Abstract
Certain variants of human papillomavirus (HPV)type 58 are associated with an increased risk of high grade squamous intraepithelial lesions and cervical cancer. However, little is known about the persistence of HPV58 E6/E7 variants in women with incident HPV58 infections. The aim of the present study was to evaluate the presence and persistence of HPV58 E6/E7 variants in 71 women with incident HPV58 infection throughout their follow-up. These women belonged to a cohort examined in a longitudinal study of 1,610 Colombian women, who were HPV-negative and had normal baseline cytology. E6/E7 DNA regions of HPV58-positive samples were amplified and sequenced using automated direct sequencing. A total of 639 samples were analyzed from the 71 women, and 117 samples (18.3%) were HPV58-positive. HPV58 E6/E7 variants were detected in 85.5% of the samples. The T307/A694/G744/A761 variant was identified in 88% of the samples, the T307/G744 variant was identified in 9% of samples and the T187/T307/A367/G744/G793/T798/A801/T840/C852 was identified in 3% of the samples. Overall, 50% of the HPV58 infections were present after 1 year of follow-up and all infections were cleared after 7 years. Women who had first sexual intercourse at >15 years of age had a lower clearance rate than those who had sexual intercourse for the first time at ≤15 years of age [hazard ratio (HR)=0.29; 95% confidence interval (CI)=0.09–0.92]. Likewise, parous women had a higher clearance rate than nulliparous women (HR=3.43, 95% CI=1.23–9.60). There was no difference in clearance rates between HPV58 E6/E7 variants. In conclusion, HPV58 variants were not associated with persistence of the infection in this group of women.
Keywords: human papilloma virus, follow-up studies, genomic variation, persistent infection
Introduction
Clinical and epidemiological studies have established that infections by certain human papillomavirus (HPV) types are associated with the development of cervical cancer and its precursor lesions (1–4). The marked geographic differences in the incidence of cervical cancer are not only the result of differences in screening patterns to detect and treat precursor lesions, but also due to differences in exposure to various risk factors. Different HPV genotypes, including variant presence, genetic alterations, host factors and environmental factors are involved in the pathways underlying cervical carcinogenesis (5–8). A persistent infection with carcinogenic types of HPV is a pivotal event in the incidence of cervical cancer. However, few long-term studies following persistent HPV infections and quantifying risk have been performed (9).
In Colombia, the incidence of cervical cancer is relatively high at 26.1/100,000, and it is the principal cause of cancer-associated mortality among women (10). In this country, a prospective cohort study was initiated in the early 1990s by the Colombian National Institute of Cancer (Bogotá, Colombia) and the International Agency for Research on Cancer (IARC; Lyon, France). The aim of the study was to investigate the natural history of HPV infections and cervical intraepithelial neoplasia (CIN) lesions among low-income Colombian women (the Bogotá cohort); however, viral factors including variant presence were not studied (11–13).
HPV type 58 (HPV58) is the second most prevalent viral type (6.2%) and has the second highest rate of incident infection (11.2%) in women with normal cytology from the Colombian population (11,12). The relative contribution of HPV58 to invasive cervical cancer and CIN 3 lesions worldwide is 2.3 and 9.0%, respectively (14), and certain variants of HPV58 have been reported to have an increased risk of development of high grade squamous intraepithelial lesions (HGSIL) and cervical cancer in certain populations (15–17). In South America there are few studies investigating HPV58 molecular variants, and worldwide there are no follow-up studies analyzing their involvement in the persistence and clearance of HPV58 incident infections in women with normal cytology. The present study focuses on this analysis, and also provides information concerning the function of other risk factors in the clearance of HPV58 infections and whether the HPV58 infection-associated risk of developing CIN 2,3 differs by variant. The aim of the present study was to evaluate the presence and persistence of incident HPV58 E6/E7 variants during a long follow-up period in Colombian women with normal baseline cytology, and then analyze variant status and other risk factors associated with clearance of HPV58 infections.
Materials and methods
Study population
Between November 1993 and November 1995 a total of 2,200 sexually active women, 13–85 years old, who presented to cervical cancer screening centres and family-planning clinics in low socioeconomic settings in Bogotá, Colombia, were invited to participate in a prospective study of cervical HPV infection (11). Methods of recruitment and data collection have been described previously (11,12). The ethical committees of the National Cancer Institute and the IARC approved the study protocol. A total of 1,610 women who were HPV-negative and had normal cytology at base line were included in the analysis of HPV incidence.
