Abstract
BRCAness breast tumors represent a group of sporadic tumors characterized by a reduction in BRCA1 gene expression. As BRCA1 is involved in double-strand breaks (DSBs) repair, dysfunctional BRCA pathway could make a tumor sensitive to DNA damaging drugs (e.g., platinum agents). Thus, accurately identifying BRCAness could contribute to therapeutic decision making in patients harboring these tumors. The purpose of this study was to identify if BRCAness tumors present a characteristic methylation profile and/or were related to specific clinicopathological features. BRCAness was measured by MLPA in 63 breast tumors; methylation status of 98 CpG sites within 84 cancer-related genes was analyzed by MS-MLPA. Protein and mRNA expressions of the selected genes were measured by quantitative real-time PCR and Western Blot. BRCAness was associated with younger age, higher nuclear pleomorfism, and triple-negative (TN) status. Epigenetically, we found that the strongest predictors for BRCAness tumors were the methylations of MLH1 and PAX5 plus the unmethylations of CCND2 and ID4. We determined that ID4 unmethylation correlated with the expression levels of both its mRNA and protein. We observed an inverse relation between the expressions of ID4 and BRCA1. To the best of our knowledge, this is the first report suggesting an epigenetic regulation of ID4 in BRCAness tumors. Our findings give new information of BRCAness etiology and encourage future studies on potential drug targets for BRCAness breast tumors.
Keywords: Breast tumors, BRCAness, ID4, Aberrant DNA methylation
Introduction
Breast cancer is the most frequent malignant disease and the leading cause of cancer death among women in both economically developed and developing countries. The incidence of breast cancer is increasing due to higher life expectancy, increased urbanization, and adoption of Western lifestyles [1].
Breast cancer can be classified as sporadic when genetic changes occur in somatic cells over time or hereditary when the tumor is originated by germline mutations principally in BRCA1 or BRCA2 genes. Notably, only 5–10 % of all breast tumors are of hereditary origin. The accepted criteria to consider a breast cancer hereditary consist of: at least three breast and/or ovarian cancers in a family; two breast cancer cases in close relatives before age 50; and Ashkenazi Jewish ancestry with breast cancer particularly triple negative (TN) before age 60 [2]. BRCA1 gene is involved in several cellular processes such as DNA repair, transcriptional regulation, and chromatin remodeling, whereas BRCA2 is principally involved in DNA recombination and repair processes [3]. Women with BRCA1 mutations typically develop breast cancer at an earlier age, present higher nuclear pleomorfism, and exhibit TN tumors [3–6].
For sporadic breast tumors, it has been proposed that one-third of them present a reduction in the expression of BRCA1 without any mutation in this gene. The term BRCAness was initially suggested by Asworth et al. and defines phenotypic traits that some sporadic tumors share with hereditary BRCA cancers [3]. This BRCAness phenotype is associated with alterations in DNA repair pathways, presents higher histological grade, expresses EGFR, and is more likely to be estrogen receptor (ER) negative [7]. The mechanisms that cause this BRCAness phenotype could be methylation in BRCA1 promoter or alterations in genes involved in the BRCA pathway [8]. So, genetic and epigenetic mechanisms can create the BRCAness phenotype in sporadic breast cancers. Evidence is accumulating that silencing of BRCA genes, or dysfunction of other genes acting in similar biochemical pathways, might be important in the pathogenesis of a significant proportion of sporadic hereditary-like cancers [3]. Therefore, it is important to identify these genes and define their role in BRCAness etiology. BRCAness phenotype may resemble BRCA1-mutated tumors by a pattern of gains and losses of specific genomic regions. This can be assessed by different methodologies such as array Comparative Genomic Hybridization (aCGH) and MLPA.
Inhibitors of differentiation proteins 1, 2, 3, and 4 (ID1–4) are dominant negative regulators of the basic helix-loop helix (bHLH) family of transcription factors [9]. In human tumors, an increased expression of ID proteins has been associated with reversion to an embryonic-like state, with loss of differentiation, high rates of proliferation, migration, and neo-angiogenesis [10, 11]. In breast cancer there are apparently controversial findings regarding the role of ID4 in tumorigenesis. For instance, the hypermethylation of ID4 promoter is a frequent event in breast tumors and associated with an increased risk of lymph node metastasis [12]. ID4 increased expression, however, has been associated with the ability of breast cancer cells to exhibit anchorage-independent growth [13]. Therefore, it is important to accurately define the function ID4 in breast tumorigenesis.
In this current study, we determined BRCAness phenotype in 63 human Invasive Ductal breast Carcinomas (IDC) and analyzed the epigenetic profile of 98 CpG sites located in 84 cancer-related genes. Interestingly, our results reveal that BRCAness breast tumors are significantly associated with the unmethylation of ID4. In addition, we show that unmethylation of ID4 correlates with the expression levels of both its mRNA and protein. Furthermore, we report the existence of an inverse relation between the expressions of ID4 and BRCA1. These combined results demonstrate that ID4 is epigenetically regulated in the determination of BRCAness human breast tumors. Therefore, ID4 regulation may have implications regarding tumor biology, prognosis, and treatment.
Materials and methods
Patients and tumors
Eighty-eight women who had invasive ductal breast carcinomas were enrolled in this study. In 63 tumors, we evaluated BRCAness phenotype status and the remaining tumors were used to perform qRT-PCR experiments; methylation analysis was performed on the 88 tumors. Ethical approval was obtained from the Ethics Committee of the School of Medical Sciences, National University of Cuyo, Mendoza, Argentina. Patients signed an informed consent based on the scientific and ethical principles of the World Medical Association’s Declaration of Helsinki. Patients were treated in the Gineco-Mamario Institute of Mendoza, Argentina.
