35). A study by Chen et al (17) also demonstrated that per2 knockdown increased the activity of the PI3K/AKT/mTOR signaling pathway, and that the overexpression of per2 reduced growth and promoted apoptosis in A549 cells.
The Bcl-2 family of proteins is a key regulator of the mitochondrial response to apoptotic signals. The Bcl-2 gene family comprises different members that regulate apoptosis either positively or negatively. In the present study, the expression of Bcl-2 was investigated. As shown in Fig. 4, there was a decrease in the Bcl-2 protein levels in per2-overexpressing cells compared with control cells.
It was also identified that per2 over-expression inhibited p21/p27 protein expression. As key regulators in cell proliferation, p21 and p27 serve important roles in DNA damage repair, cell differentiation and senescence (36). Per2 may inhibit MNNG/HOS cell proliferation through inhibiting p21/p27 expression.
Cyclin-dependent kinases (CDKs) are oncogenes in a range of cancer types (37). Components of the CDK pathway are deregulated in the majority of human tumors (38). In the present study, there was no alteration of CDK4 expression. The expression of the apoptosis-associated protein cleaved caspase-3 was also detected. Per2 overexpression may have increased cleaved caspase-3 expression to induce MNNG/HOS cell apoptosis.
In the present study, the rates of MNNG/HOS cell proliferation and apoptosis at different per2 expression levels were observed, confirming that the per2 level serves an important role in osteosarcoma cell function. In a previous study (34), another group reported that the overexpression of per2 increased apoptosis and decreased proliferation in MG-63 cells. However, they did not further explore the influence of a lower expression level of per2 on MG-63 function, and did not explore the p53-dependent apoptosis signaling pathway.
In summary, the present study demonstrated that the overexpression of per2 resulted in reduced proliferation and increased apoptosis in MNNG/HOS osteosarcoma cells. Overexpressed per2 altered the expression of apoptosis-associated proteins that were involved directly or indirectly in apoptosis; however, the exact mechanisms for the effects of per2 require further elucidation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81600344, 81500339, 81302939 and 81402218), Shandong Provincial Natural Science Foundation, China (grant nos. ZR2016HQ29 and ZR2016HQ01), and Shandong Province Outstanding Young Scientists Research Award Fund (grant no. BS2015YY026).
Availability of data and materials
The data sets generated and analyzed during the study are available from the corresponding author, on reasonable request.
Authors' contributions
All authors read and approved the manuscript. YYS made substantial contributions to the conception and design. TQ made substantial contributions to the acquisition of data. XTL, YGL, YL, WY and NL made substantial contributions to the analysis and interpretation of data. WY was involved in drafting the manuscript and revising it critically for important intellectual content. YYS gave final approval of the version to be published.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Whelan J, McTiernan A, Cooper N, Wong YK, Francis M, Vernon S, Strauss SJ. Incidence and survival of malignant bone sarcomas in England 1979–2007. Int J Cancer. 2012;131:E508–E517. doi: 10.1002/ijc.26426. [DOI] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Admassi D. Osteosarcoma of medial cuniform bone. Ethiop Med J. 2009;47:305–308. [PubMed] [Google Scholar]
- 4.Heaton TE, Hammond WJ, Farber BA, Pallos V, Meyers PA, Chou AJ, Price AP, LaQuaglia MP. A 20-year retrospective analysis of CT-based pre-operative identification of pulmonary metastases in patients with osteosarcoma: A single-center review. J Pediatr Surg. 2017;52:115–119. doi: 10.1016/j.jpedsurg.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gelderblom H, Jinks RC, Sydes M, Bramwell VH, van Glabbeke M, Grimer RJ, Hogendoorn PC, McTiernan A, Lewis IJ, Nooij MA, et al. Survival after recurrent osteosarcoma: Data from 3 European Osteosarcoma Intergroup (EOI) randomized controlled trials. Eur J Cancer. 2011;47:895–902. doi: 10.1016/j.ejca.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 6.Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254. doi: 10.1155/2012/627254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Zhang W. Newmolecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25:398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 8.Gubin DG, Weinert D, Rybina SV, Danilova LA, Solovieva SV, Durov AM, Prokopiev NY, Ushakov PA. Activity, sleep and ambient light have a different impact on circadian blood pressure, heart rate and body temperature rhythms. Chronobiol Int. 2017;34:632–649. doi: 10.1080/07420528.2017.1288632. [DOI] [PubMed] [Google Scholar]
- 9.Bueno C, Menna-Barreto L. Development of sleep/wake, activity and temperature rhythms in newborns maintained in a neonatal intensive care unit and the impact of feeding schedules. Infant Behav Dev. 2016;44:21–28. doi: 10.1016/j.infbeh.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Kelleher FC, Rao A, Maguire A. Circadian molecular clocks and cancer. Cancer Lett. 2014;342:9–18. doi: 10.1016/j.canlet.2013.09.040. [DOI] [PubMed] [Google Scholar]
- 11.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene MW. Circadian rhythms and tumor growth. Cancer Lett. 2012;318:115–123. doi: 10.1016/j.canlet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Kelly-Hayes M, Wolf PA, Kase CS, Brand FN, McGuirk JM, D'Agostino RB. Temporal patterns of stroke onset. The Framingham Study. Stroke. 1995;26:1343–1347. doi: 10.1161/01.STR.26.8.1343. [DOI] [PubMed] [Google Scholar]