The analysis described in the present study was performed in a subset of the study cohort of 71 women with incident HPV58 infections, together with their respective longitudinal follow-up every 6–9 months. A total of 639 samples were available for analysis over 7 years.
Biological specimens
The cell pellets used in the present study were obtained from women who were HPV58 positive, and were suspended in 1 ml Tris-HCl buffer (10 mM; pH 8.3) and stored at−70°C. For analysis, 100 µl aliquots were boiled for 10 min at 100°C, cooled on ice and centrifuged for 1 min at 3,000 × g at 4°C. A total of 10 µl of these pre-treated crude cell suspensions were used for polymerase chain reaction (PCR) analysis. To assess the quality of the target DNA, a 209 base pair amplifying β-globin PCR was performed using the primer combination BGPCO3 and BGPCO5, as described previously (18).
E6 and E7 HPV58 PCR amplification
The E6 and E7 regions of HPV58 positive samples were amplified using E6 forward, (5′-CAGACATTTTTTGGTAGGCTA-3′); E7 reverse 1, (5′-TCTTAGCGTTGGGTTGTT-3′); E7 forward 1, (5′-CTGTAACAACGCCATGAGAG-3′); and E7 reverse 2, (5′-TCAGGGTCATCCATTGCAGA-3′) primers, which amplify fragments of 521 and 335 bp, respectively. The PCR amplifications were performed in a final volume of 50 µl containing 10 mM Tris HCl, 2.5 mM MgCl2, 0.2 mM of each dNTP, 25 pmol of each primer (Integrated DNA Technologies, Inc., Coralville, IA, USA) 1.25 U Taq polymerase (Amplitaq; Cetus; PerkinElmer, Inc., Waltham, MA, USA) and 10 µl isolated nucleic acid. The optimal amplification conditions for the PCR were as follows: Denaturation for 4 min at 94°C followed by 40 cycles of amplification. Each cycle consisted of 1 min at 94°C, 1 min at 50°C and 1.5 min at 72°C. The final elongation step was extended for a further 10 min. The amplified DNA was analysed by 1.5% agarose gel electrophoresis and ethidium bromide staining (0.5 µg/ml) and viewed with the Gel Doc XR Gel Documentation System (Image Lab™ software; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Negative controls, with PCR mix and water, and positive controls with 100 fg and 1 pg cloned HPV58 DNA in a background of 100 ng human placental DNA were included in each experiment. These controls were obtained from the World Health Organisation HPV DNA proficiency panel [HPV LabNet Global Reference Laboratory (GRL), Sweden].
E6 and E7 HPV58 sequence analysis
The E6 and E7 HPV58 amplicons were purified using the commercial GFX PCR kit (GE Healthcare, Chicago, IL, USA) according the manufacturer's protocol. Cycle sequencing was performed using the 7-Deaza-dGTP-Cy5.5 Dye Primer Cycle Sequencing kit (Visible Genetics, Inc., Toronto, Canada) according to the manufacturer's protocol, and was run on an automated DNA sequencer (SEQ 4×4; GE Healthcare). For the sequencing reaction, the same primers were used as for the PCR. Sequences of sense, antisense and combined strands of the PCR products were generated. The sequences were analyzed using the Basic Local Alignment Search Tool function (BLAST + 2.2.28; National Center for Biotechnology Information; U.S. National Library of Medicine, Bethesda, MD, USA), and variants were interpreted visually in comparison with the HPV58R prototype sequence (GenBank no. D90400.1) (19).
Statistical methods
Frequencies and percentages were used to analyse HPV58 E6/E7 variants presence and viral load. An incident HPV58 infection was defined as the first positive type-specific result following a negative result for the same type of HPV. Correspondingly, type-specific HPV58 clearance was defined as the first negative PCR result following an incident HPV58 infection. Duration of an HPV58 incident infection was defined as the elapsed time from the date of infection to the date of clearance. The survival function, which describes the probability that an HPV infection has cleared as a function of time, was estimated using Kaplan-Meier analysis of interval-censored data, followed by the log rank test. Cox regression analysis was performed to estimate the age-adjusted hazard ratios (HRs) and 95% confidence intervals (CIs) on clearance of HPV58 infection according to different risk factors. All analyses were performed using SPSS software (version 17; SPSS Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference..