BRCAness phenotype determination by Multiplex Ligation-dependent Probe Amplification assay (MLPA)
BRCAness was determined by MLPA analysis on DNA extracted from fresh tumors tissue. DNA was isolated using CTAB (Cetyltrimethyl Ammonium Bromide) as previously described [14, 15]. Classification of BRCAness phenotype was performed using the MLPA assay, probemix P376-B2 BRCA1ness according to manufacturer’s recommendations (MRC-Holland, Amsterdam, the Netherlands) [16]. In brief, gains and losses of specific chromosome regions detected by aCGH assays were proposed as markers to distinguish between hereditary-BRCA1-mutated and sporadic breast tumors. The MLPA panel of involved genomic regions was denominated as “BRCAS1-like classifier.” Thirty-four of the most important regions of this classifier were included in the MLPA probemix [17]. Gains and losses were represented by changes in peak areas on an electropherogram after capillary electrophoresis and values from all the 34 target-specific probes were used for Prediction analysis for microarrays (PAM) which is a statistical technique that performs sample classification from gene expression data using shrunken centroids [16]. A gains-and-losses pattern was afterward established by R-based language analysis and a score was determined. Based on the concordance with previous aCGH determinations, the BRCAness positivity was established as follows: samples with a score ≥ 0.5 were classified as ‘‘BRCAness-like,’’ and below this score, a sample was classified as ‘‘non BRCAness-like’’ [16, 18].
Methylation profiles determination by methyl specific-multiplex ligation-dependent probe amplification assay (MS-MLPA)
To assess the méthylation status of 98 CpG sites located in 84 cancer-related genes, the MS-MLPA kits ME001, ME002, ME003, and ME011 were used on DNA obtained, as mentioned before, from 88 IDC and from the following cancer cell lines: MDA-MB231, MCF-7, K562, and HeLa. The MS-MLPA assays were performed basically according to manufacturer’s recommendations (MRC-Holland, Amsterdam, the Netherlands (www.mrcholland.com) [19], introducing subtle modifications [20]. A CpG site was considered to be methylated when the methylation dosage ratio between digested and undigested sample was superior to the cut-off threshold of 8 %. [14, 15, 21]. Afterward, DNA methylation data were dichotomized in unmethylated and methylated status.
Gene expression analyses by real-time polymerase chain reaction
RNA was extracted from fresh tumors and from the human breast cancer cell lines MDA-MB231, MCF-7, K-562, and HeLa cell lines with Trizol Reagent (Life Technologies, USA). One hundred ng of cDNA was used to perform realtime PCR using specific primers for 32 cancer-related genes (Table 1) as previously described [21].
Table 1.
Tumor suppressor genes expression analyzed by qPCR
| Gene symbol* | Gene name |
|---|---|
| APC | Adenomatous polyposis coli |
| ATM | Ataxia telangiectasia mutated |
| BRCA1 | Breast cancer 1 |
| BRCA2 | Breast cancer 2 |
| CACNA1G | Calcium channel, voltage-dependent, P/Q type, alpha 1B subunit |
| CCND2 | Cyclin D2 |
| CD44 | CD44 molecule |
| CDH13 | Cadherin 13 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B |
| DAPK1 | Death-associated protein kinase 1 |
| DLC1 | DLC1 Rho GTPase activating protein |
| ESR | Estrogen receptor 1 |
| ID4 | Inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| MGMT | O-6-methylguanine-DNA methyltransferase |
| MLH1 | MutL homolog 1 |
| MLH3 | MutL Homolog 3 |
| MSH2 | MutS homolog 2 |
| MSH6 | MutS homolog 6 |
| PAX6 | Paired box 6 |
| PMS2 | PMS2 postmeiotic segregation increased 2 |
| PTEN | Phosphatase and tensin homolog |
| RARb | Retinoic acid receptor beta |
| RASSF1 | Ras association domain family member 1A |
| RB1 | Retinoblastoma 1 |
| SCGB3 | Secretoglobin, family 3A, member 1 |
| SFRP4 | Secreted frizzled-related protein 4 |
| THBS1 | Thrombospondin 1 |
| TP53 | Tumor protein p53 |
| TP73 | Tumor protein p73 |
| TWIST | Twist family bHLH transcription factor 1 |
According to the Human Gene Nomenclature Committee (HGNC)
ID4 expression analyses by Western blotting
Proteins were extracted from MDA-MB231, MCF-7, HeLa, and K-562 cell lines with Trizol Reagent (Life Technologies, USA) according to manufacturer’s instruction, and quantification was performed using Bradford assay. Proteins were separated on 10 % Tris-glycine SDS gels, and Western blot assay was performed as previously described [9] using an anti-ID4 (AVIVA) primary antibody. Detection was performed with chemiluminescence (Thermo Scientific Pierce) on a Luminescent Image Analyzer LAS-4000 (Fujifilm, Tokyo, Japan). Quantification was carried out with ImageJ.
Statistical analyses
The association of BRCAness phenotype with clinical variables or methylation of CpG sites was assessed by fitting a multivariate linear regression model. The associations were considered statistically significant when p values were less than 0.05.
The association between relative gene expression values was performed by singular value decomposition (SVD) and total least squares-least squares (TLS-LS). SVD represents an appropriate tool for lineal models with fewer equations than unknowns (as in the analysis of ID4 gene expression with respect to the expression of 32 genes, Table 1), and TLS-LS was applied when performing regression analysis considering error in both variables. Because SVD and TLS-LS are statistical methods infrequently used in biomedical research, more specifications can be found in Supplemental Material “S1.”
Multivariable linear regression models were carried out by using SPSS v17 software (SPSS Inc, Chicago, IL, USA). SVD and TLS-LS were performed using Matlab (MathWorks®).