- 14.Muller JE, Stone PH, Turi ZG, Rutherford JD, Czeisler CA, Parker C, Poole WK, Passamani E, Roberts R, Robertson T, et al. Circadian variation in the frequency of onset of acute myocardial infarction. N Engl J Med. 1985;313:1315–1322. doi: 10.1056/NEJM198511213132103. [DOI] [PubMed] [Google Scholar]
- 15.Hansen AB, Stayner L, Hansen J, Andersen ZJ. Night shift work and incidence of diabetes in the Danish Nurse Cohort. Occup Environ Med. 2016;73:262–268. doi: 10.1136/oemed-2015-103342. [DOI] [PubMed] [Google Scholar]
- 16.Jensen LD, Cao Z, Nakamura M, Yang Y, Bräutigam L, Andersson P, Zhang Y, Wahlberg E, Länne T, Hosaka K, Cao Y. Opposing effects of circadian clock genes bmal1 and period2 in regulation of VEGF-dependent angiogenesis in developing zebrafish. Cell Rep. 2012;2:231–241. doi: 10.1016/j.celrep.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Chen B, Tan Y, Liang Y, Li Y, Chen L, Wu S, Xu W, Wang Y, Zhao W, Wu J. Per2 participates in AKT-mediated drug resistance in A549/DDP lung adenocarcinoma cells. Oncol Lett. 2017;13:423–428. doi: 10.3892/ol.2016.5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 19.Ripperger JA, Albrecht U. The circadian clock component PERIOD2: From molecular to cerebral functions. Prog Brain Res. 2012;199:233–245. doi: 10.1016/B978-0-444-59427-3.00014-9. [DOI] [PubMed] [Google Scholar]
- 20.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–1246. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 21.Lin YM, Chang JH, Yeh KT, Yang MY, Liu TC, Lin SF, Su WW, Chang JG. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog. 2008;47:925–933. doi: 10.1002/mc.20446. [DOI] [PubMed] [Google Scholar]
- 22.Štorcelová M, Vicián M, Reis R, Zeman M, Herichová I. Expression of cell cycle regulatory factors hus1, gadd45a, rb1, cdkn2a and mre11a correlates with expression of clock gene per2 in human colorectal carcinoma tissue. Mol Biol Rep. 2013;40:6351–6361. doi: 10.1007/s11033-013-2749-2. [DOI] [PubMed] [Google Scholar]
- 23.Relles D, Sendecki J, Chipitsyna G, Hyslop T, Yeo CJ, Arafat HA. Circadian gene expression and clinicopathologic correlates in pancreatic cancer. J Gastrointest Surg. 2013;17:443–450. doi: 10.1007/s11605-012-2112-2. [DOI] [PubMed] [Google Scholar]
- 24.Neiva KG, Zhang Z, Miyazawa M, Warner KA, Karl E, Nör JE. Cross talk initiated by endothelial cells enhances migration and inhibits anoikis of squamous cell carcinoma cells through STAT3/Akt/ERK signaling. Neoplasia. 2009;11:583–593. doi: 10.1593/neo.09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sgourakis G, Gockel I, Lyros O, Hansen T, Mildenberger P, Lang H. Detection of lymph node metastases in esophageal cancer. Expert Rev Anticancer Ther. 2011;11:601–612. doi: 10.1586/era.10.150. [DOI] [PubMed] [Google Scholar]
- 26.Mazzoccoli G, Panza A, Valvano MR, Palumbo O, Carella M, Pazienza V, Biscaglia G, Tavano F, Di Sebastiano P, Andriulli A, Piepoli A. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int. 2011;28:841–851. doi: 10.3109/07420528.2011.615182. [DOI] [PubMed] [Google Scholar]
- 27.Gu X, Xing L, Shi G, Liu Z, Wang X, Qu Z, Wu X, Dong Z, Gao X, Liu G, et al. The circadian mutation PER2(S662G) is linked to cell cycle progression and tumorigenesis. Cell Death Differ. 2012;19:397–405. doi: 10.1038/cdd.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karantanos T, Theodoropoulos G, Pektasides D, Gazouli M. Clock genes: Their role in colorectal cancer. World J Gastroenterol. 2014;20:1986–1992. doi: 10.3748/wjg.v20.i8.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazaki K, Wakabayashi M, Hara Y, Ishida N. Tumor growth suppression in vivo by overexpression of the circadian component, PER2. Genes Cells. 2010;15:351–358. doi: 10.1111/j.1365-2443.2010.01384.x. [DOI] [PubMed] [Google Scholar]
- 30.Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang C, Wang X, Wang Z, Cornelissen-Guillaume G, Halberg F. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006;97:589–596. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu L, Pelicano H, Liu JS, Huang P, Lee CC. The circadian gene period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/S0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Wood PA, Oh EY, Du-Quiton J, Ansell CM, Hrushesky WJ. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat. 2009;117:423–431. doi: 10.1007/s10549-008-0133-z. [DOI] [PubMed] [Google Scholar]
- 33.Koyanagi S, Kuramoto Y, Nakagawa H, Aramaki H, Ohdo S, Soeda S, Shimeno H. A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res. 2003;63:7277–7283. [PubMed] [Google Scholar]
- 34.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompso CB. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, He X, Yang Z, Jabbari E. Mammalian PER2 regulates AKT activation and DNA damage response. Biochem Cell Biol. 2012;90:675–682. doi: 10.1139/o2012-025. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Yépez EA, Saldívar-Cerón HI, Villamar-Cruz O, Pérez-Plasencia C, Arias-Romero LE. p21 activated kinase 1: Nuclear activity and its role during DNA damage repair. DNA Repair (Amst) 2018;65:42–46. doi: 10.1016/j.dnarep.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Williams GH, Stoeber K. The cell cycle and cancer. J Pathol. 2012;226:352–364. doi: 10.1002/path.3022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and analyzed during the study are available from the corresponding author, on reasonable request.