Results
Frequency of HPV58 E6/E7 variants in women with incident HPV58 infections
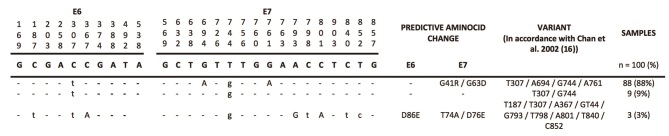
A total of 71 women had incident HPV58 infections during the follow-up period. The median age of the women was 33.6 years (range, 16–67.72). The follow-up period lasted between 1 and 7 years, with a total of 2–12 visits (average, 8.72 visits). Of a total of 639 cervical scrapes obtained during follow-up, 117 samples were HPV58-positive. However, variants were identified in 100 samples (85.5%). Three different variants were identified: 88% of the samples were positive for the T307/A694/G744/A761 variant, 9% were positive for the T307/G744 variant and 3% were positive for the T187/T307/A367/G744/G793/T798/A801/T840/C852 variant. None of the samples were infected with multiple variants or the reference sequence of HPV58. The E6 and E7/HPV58 sequence variants detected during follow-up in women with incident HPV58 infections are presented in Fig. 1.
Figure 1.
E6 and E7/HPV58 sequence variants detected during follow-up in women with incident HPV58 infections from the Colombian cohort. Upper-case letters in bold indicate HPV58 R (reference sequence; Gen Bank no. D90400). Dashes indicate no nucleotide change, upper-case letters indicate mutations that produce an amino acid change and lower-case letters indicate silent mutations. HPV, human papillomavirus.
Clearance of incident HPV58 infections
From the 71 women who had incident HPV58 infections, 50 (70.4%) were transient, whilst 21 (29.6%) were persistent at more than one visit. The persistence of HPV58 infections as a function of time is presented in Fig. 2. Overall, 50% of HPV58 infections were still present at 1 year of follow-up, 15% at 2 years, 3% at 4 years and 0% at 7 years of follow-up. There was no difference in HPV58 persistence when the HPV58 infection was identified in single or in multiple infections (co-infection; Fig. 3). The mean duration of HPV58 infection as a single infection was 16.03 months and in multiple infections was 15.53 months (P=0.84; log rank test).
Figure 2.
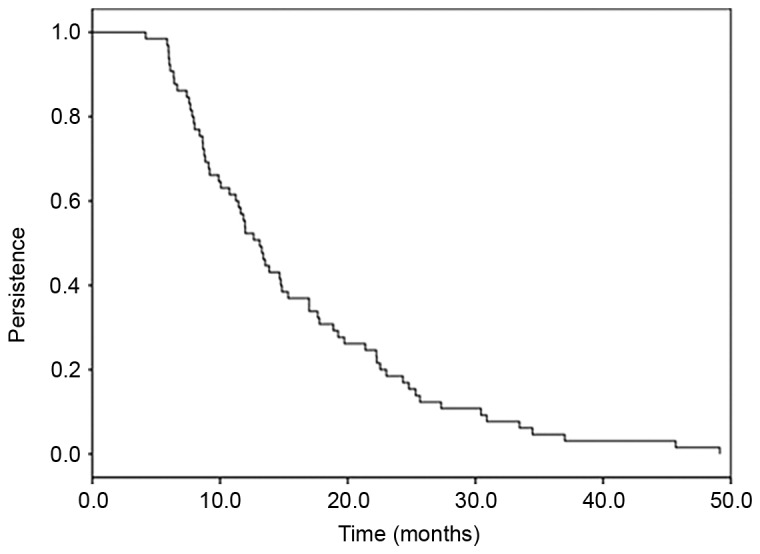
Persistence of HPV58 incident infections. The mean duration of HPV58 incident infections was 16.85 months (95% CI=14.02–19.68). HPV, human papillomavirus; CI, confidence intervals.
Figure 3.