Results
BRCAness phenotype determination by MLPA
We determined BRCAness status in a cohort of 63 IDC by MLPA, which has been demonstrated to be robust in this determination and therefore supports its application to both clinical genetic testing and as a predictor of treatment benefit [16, 22]. The robustness of MLPA is based on its concordance with aCGH results performing equally well in identifying BRCAness. Since there is no gold standard for the determination of BRCAness, MLPA has shown to be a less complex method with high sensitivity, specificity, and reproducibility and is therefore proposed as a surrogate for aCGH. Our analysis revealed that 10 out of 63 patients had tumors with a BRCAness profile (Table 2). The cut-off threshold to consider a tumor as BRCAness was established to 0.5. It is interesting to notice that all BRCAness tumors presented index above 0.6, a considerable distance from the threshold (Fig. 1). To identify clinical characteristics associated with BRCAness tumors, we performed regression analyses using clinico-pathological variables as the covariates and BRCAness status as the outcome variable. The strongest predictor of BRCAness status was higher nuclear pleomorphism (r = 0.183, p = 0.0258), and there was a statistical tendency toward younger age (r = −0.315, p = 0.078) (both p values were adjusted with Bonferroni multiple testing correction). There was no significant difference between the groups of subjects when compared for axillar lymph node status (p = 0.37). Since TN tumors may present a BRCA defective pathway [17], we sought to determine if TN tumors displayed the BRCAness phenotype. Our results show that, indeed, TN status was significantly associated with BRCAness phenotype. (p = 0.036). In summary, BRCAness tumors can be defined by higher nuclear pleomorfism and enhanced triple negativity.
Table 2.
Clinical-pathological features of BRCAness-tested patients
| BRCAness N (%) | Non-BRCAness N (%) | |
|---|---|---|
| Total patients | 10 (15.8) | 53 (84.1) |
| Age (years) | ||
| <40 | 3 (4.7) | 46 (73) |
| >40 | 7(11.1) | 4 (6.3) |
| NA | 3 (4.7) | |
| Stage* | ||
| I | 1 (1.5) | 24 (38) |
| II | 8 (12.6) | 14 (22.2) |
| II | 1 (1.5) | 13 (20.6) |
| NA | 2 (3.1) | |
| Tumor grade | ||
| I | 0 (0) | 9 (14.2) |
| II | 3 (4.7) | 23 (36.5) |
| III | 7(11.1) | 19 (30.1) |
| NA | 2 (3.1) | |
| Nuclear pleomorfism | ||
| I | 0 (0) | 6 (9.5) |
| II | 1 (1.5) | 25 (39.6) |
| III | 9 (14.2) | 20 (31.7) |
| NA | 2 (3.1) | |
| Molecular subtypes | ||
| Luminal A | ||
| Positive | 1 (1.5) | 10 (15.8) |
| Negative | 9 (14.2) | 42 (66.6) |
| NA | 1 (1.5) | |
| Luminal B | ||
| Positive | 4 (6.3) | 23 (36.5) |
| Negative | 6 (9.5) | 29 (46) |
| NA | 1 (1.5) | |
| HER2+ | ||
| Positive | 0(0) | 5 (7.9) |
| Negative | 9 (14.2) | 47 (74.6) |
| NA | 1 (1.5) | 1 (1.5) |
| TN | ||
| Positive | 6 (9.5) | 10 (15.8) |
| Negative | 4 (6.3) | 41 (65) |
| NA | 0(0) | 2 (3.1) |
Stage according to AJCC (American Joint Committee on Cancer)
Fig. 1.
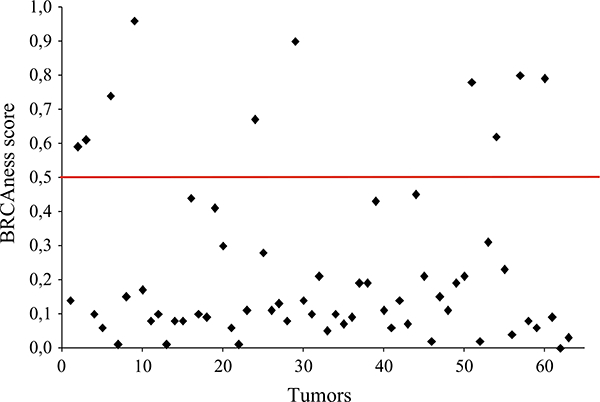
BRCAness analyses on breast tumors. In the Y axis, the obtained BRCAness scores are represented. Each dot represents a tumor. The red horizontal line indicates the 0.5 cut-off value established for BRCAness determination. Dots above the red line show BRCAness tumors, and dots below the red line represent non-BRCAness tumors. 10/63 tumors revealed BRCAness scores >0.6, a considerable distance from the scores of non-BRCAness tumors
Methylation profile of BRCAness tumors
To determine if BRCAness tumors present a characteristic methylation profile, we performed MS-MLPA analyses on 98 CpG sites in the 63 tumors. For the statistical analysis, we considered only CpG sites which were methylated in more than 10 % of the tumors, to decrease the influence unmethylation events. This selection of the 98 CpG sites initially being considered was reduced to a final number of 43 regions. Next, in a linear regression analysis, considering the 43 CpG sites as predictor variables and the BRCAness status as outcome variable, we determined that the strongest predictors for BRCAness tumors were the methylations of MLH1 at −383 nt before the transcription start site (TSS) and PAX5 (−236 nt before TSS) (Fig. 2a, b) plus the unmethylations of CCND2 (—1289 nt before TSS) and ID4 (—367 nt before TSS) (Fig. 2c, d). Regression coefficients and standard errors of these four regions are summarized in Table 3.
Fig. 2.