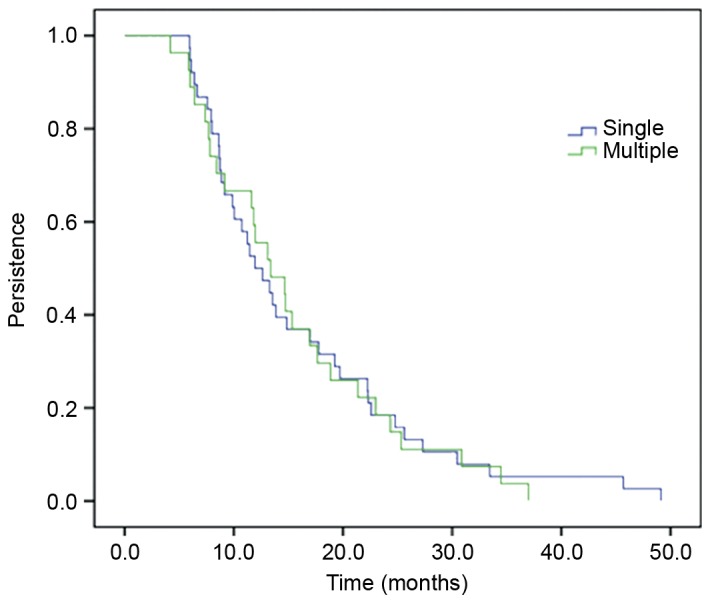
Persistence of single and multiple HPV58 infections. The mean durations of HPV58 incident infections (single and multiple) were 16.03 months (95% CI=12.70–19.37) and 15.53 months (95% CI=12.17–18.89), respectively. Data were analyzed using the log rank test and there was no significant difference (P=0.84). HPV, human papillomavirus; CI, confidence intervals.
Clearance of HPV58 E6/E7 variants
The probability of persistence of HPV58 E6/E7 variants in women with incident HPV58 infections is presented in Fig. 4. The same variant was identified in persistent cases at all visits for all women. The T307/A694/G744/A761 variant had a mean duration of infection of 16.65 months, while the mean duration for the T307/G744 variant was 16.76 months and for the T187/T307/A367/G744/G793/T798/A801/T840/C852 variant 17.91 months (P=0.98; log rank test).
Figure 4.
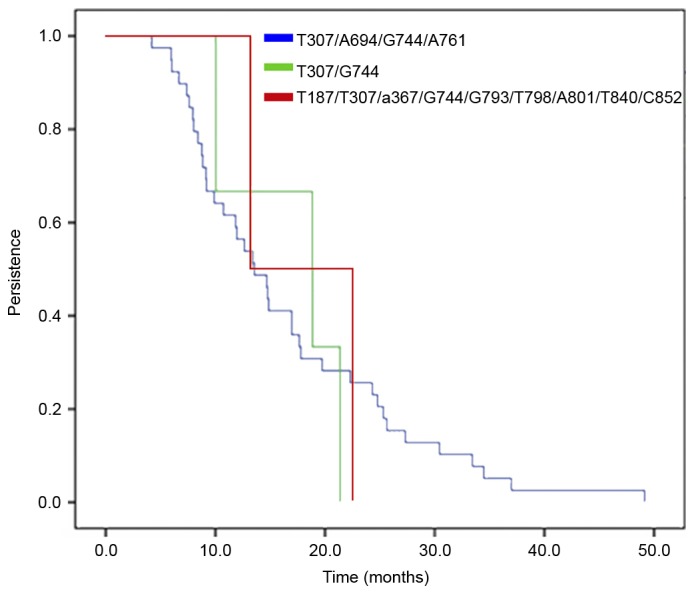
Persistence of human papillomavirus 58 infection stratified by E6/E7 variants in women with incident infections. Data were analyzed using the log rank test and there was no significant difference (P=0.98).
Risk factors associated with clearance of HPV58 incident infections
Table I presents the age-adjusted and multivariate rate ratios of HPV58 clearance by several viral and host characteristics. During follow-up, women that first engaged sexual intercourse at >15 years of age had a lower clearance rate than those who first engaged in sexual intercourse at ≤15 years of age [hazard ratio (HR)=0.29; 95% confidence interval (CI)=0.09–0.92]. In contrast, parous women were associated with a higher clearance rate than nulliparous women (HR=3.43, 95% CI=1.23–9.60). There was no difference in clearance rates between the HPV58 variants.
Table I.
HR and 95% CI for clearance of HPV58 incident infections.
| Clinicopathological characteristic | No. | % | HR | 95% CI |
|---|---|---|---|---|
| Age, years | ||||
| <30 | 23 | 35.4 | 1.00 | |
| ≥30 | 42 | 64.6 | 0.78 | 0.35–1.73 |
| Age at point of first sexual intercourse, years | ||||
| ≤15 | 15 | 23.1 | 1.00 | |
| >15 | 50 | 76.9 | 0.29 | 0.09–0.92 |
| Parity | ||||
| None | 11 | 16.9 | 1.00 | |
| ≥1 | 54 | 83.1 | 3.43 | 1.23–9.60 |
| Use of oral contraceptives | ||||
| No | 48 | 75.0 | 1.00 | |
| Yes | 16 | 25.0 | 0.63 | 0.27–1.47 |
| Type of infection | ||||
| Single | 41 | 63.1 | 1.00 | |
| Multiple | 24 | 36.9 | 0.72 | 0.32–1.00 |
| Viral load, RLU | ||||
| Low, <0.5 | 16 | 24.6 | 1.00 | |
| Medium, ≥0.5-<1.5 | 14 | 21.5 | 2.58 | 0.78–8.52 |
| High, ≥1.5 | 35 | 53.9 | 1.67 | 0.71–3.93 |
| HPV58 variant | ||||
| 1a | 54 | 87.1 | 1.00 | |
| 2b | 5 | 8.1 | 0.54 | 0.12–3.29 |
| 3c | 3 | 4.8 | 0.77 | 0.15–3.94 |
T307 A694 G744 A761.
T307 G744.
T187 T307 A367 G744 G793 T798 A801 T840 C852. HR were adjusted for age and cofactors. Multiple infections refer to the presence of HPV58 with other types of HPV. HR, hazard ratio; CI, confidence interval; HPV, human papillomavirus.
Characteristics of the women with incident HPV58 infections that developed HGSIL during follow-up
Table II lists the characteristics of 2 women with incident HPV58 infections that developed HGSIL during the follow-up. In the two women the HGSIL was diagnosed at the second visit. The women were aged >35 years and had multiple HPV infections.
Table II.
Characteristics of the patients with incident HPV58 infections that developed high grade squamous intraepithelial lesions during follow-up.
| Case | Visit | Age (years) | Type of infection | HPV58 variant |
|---|---|---|---|---|
| 1 | 2 | 36 | Multiple (HPV31-33-58) | No amplification |
| 2 | 2 | 44 | Multiple (HPV58-81) | T307/A694/G744/A761 |
HPV, human papillomavirus.
Discussion
A number of studies have demonstrated that certain HPV58 E6/E7 variants are associated with a higher risk of cervical cancer (15,16,20–27). In the present study, the presence and persistence of incident HPV58 E6/E7 variants was assessed during a long follow-up period in women with normal baseline cytology, as well as other risk factors associated with clearance of HPV58 infections.
At present, information concerning sequence variability of HPV58 is limited. The majority of studies have been performed in Asian populations where higher rates of HPV58 prevalence have been reported compared with the rest of the world (15,16,20–22). Only a few studies in Latin-American countries have evaluated the presence of HPV58 variants (23–29).
Two previous publications by an Asian group analyzed samples from 15 different countries (30,31). These studies reported sequence variability of the E6, E7, E2, E5, L1 and LCR regions of the HPV58 genome, and proposed a novel classification of HPV58 variants into 4 lineages. They also identified specific variants that are associated with an increased risk of cervical intraepithelial neoplasia grade III and invasive cervical cancer (30,31). However, there is only one follow-up study that analyzes the function of HPV58 variants in predicting persistence of HPV58 infections in women with low grade squamous intraepithelial lesions and atypical squamous cells of undetermined significance (32). The present study included women with normal baseline cytology who developed incident HPV58 infections during follow-up.
Three different variants were identified in the E6/E7 regions, with the T307/A694/G744/A761 variant demonstrating the highest frequency (88% of the samples); which is in agreement with a previous study examining the same cohort of women with prevalent HPV58 variants (27) as well as the study published by Chan et al (16) in 2002, where this variant was the most frequent in women with normal cytology.
To the best of our knowledge there are no follow-up studies analyzing the involvement of incident HPV58 variants in the clearance of HPV58 infections and other risk factors in women with normal cytology. The variant analysis demonstrated that for those women with persistent HPV58 infections, the same variant was detected over time. This confirmed the real persistence of HPV58 infections in the study population (27). In total, 50% of the HPV58 incident infections were still present after 1 year of follow-up and 3% after 4 years; a similar result to those reported by others (33). The present study also revealed no difference in HPV58 clearance rates when the infection was a single event or comprised of multiple infections. In a follow-up study of 2,462 women enrolled in the Ludwig-Mc Gill cohort, Trottier et al (34) revealed differences in HPV58 infection duration when the infection was analyzed as mono-infection or co-infection with other HPV genotypes. Differences between studies may be explained by study design, the study population, and differences in data analysis and methods of sampling and genotyping.
One of the aims of the present study was to analyze whether there were differences in clearance rates between HPV58 variants in women with normal baseline cytology. The present study revealed that there was no significant difference in clearance rates and the variants detected had similar time of median duration, suggesting that these variants did not serve a direct function in the clearance of the HPV58 infection in this cohort of women.