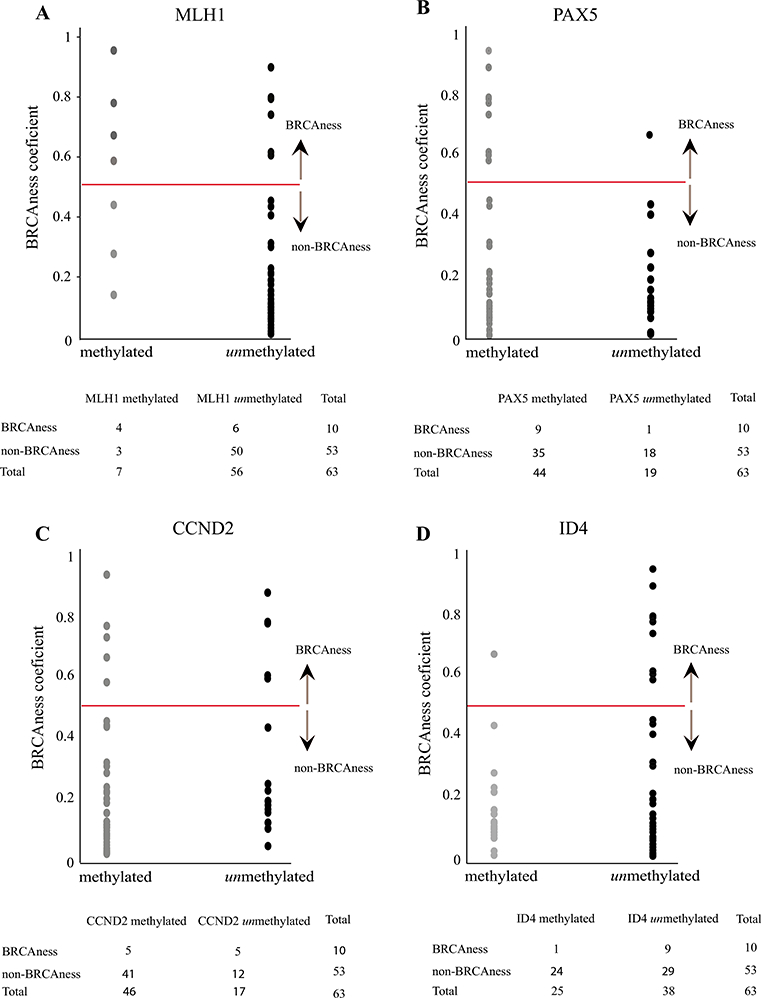
Epigenetic predictors of BRCAness phenotype. Y axis represents BRCAness scores. The red horizontal line indicates the 0.5 cut-off value established for BRCAness determination. Each dot represents a tumor. X axis represents methylated and unmethylated status of each gene. a MLH1 representation of methylated and unmethylated tumors with respect to BRCAness scores. b PAX5 representation of methylated and unmethylated tumors with respect to BRCAness scores. c CCND2 representation of methylated and unmethylated tumors with respect to BRCAness scores. d ID4 representation of methylated and unmethylated tumors with respect to BRCAness scores
Table 3.
Pearson correlation coefficients (r), standard errors, and nominal significance levels of predictive epigenetic factors for BRCAness status
| Gene | r | Standard error | p value* |
|---|---|---|---|
| MLH1 | 0.502 | 0.126 | 0.0004 |
| PAX5 | 0.290 | 0.084 | 0.008 |
| CCND2 | −0.310 | 0.086 | 0.023 |
| ID4 | −0.285 | 0.079 | 0.035 |
Bonferroni corrected p values
ID4 (−367nt to TSS) CpG site defines gene expression
Given that ID4 is a negative regulator of BRCA1 [13] and that the unmethylation of the ID4 CpG site −367 correlated with BRCAness tumors, we sought to analyze if this region had a role in the regulation of gene expression. To test this, we analyzed by MS-MLPA, the methylation status of two CpG sites in the ID4 promoter in MDA-MB231, MCF-7, K-562, and HeLa cell lines. The MS-MLPA ME003 panel contains 27 probes two of which hybridize at two different CpG sites in the ID4 promoter, one at −956 nt and the other one at −367 nt both relative to the TSS. This allowed us to analyze separately the methylation percentages of each site and correlate these results with mRNA and protein expression. We found that the levels of both mRNA and protein were impaired when the methylation percentage was higher or equal than 13 % in the −367 CpG site. In contrast, we found no association between methylation of the upstream −956 CpG site with gene expression. For instance, in the K-562 cell line, the methylation of −956 CpG site was 7 % and yet we found no evidence of protein or gene expression, and on the contrary, in MDA-MB231 cells, the methylation value was of 27 % and there was an increase in gene and protein expression. We hypothesize that the methylation of the site −367 affected gene expression, whereas the methylation of the CpG site −956 did not (Fig. 3). Given that BRCAness tumors present the CpG −367 site unmethylated, and based on the observations that this site regulates gene expression, we can infer that BRCAness tumors express ID4.
Fig. 3.