Clinicopathological characteristics, including age at the point of first sexual intercourse and parity, were associated with HPV58 incident infection clearance time. These factors may therefore be added to the list of other previously reported host factors that affect the clearance of HPV infections (12).
In the present study only 2 women infected by HPV58 incident infections developed HGSIL. It was not possible to amplify the E6 and E7 region in 1 patient, and the other presented with the variant T307/A694/G744/A761, which has been previously reported in other studies in women with normal histology and HGSIL (31). Other molecular HPV58 variants isolated from HGSIL have been demonstrated to exhibit different transcriptional activity that may be important in the expression of E6 and E7 viral oncogenes (35).
The E6 and E7 regions of HPV58 positive samples were amplified, but in a small number of samples it was not possible to amplify these regions and therefore E6/E7 variants failed to be identified. A potential reason for this may be low efficiency in the amplification process due to the fragment size amplified (36). Other reasons may involve DNA degradation or DNA fragmentation in the samples analyzed, low viral loads, viral integration or disruption in the HPV58 genome (20).
The sample size in the present study was small, which was a limitation to the analysis of the function of HPV58 variants in the development of HGSIL. Further studies are required to examine the persistence of HPV58 E6/E7 variants in women with abnormal cytology to analyze their pathological implications.
Acknowledgements
The authors would like to thank the National Cancer Institute HPV study group: Dr Mauricio González, Dr Natasha Ortiz, Dr Gustavo Hernández, Dr Oscar Gamboa, Dr Raul H Murillo, Dr Joaquín Luna, Dr Hector Posso, Dr Margarita Ronderos, Dr Gilberto Martínez, Dr Edmundo Mora, Dr Gonzalo Pérez, Dr José María Fuentes, Dr Constanza Gómez, Dr Eva Klaus, Mrs Constanza Camargo, Mrs Cecilia Tobón, Mrs Teodolinda Palacio, Dr Carolina Suarez, Mrs Claudia Molina and Mr Alex Torres. The present study was supported by the National Cancer Institute, Bogotá, Colombia (DNP grant no. 41030310-30).
References
- 1.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 2.Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJ, Meijer CJ. International Agency for Research on Cancer Multicenter Cervical Cancer Study Group: Epidemiologic classification of human papillomavirus types associated with cervical cancer. New Eng J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 3.de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Onco. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 4.Guan P, Howell-Jones R, Li N, Bruni L, de Sanjosé S, Franceschi S, Clifford GM. Human papillomavirus (HPV) types in 115,789 HPV-positive women: A meta-analysis from cervical infection to cancer. Int J Cancer. 2012;131:2349–2359. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- 5.Bosch FX, Manos MM, Muñoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. Prevalence of human papillomavirus in cervical cancer: A worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 6.Palefsky JM, Minkoff H, Kalish LA, Levine A, Sacks HS, Garcia P, Young M, Melnick S, Miotti P, Burk R. Cervicovaginal human papillomavirus infection in human immunodeficiency virus-1 (HIV)-positive and high-risk HIV-negative women. J Natl Cancer Inst. 1999;91:226–236. doi: 10.1093/jnci/91.3.226. [DOI] [PubMed] [Google Scholar]
- 7.Pett M, Coleman N. Integration of high-risk human papillomavirus: A key event in cervical carcinogenesis? J Pathol. 2007;212:356–367. doi: 10.1002/path.2192. [DOI] [PubMed] [Google Scholar]
- 8.Doeberitz MV, Vinokurova S. Host factors in HPV-related carcinogenesis: Cellular mechanisms controlling HPV infections. Arch Med Res. 2009;40:435–442. doi: 10.1016/j.arcmed.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Chen HC, Schiffman M, Lin CY, Pan MH, You SL, Chuang LC, Hsieh CY, Liaw KL, Hsing AW, Chen CJ. CBCSP-HPV Study Group: Persistence of type-specific human papillomavirus infection and increased long-term risk of cervical cancer. J Natl Cancer Inst. 2011;103:1387–1396. doi: 10.1093/jnci/djr283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pardo C, Cendales R. Incidencia, Mortalidad y Prevalencia de Cáncer en Colombia, 2007–2011. First edtion. Vol. 1. Instituto Nacional de Cancerología; Bogotá, DC: 2015. p. 148. (In Spanish) [Google Scholar]
- 11.Molano M, Posso H, Weiderpass E, van den Brule AJ, Ronderos M, Franceschi S, Meijer CJ, Arslan A, Munoz N. HPV Study Group HPV Study: Prevalence and determinants of HPV infection among Colombian women with normal cytology. Br J Cancer. 2002;87:324–333. doi: 10.1038/sj.bjc.6600442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz N, Méndez F, Posso H, Molano M, van den Brule AJ, Ronderos M, Meijer C, Muñoz A. Instituto Nacional de Cancerologia HPV Study Group: Incidence, duration and determinants of cervical human papillomavirus infection in a cohort of Colombian women with normal cytological results. J Infect Dis. 2004;190:2077–2087. doi: 10.1086/425907. [DOI] [PubMed] [Google Scholar]
- 13.Mendez F, Munoz N, Posso H, Molano M, Moreno V, van den Brule AJ, Ronderos M, Meijer C, Munoz A. Instituto Nacional de Cancerologia Human Papillomavirus Study Group: Cervical coinfection with human papillomavirus (HPV) types and possible implications for the prevention of cervical cancer by HPV vaccines. J Infect Dis. 2005;192:1158–1165. doi: 10.1086/444391. [DOI] [PubMed] [Google Scholar]
- 14.Serrano B, Alemany L, Tous S, Bruni L, Clifford GM, Weiss T, Bosch FX, de Sanjosé S. Potential impact of a nine-valent vaccine in human papillomavirus related cervical disease. Infect Agent Cancer. 2012;7:38. doi: 10.1186/1750-9378-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin CY, Matsumoto K, Yoshikawa H, Yasugi T, Onda T, Nakagawa S, Yamada M, Nozawa S, Sekiya S, Hirai Y, et al. Analysis of E6 variants of human papillomavirus type 33, 52 and 58 in Japanese women with cervical intraepithelial neoplasia/cervical cancer in relation to their oncogenic potential. Cancer Lett. 2001;170:19–24. doi: 10.1016/S0304-3835(01)00570-5. [DOI] [PubMed] [Google Scholar]
- 16.Chan PK, Lam CW, Cheung TH, Li WW, Lo KW, Chan MY, Cheung JL, Cheng AF. Association of human papillomavirus type 58 variant with the risk of cervical cancer. J Natl Cancer Inst. 2002;94:1249–1253. doi: 10.1093/jnci/94.16.1249. [DOI] [PubMed] [Google Scholar]
- 17.Calleja-Macias IE, Villa LL, Prado JC, Kalantari M, Allan B, Williamson AL, Chung LP, Collins RJ, Zuna RE, Dunn ST, et al. Worldwide genomic diversity of the high-risk human papillomavirus types 31, 35, 52 and 58, four close relatives of human papillomavirus type 16. J Virol. 2005;79:13630–13640. doi: 10.1128/JVI.79.21.13630-13640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76:1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 19.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu EQ, Zha X, Yu XH, Zhang GN, Wu YG, Fan Y, Ren Y, Kong LQ, Kong W. Profile of physical status and gene variation of human papillomavirus 58 genome in cervical cancer. J Gen Virol. 2009;90:1229–1237. doi: 10.1099/vir.0.008227-0. [DOI] [PubMed] [Google Scholar]
- 21.Chang YJ, Chen HC, Lee BH, You SL, Lin CY, Pan MH, Chou YC, Hsieh CY, Chen YM, Cheng YJ, et al. Unique variants of human papillomavirus genotypes 52 and 58 and risk of cervical neoplasia. Int J Cancer. 2011;129:965–973. doi: 10.1002/ijc.25724. [DOI] [PubMed] [Google Scholar]
- 22.Chan PK. Human papillomavirus type 58: The unique role in cervical cancers in East Asia. Cell Biosci. 2012;2:17. doi: 10.1186/2045-3701-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veras VS, Cerqueira DM, Martins CR. L1 sequence of a new human papillomavirus type-58 variant associated with cervical intraepithelial neoplasia. Braz J Med Biol Res. 2005;38:1–4. doi: 10.1590/S0100-879X2005000100001. [DOI] [PubMed] [Google Scholar]