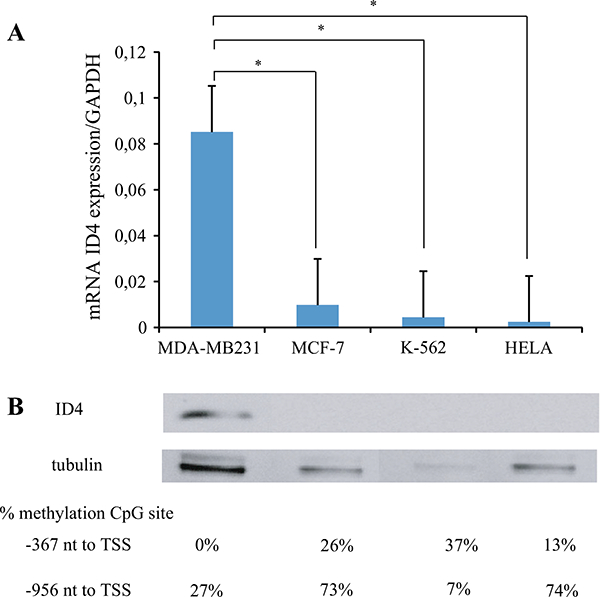
Epigenetic regulation of ID4 for mRNA and protein expression. a Relative expression of ID4 in different cell lines is shown in the bar diagram. Y axis represents ID4/GAPDH relative expression, and X axis represents the cell lines analyzed. Significant differences are shown between MDA-MB231, MCF7, K-562, and HeLa cell lines. b Representative immunoblots of ID4 and β-tubulin in MDA-MB231, MCF-7, K-562, and HeLa cell lines. Significant differences are observed between MDA-MB231 and MCF7, K-562, and HeLa cell lines; there only the former showed ID4 expression. The methylation percentage of the two CpG sites of ID4 is shown below each sample. These values infer that the methylation of at least 13 % of the −367 CpG site is regulating the expression of ID4 at a mRNA and protein level, in the four studied cancer cell lines. In addition, −956 CpG site does not present any regulation on the gene expression
ID4 expression is associated with BRCA1 and MGMT expression
Since ID proteins are associated with loss of differentiation, unrestricted proliferation, and neo-angiogenesis in diverse human cancers [23], we sought to identify genes associated with ID4 expression. We performed qRT-PCR experiments in a different cohort of 25 breast cancer samples and determined the expression of 32 genes involved in tumorigenesis (Table 1). The association between the expressions of ID4 and the 32 genes was evaluated by the SVD method. Using this approach, we determined that ID4 expression significantly associates with that of two other genes: BRCA1 and MGMT (Fig. 4a). As this use of the SVD is equivalent to a principal component analysis (PCA) but, unlike the latter, applicable to underdetermined systems, we could observe (Fig. 4a) that the variances associated with BRCA1 and MGMT are more significant in explaining ID4 expression than the variances of other variables. Between these two genes, BRCA1 is the variable with greater statistical significance to predict the expression of ID4 (Fig. 4a, the highest point in the diagram). The remaining variables show little negligible statistical implication to ID4 expression.
Fig. 4.
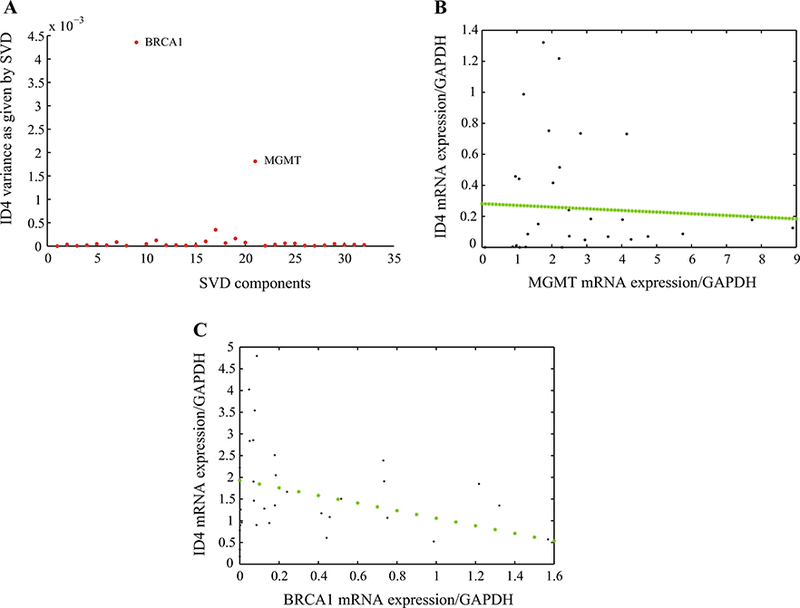
SVD Model of ID4 variances relative to the expression of 32 Genes involved in fresh tumors. a Y axis represents ID4 variance, and X-axis represents SVD components. The two upper points represent the two genes most significant related to ID4 expression: BRCA1 and MGMT. b Negative correlation of ID4 gene expression and MGMT gene expression measured by qRT-PCR. The green dashed line represents TLS-LS analysis. c Negative correlation of ID4 gene expression and BRCA1 gene expression measured by qRT-PCR. The green dashed line represents TLS-LS analysis
To determine the association between the expressions of ID4 with BRCA1 and with MGMT separately, we performed a TLS-LS analysis, an association test that considers error in both variables. TLS-LS revealed an inverse association between ID4 and BRCA1 expression levels indicating that increased ID4 expression correlated with decreased levels of BRCA1 mRNA. A similar inverse association was observed between ID4 and MGMT (Fig. 4b, c). An inverse association between the expressions of ID4 and BRCA1 was also observed in MDA-MB231 and MCF-7 cell lines (Fig. 5). We performed qRT-PCR analysis in both cell lines and determined that when the expression of ID4 is high, as in the case of MDA-MB231 cells, BRCA1 is expressed at low levels; an inverse behavior was seen in MCF-7 cells. It is important to mention that none of the cell lines presented BRCA1 methylation, an observation that suggests that the reduced BRCA1 expression could be due to the increased ID4 expression.
Fig. 5.