- 24.Raiol T, Wyant PS, de Amorim RM, Cerqueira DM, Milanezi Nv, Brígido Mde M, Sichero L, Martins CR. Genetic variability and phylogeny of the high-risk HPV-31, −33, −35, −52 and −58 in central Brazil. J Med Virol. 2009;81:65–92. doi: 10.1002/jmv.21432. [DOI] [PubMed] [Google Scholar]
- 25.Canche JC, López IR, Suárez NG, Acosta GC, Conde-Ferráez L, Cetina TC, Losa MR. High prevalence and low E6 genetic variability of human papillomavirus 58 in women with cervical cancer and precursor lesions in Southeast Mexico. Mem Inst Oswaldo Cruz. 2010;105:144–148. doi: 10.1590/S0074-02762010000200006. [DOI] [PubMed] [Google Scholar]
- 26.Godínez JM, Heideman DA, Gheit T, Alemany L, Snijders PJ, Tommasino M, Meijer CJ, de Sanjosé S, Bosch FX, Bravo IG. Differential presence of Papillomavirus variants in cervical cancer: An analysis for HPV33, HPV45 and HPV58. Infect Genet Evol. 2013;13:96–104. doi: 10.1016/j.meegid.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Buitrago O, Morales N, Martín C, Huertas A, Moreno P, Martínez T, Molano M. Presencia y persistencia de variantes moleculares del ORF E6 y ORF E7 del VPH58 en mujeres con citología normal que pertenecen a la cohorte de Bogotá, Colombia. Rev Colomb Cancerol. 2012;16:40–48. (In Spanish) [Google Scholar]
- 28.Mejía L, Muñoz D, Trueba G, Tinoco L, Zapata S. Prevalence of human papillomavirus types in cervical cancerous and precancerous lesions of Ecuadorian women. J Med Virol. 2016;88:144–152. doi: 10.1002/jmv.24310. [DOI] [PubMed] [Google Scholar]
- 29.Gurgel AP, Chagas BS, do Amaral CM, Nascimento KC, Leal LR, Silva Neto Jda C, Muniz Cartaxo MT, de Freitas AC. Prevalence of human papillomavirus variants and genetic diversity in the L1 gene and long control region of HPV16, HPV31 and HPV58 found in North-East Brazil. Biomed Res Int. 2015;2015:130828. doi: 10.1155/2015/130828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan PK, Luk AC, Park JS, Smith-McCune KK, Palefsky JM, Konno R, Giovannelli L, Coutlée F, Hibbitts S, Chu TY, et al. Identification of human papillomavirus type 58 lineages and the distribution worldwide. Infect Dis. 2011;203:1565–1573. doi: 10.1093/infdis/jir157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan PK, Zhang C, Park JS, Smith-McCune KK, Palefsky JM, Giovannelli L, Coutlée F, Hibbitts S, Konno R, Settheetham-Ishida W, et al. Geographical distribution and oncogenic risk association of human papillomavirus type 58 E6 and E7 sequence variations. Int J Cancer. 2013;132:2528–2536. doi: 10.1002/ijc.27932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xi LF, Schiffman M, Koutsky LA, Hughes JP, Hulbert A, Shen Z, Galloway DA, Kiviat NB. Variant-specific persistence of infections with human papillomavirus Types 31, 33, 45, 56 and 58 and risk of cervical intraepithelial neoplasia. Int J Cancer. 2016;139:1098–1105. doi: 10.1002/ijc.30164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Insinga RP, Perez G, Wheeler CM, Koutsky LA, Garland SM, Leodolter S, Joura EA, Ferris DG, Steben M, Hernandez-Avila M, et al. Incident cervical HPV infections in young women: Transition probabilities for CIN and infection clearance. Cancer Epidemiol Biomarkers Prev. 2011;20:287–296. doi: 10.1158/1055-9965.EPI-10-0791. [DOI] [PubMed] [Google Scholar]
- 34.Trottier H, Mahmud S, Prado JC, Sobrinho JS, Costa MC, Rohan TE, Villa LL, Franco EL. Type-specific duration of human papillomavirus infection: Implications for human papillomavirus screening and vaccination. J Infect Dis. 2008;197:1436–1447. doi: 10.1086/587698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raiol T, de Amorim RM, Galante P, Martins CR, Villa LL, Sichero L. HPV-58 molecular variants exhibit different transcriptional activity. Intervirology. 2011;54:146–150. doi: 10.1159/000320199. [DOI] [PubMed] [Google Scholar]
- 36.van den Brule AJ, Meijer CJ, Bakels V, Kenemans P, Walboomers JM. Rapid detection of human papillomavirus in cervical scrapes by combined general primer-mediated and type-specific polymerase chain reaction. J Clin Microbiol. 1990;28:2739–2743. doi: 10.1128/jcm.28.12.2739-2743.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]