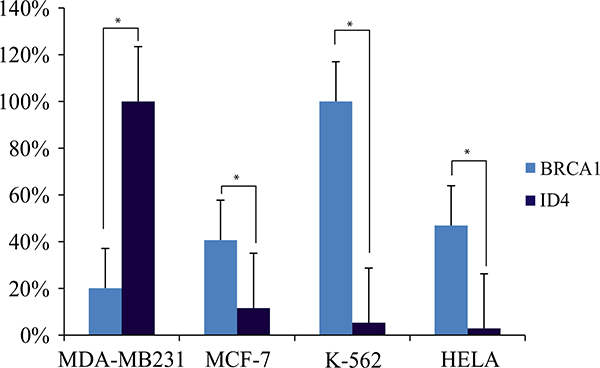
Inverse association of ID4 and BRCA1 gene expression measured by qRT-PCR in cell lines. Gene expression values were normalized by considering the highest value of ID4 and BRCA1 expression of MDA-MB231, MCF7, K562, and HeLa cell lines as 100 %. The remaining values were normalized to 100 %. Asterisk indicates p values <0.05
Discussion
The BRCAness phenotype was initially described to refer to phenotypic traits that some sporadic tumors share with BRCA hereditary tumors [3]. The mechanisms that originate this BRCAness phenotype in sporadic tumors rely on the dysfunction of BRCA1 pathway, which may be due to BRCA1 promoter methylation or genetic and/or epigenetic alterations in genes that affect directly or indirectly the BRCA pathway. At a cellular level, the BRCAness phenotype is characterized by defective homologous recombination. Consequently, patients who harbor BRCAness phenotype may benefit from DNA damaging agents, making therefore the determination of BRCAness phenotype an interesting predictive marker for treatment response.
The determination of the BRCAness phenotype can be performed by either array comparative genomic hybridization (aCGH) or MLPA [16, 17]. Between these methods, MLPA is more rapid and similarly robust in identifying BRCAness tumors and could be applied both for clinical genetic testing and as a predictor of treatment benefit [16, 22]. Our results reveal that 10 of the 63 tumors analyzed presented BRCAness phenotype. When crossed with clinico-pathological variables, we established that higher nuclear pleomorfism is a predictor for BRCAness in breast tumors. There also was a tendency toward younger age for this tumor phenotype. There is some controversy regarding the correlation between BRCAness and axillar lymph node status. In this regard, for instance, a report published by Foulkes et al. shows that patients with a BRCA1 mutated tumors do not have correlation with nodal status. On the contrary, Oonks et al. found that BRCAness tumors display a significantly higher number of patients without tumor positive lymph nodes [18]. When we analyzed nodal status among BRCAness and non-BRCAness tumors, we did not find significant difference between the two groups (p = 0.37). Even though BRCA1 mutated and BRCAness tumors are not the same, our observations are revealing that BRCAness tumors could behave as BRCA1 mutated tumors with regard to nodal involvement in line with Foulkes et al. observations.
Several studies have hypothesized that BRCA1 inactivation might also have a role in sporadic TN breast cancers [24–26]. We show here that TN tumors were principally BRCAness-like (p = 0.036), suggesting that they may have an underlying homologous recombination (HR) defect and therefore may benefit from PARPi therapies, when classified as BRCAness.
We expected that BRCAness tumors would show a distinct methylation profile; our experiments revealed 4 cancer-related genes which had a differential methylation profile in BRCAness tumors (Fig. 2a-d; Table 3). The epigenetic dysregulation of these genes has been implicated in several tumor types, but to the best of our knowledge, they have not been previously associated with BRCAness phenotype. Even though BRCAness phenotype has been proposed to be in part explainable by the methylation of BRCA1 (Lips et al.), we do not observe this association in our study probably due to analyses on different CpG sites. In fact, Lips et al. have shown that other still unknown mechanisms besides the methylation of BRCA1 could explain BRCAness phenotype. The methylation of MLH1 has been observed in colorectal and ovarian cancers. Particularly in ovarian cancer, MLH1 methylation associates with poor survival and chemoresistance [27]. The observation that MLH1 is methylated in BRCAness tumors suggests that they may have not only affected the HR repair pathway but also the MMR (mismatch repair) machinery. Due to the fact that BRCAness tumors are impaired in DNA repair, they may likely be more sensitive to DNA damaging agents. However, the absence of MMR repair proteins has been associated with resistance to damaging agents because of the dysregulation of MMR system. Therefore, the observation that MLH1 is methylated in BRCAness tumors is controversial. Perhaps the CpG site methylated in BRCAness tumors does not affect gene expression or maybe more CpG sites in the MLH1 promoter are needed to silence gene expression. Thus, additional experiments should be conducted to determine MLH1 gene and protein status in BRCAness tumors.
Promoter methylation of PAX5 seems to be an early event in breast tumorigenesis and high expression levels of CCND2 have been detected in ovarian tumors [28, 29].
It is interesting to remark that neither BRCA1 nor BRCA2 was part of the BRCAness methylation profile, but instead the negative regulator of BRCA1, ID4, was significantly unmethylated in BRCAness tumors. The role of ID4 in breast tumors is debated, where both tumor suppressor and oncogenic functions for this gene have been proposed [ 12, 24, 30]. The observation that ID4 is significantly unmethylated in BRCAness tumors could be indicating that there may exist an epigenetic dysregulation of ID4 in these tumors favoring an oncogenic function for this gene.
We determined the methylation status of two CpG sites in the ID4 promoter, one at −956 nt and another at −367 nt. Only one of them (−367 nt) was associated with BRCAness phenotype and appeared to impact on the regulation of gene expression (Fig. 3). The fact that the CpG site −956 is at a further distance from the TSS could suggest that this site does not affect gene expression. It has been shown that binding and transcriptional repression of methyl-CpG binding domain (MBD) proteins weakens as the distance between binding site and TSS increases [31]. But since the ID4 promoter has many CpG sites near the TSS, we consider that further research should be carried to accurately identify other CpG sites involved in ID4 gene expression, as these data will be important to make accurate experimental conclusions.
To enlighten the role of ID4 in breast tumorigenesis, we correlated the expression of ID4 with that of 32 tumor suppressor genes (Table 1). Of the 32 genes analyzed, we find that only two of them show a significant correlation with ID4 expression: MGMT and BRCA1. The expression of these genes showed an inverse correlation, indicating that when ID4 expression augmented the expressions of MGMT and BRCA1 diminished. A high correlation between ID4 and MGMT expressions has been proposed in Glioblastomas (GBM). In GBM, ID4 diminished expression identifies a subgroup of patients with better prognosis due to inhibition of angiogenesis [32]. In line with this, the observation that TN tumors have enhanced angiogenesis may likely be due to higher expression levels of ID4. In addition, GBM patients showing MGMT methylation had significantly longer overall survival than patients whose tumors presented unmethylated MGMT [32]. Our group has previously shown that MGMT is significantly methylated and ID4 significantly unmethylated in TN breast tumors [14]. Thus, both, ID4 unmethylation and MGMT methylation, could explain enhanced angiogenesis and poor overall survival characteristics of TN breast cancer patients.
The correlation between ID4 and BRCA1 gene expressions was initially described by Berger et al., who demonstrated that ID4 down-regulated BRCA1 expression in vitro [13]. Roldan and colleagues also observed an inverse correlation between the mRNA expressions of BRCA1 and ID4 in sporadic breast cancer [33]. Some authors have hypothesized a regulatory loop between ID4 and BRCA1 that permits the correct cycling levels of expression of both genes during normal cell division, a mechanism that may become disfunctional in many breast cancers [33–36]. Based on the data presented in this study, we propose that this regulatory loop between ID4 and BRCA1 may be regulated trough an epigenetic mechanism involving promoter methylation of ID4.
In conclusion, our experiments lead us to report for the first time that the unmethylation of ID4 is an epigenetic predictor of BRCAness phenotype in breast tumors. Our observations on 63 breast tumors are statistically solid, and we consider that future analyses should focalize on BRCAness tumors and/or cell lines, in order to better understand the role of ID4 in the phenotype regulation.
This knowledge is biomedically important because the accurate identification of BRCAness phenotype can have clinical implications, as patients with BRCA deficiency may benefit from cisplatin and PARPi therapies [37]. In this context, identifying a methylation profile that correlates with BRCAness status might eventually offer PARP inhibitors or DNA damaging agents to breast cancer patients, regardless of their BRCA mutation status.
Supplementary Material
Acknowledgments
This study was supported in part by the National Institute of Cancer (‘‘Instituto Nacional del Cancer’’), Argentina. RU was supported by funding from the National Institutes of Health (grant DK52913), the Mayo Clinic SPORE in Pancreatic Cancer (P50 CA102701), and the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567 to GL).
Compliance with ethical standards
Conflict of interest The authors declare no conflict of interest.
Ethical approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Human and animal rights This article does not contain any studies with animals performed by any of the authors.
Electronic supplementary material The online version of this article (doi:10.1007/s10549-015-3648-0) contains supplementary material, which is available to authorized users.
References
- 1.Larsen MJ, Thomassen M, Gerdes AM, Kruse TA (2014) Hereditary breast cancer: clinical, pathological and molecular characteristics. Breast Cancer (Auckl.) 8:145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domchek SM (2015) Evolution of genetic testing for inherited susceptibility to breast cancer. J Clin Oncol 33:295–296 [DOI] [PubMed] [Google Scholar]
- 3.Turner N, Tutt A, Ashworth A (2004) Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 4:814–819 [DOI] [PubMed] [Google Scholar]
- 4.Brekelmans CT, Seynaeve C, Menke-Pluymers M, Bruggenwirth HT, Tilanus-Linthorst MM, Bartels CC, Kriege M, Geel AN, Crepin CM, Blom JC, Meijers-Heijboer H, Klijn JG (2006) Survival and prognostic factors in BRCA1-associated breast cancer. Ann Oncol 17:391–400 [DOI] [PubMed] [Google Scholar]
- 5.Arnes JB, Brunet JS, Stefansson I, Begin LR, Wong N, Chappuis PO, Akslen LA, Foulkes WD (2005) Placental cadherin and the basal epithelial phenotype of BRCA1-related breast cancer. Clin Cancer Res 11:4003–4011 [DOI] [PubMed] [Google Scholar]
- 6.Apostolou P, Fostira F (2013) Hereditary breast cancer: the era of new susceptibility genes. Biomed Res Int 2013:747318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalasani P, Livingston R (2013) Differential chemotherapeutic sensitivity for breast tumors with ‘‘BRCAness’’: a review. Oncologist 18:909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedenson B (2005) BRCA1 and BRCA2 pathways and the risk of cancers other than breast or ovarian. MedGenMed 7:60. [PMC free article] [PubMed] [Google Scholar]
- 9.Carey JP, Knowell AE, Chinaranagari S, Chaudhary J (2013) Id4 promotes senescence and sensitivity to doxorubicin-induced apoptosis in DU145 prostate cancer cells. Anticancer Res 33:4271–4278 [PMC free article] [PubMed] [Google Scholar]
- 10.Fontemaggi G, Dell’Orso S, Trisciuoglio D, Shay T, Melucci E, Fazi F, Terrenato I, Mottolese M, Muti P, Domany E, Del BD, Strano S, Blandino G (2009) The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat Struct Mol Biol 16:1086–1093 [DOI] [PubMed] [Google Scholar]
- 11.Galatro TF, Uno M, Oba-Shinjo SM, Almeida AN, Teixeira MJ, Rosemberg S, Marie SK (2013) Differential expression of ID4 and its association with TP53 mutation, SOX2, SOX4 and OCT-4 expression levels. PLoS ONE 8:e61605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umetani N, Mori T, Koyanagi K, Shinozaki M, Kim J, Giuliano AE, Hoon DS (2005) Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene 24:4721–4727 [DOI] [PubMed] [Google Scholar]
- 13.Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P, Welch PJ, Welte K, King MC, Barber JR, Wong-Staal F (2001) Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA 98:130–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branham MT, Marzese DM, Laurito SR, Gago FE, Orozco JI, Tello OM, Vargas-Roig LM, Roque M (2012) Methylation profile of triple-negative breast carcinomas. Oncogenesis 1:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marzese DM, Gago FE, Vargas-Roig LM, Roque M (2010) Simultaneous analysis of the methylation profile of 26 cancer related regions in invasive breast carcinomas by MS-MLPA and drMS-MLPA. Mol Cell Probes 24:271–280 [DOI] [PubMed] [Google Scholar]
- 16.Lips EH, Laddach N, Savola SP, Vollebergh MA, Oonk AM, Imholz AL, Wessels LF, Wesseling J, Nederlof PM, Rodenhuis S (2011) Quantitative copy number analysis by multiplex ligation dependent probe amplification (MLPA) of BRCA1-associated breast cancer regions identifies BRCAness. Breast Cancer Res 13:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joosse SA, Beers EH, Tielen IH, Horlings H, Peterse JL, Hoogerbrugge N, Ligtenberg MJ, Wessels LF, Axwijk P, Verhoef S, Hogervorst FB, Nederlof PM (2009) Prediction of BRCA1-association in hereditary non-BRCA1/2 breast carcinomas with array-CGH. Breast Cancer Res Treat 116:479–489 [DOI] [PubMed] [Google Scholar]
- 18.Oonk AM, van Rijn C, Smits MM, Mulder L, Laddach N, Savola SP, Wesseling J, Rodenhuis S, Imholz AL, Lips EH (2012) Clinical correlates of ‘BRCAness’ in triple-negative breast cancer of patients receiving adjuvant chemotherapy. Ann Oncol 23:2301–2305 [DOI] [PubMed] [Google Scholar]
- 19.Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A (2005) Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res 33:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marzese DM, Gago FE, Vargas-Roig LM, Roque M (2010) Simultaneous analysis of the methylation profile of 26 cancer related regions in invasive breast carcinomas by MS-MLPA and drMS-MLPA. Mol Cell Probes 24:271–280 [DOI] [PubMed] [Google Scholar]
- 21.Urrutia G, Laurito S, Marzese DM, Gago F, Orozco J, Tello O, Branham T, Campoy EM, Roque M (2015) Epigenetic variations in breast cancer progression to lymph node metastasis. Clin Exp Metastasis 32:99–110 [DOI] [PubMed] [Google Scholar]
- 22.Lips EH, Mulder L, Oonk A, Kolk LE, Hogervorst FB, Imholz AL, Wesseling J, Rodenhuis S, Nederlof PM (2013) Triplenegative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br J Cancer 108:2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dell’Orso S, Ganci F, Strano S, Blandino G, Fontemaggi G (2010) ID4: a new player in the cancer arena. Oncotarget 1:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noetzel E, Veeck J, Niederacher D, Galm O, Horn F, Hartmann A, Knuchel R, Dahl E (2008) Promoter methylation-associated loss of ID4 expression is a marker of tumour recurrence in human breast cancer. BMC Cancer 8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De NA, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE (2010) Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol 28:1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, Savage K, Gillett CE, Schmitt FC, Ashworth A, Tutt AN (2007) BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 26:2126–2132 [DOI] [PubMed] [Google Scholar]
- 27.Suh DH, Kim MK, Kim HS, Chung HH, Song YS (2013) Epigenetic therapies as a promising strategy for overcoming chemoresistance in epithelial ovarian cancer. J. Cancer Prev 18:227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JY, Jeong W, Kim JH, Kim J, Bazer FW, Han JY, Song G (2012) Distinct expression pattern and post-transcriptional regulation of cell cycle genes in the glandular epithelia of avian ovarian carcinomas. PLoS ONE 7:e51592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moelans CB, Verschuur-Maes AH, van Diest PJ (2011) Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J Pathol 225:222–231 [DOI] [PubMed] [Google Scholar]
- 30.Crippa E, Lusa L, De CL, Marchesi E, Calin GA, Radice P, Manoukian S, Peissel B, Daidone MG, Gariboldi M, Pierotti MA (2014) miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PLoS ONE 9:e87039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatagnon A, Perriaud L, Nazaret N, Croze S, Benhattar J, Lachuer J, Dante R (2011) Preferential binding of the methyl-CpG binding domain protein 2 at methylated transcriptional start site regions. Epigenetics 6:1295–1307 [DOI] [PubMed] [Google Scholar]
- 32.Martini M, Cenci T, D’Alessandris GQ, Cesarini V, Cocomazzi A, Ricci-Vitiani L, De MR, Pallini R, Larocca LM (2013) Epigenetic silencing of Id4 identifies a glioblastoma subgroup with a better prognosis as a consequence of an inhibition of angiogenesis. Cancer 119:1004–1012 [DOI] [PubMed] [Google Scholar]
- 33.Roldan G, Delgado L, Muse IM (2006) Tumoral expression of BRCA1, estrogen receptor alpha and ID4 protein in patients with sporadic breast cancer. Cancer Biol Ther 5:505–510 [DOI] [PubMed] [Google Scholar]
- 34.de Candia P, Benera R, Solit DB (2004) A role for Id proteins in mammary gland physiology and tumorigenesis. Adv Cancer Res 92:81–94 [DOI] [PubMed] [Google Scholar]
- 35.Perk J, Iavarone A, Benezra R (2005) Id family of helix-loophelix proteins in cancer. Nat Rev Cancer 5:603–614 [DOI] [PubMed] [Google Scholar]
- 36.Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, Warrington JA, King MC (2002) BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci USA 99:7560–7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinbolt RE, Hays JL (2013) The role of PARP inhibitors in the treatment of gynecologic malignancies. Front Oncol 3:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Branham RL Jr (1990) Scientific data analysis. Springer, New York [Google Scholar]
- 39.Van Huffel S, Wandewalle J (1991) The total least squares problem: computational aspects and analysis. SIAM, Philadelphia [Google Scholar]
- 40.Branham RL Jr (2001) Astronomical data reduction with total least squares. New Astron Rev 45:649–661 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


