Abstract
Proteotoxic stress, that is, stress caused by protein misfolding and aggregation, triggers the rapid and global reprogramming of transcription at genes and enhancers. Genome-wide assays that track transcriptionally-engaged RNA polymerase II (Pol II) at nucleotide resolution have provided key insights into the underlying molecular mechanisms that regulate transcriptional responses to stress. In addition, recent kinetic analyses on transcriptional control under heat stress have shown how cells ‘prewire’ and rapidly execute genome-wide changes in transcription while concurrently becoming poised for recovery. The regulation of Pol II at genes and enhancers in response to heat stress is coupled to chromatin modification and compartmentalization, as well as to co-transcriptional RNA processing. These mechanistic features seem to apply broadly to other coordinated genome-regulatory responses.
Introduction
Cells and organisms encounter external and internal conditions that can disrupt homeostasis. Such stress conditions include elevated temperatures, toxins, hypoxia, cancer and aging, each of which compromise cellular functions by affecting proteins, membranes, and DNA (reviewed in1,2). To combat these adverse conditions, cells activate a rapid and transient programme that adjusts RNA and protein synthesis, cytoskeletal and membrane integrity, and the metabolic state of the cell (reviewed in1,2). In each cell, organelles employ autonomous signalling strategies that sense and communicate the stress to the cytosol, inducing stress-specific trans-activators and both specific and global transcriptional responses (reviewed in3–6) (FIG. 1). Besides cell-internal coordination, organisms integrate cellular stress responses and metabolic states of the animal (reviewed in7–8) (FIG. 1). Throughout the decades, the profound molecular and physiological changes in stressed cells have provided robust models for elucidating mechanisms of key cellular processes, including the genome-wide coordination of transcription upon heat stress9–11 (reviewed in6,8,12), the molecular basis for inhibition of translation during Unfolded Protein Response (UPR)13–14 (reviewed in 3,15), and the organismal control over cellular stress responses16–17 (reviewed in7,8). Additionally, properly regulated stress responses are vital for organismal health, as protein mis-folding and aggregation underlies neurodegeneration, while cancer cells can harness stress responses for improved cell survival and metastasis18–22 (reviewed in6,8,23).
Figure 1. Sensing, communicating and transcriptionally responding to protein-damaging stress.

Stress conditions in different compartments of the cell need distinct trajectories to communicate with the nucleus to provoke a transcriptional change. a) In the cytosol, heat stress causes heat shock proteins (HSPs) to release heat shock factor 1 (HSF1), which then can trimerize, bind to its target DNA elements and transactivate genes that encode chaperone machineries and polyubiquitin (reviewed in6,8). b) Oxidative stress inactivates prolyl hydroxylase domain-containing proteins (PHDs), which reduces the degradation of hypoxia-induced factor 1α (HIF1α), allowing HIF1α to dimerize with HIF1β and activate transcriptional programmes for induced angiogenesis and oxygen supply (reviewed in5). c) In the endoplasmic reticulum (ER), protein misfolding triggers the unfolded protein response (UPRER), which communicates from the ER lumen to the cytosol via the membrane-embedded proteins inositol-requiring protein 1 (IRE1), protein kinase R (PKR)-like ER kinase (PERK), and cyclic AMP-dependent transcription factor ATF6 (3,28,124–126 and references therein). Each of these three pathways leads to the generation of distinct transactivators (by activated splicing and translation of X-box binding protein 1 (XBP1), by translation control of ATF4 or by proteolytic processing of ATF6, respectively)3. d) In the mitochondria, misfolded proteins are first processed by protease complexes (grey sliced circle). The peptides move to the cytosol via ATP-binding cassette (ABC) transporters (white barrels), subsequently activating the mitochondrial UPR (UPRMT)-responsive transactivators ATF5 and DNA damage-inducible transcript 3 protein (DDIT3; also known as CHOP)4,127,128. In a and b, heat and hypoxia lead to a rapid activation of constitutively expressed transactivators that are maintained inactive by the complexes that sense misfolded proteins or levels of oxygen, respectively (reviewed in5,6,8). In c and d, compartment-restricted stresses are communicated over membrane barriers, which likely delays the transcriptional responses.
The cellular response to stress was first described in 1962 when chromosomal puffs, a hallmark of active transcription, were rapidly generated in heat-treated salivary glands of Drosophila busckii24. These heat-activated loci encode molecular chaperones [G] termed heat shock proteins (HSPs), that are produced at large scale in stressed cells, while global transcription and translation are reduced25–26 (reviewed in6). The HSPs, in turn, maintain homeostasis in stressed and unstressed cells by chaperoning protein folding, targeting damaged proteins for degradation, solubilizing protein aggregates, and preserving cellular structures and molecular functions6.
Mechanisms of transcriptional control have been elucidated by focused studies on model genes. However, recent advances in genome-wide methods have transformed conventional gene-centric analyses to characterization of regulatory mechanisms across the genome. Consequently, measures of the exact location of transcribing Pol II complexes across the genome have revealed the breath of gene and enhancer regulation upon heat and celastrol stresses, as well as the mechanistic steps at which the transcriptional reprogramming is executed9–11,27–28. Moreover, high-resolution chromatin analyses have uncovered stress-induced changes in nucleosome dynamics29, trans-activator binding10,21,30–32, and chromatin states11,27,29, both at transcribed and untranscribed loci.
In this Review, we summarize how transcriptional programmes are executed upon acute stress, focusing on mechanisms that globally regulate transcription from genes and enhancers [G]. Since the rapidly executed heat shock response has enabled detailed kinetic analyses on transcriptional control, we describe how RNA synthesis is orchestrated primarily in heat-stressed cells. We begin by outlining genome-wide changes upon heat stress in distinct species, before focusing on stress-induced kinetics and the molecular mechanisms that regulate RNA polymerase (Pol) II at promoters [G]. We then discuss the stress response at enhancers and the effects of stress on chromatin, before concluding with recent discoveries that couple transcriptional regulation to RNA processing in stressed cells.
Stress-induced genome-wide reprogramming
Dynamic coordination of transcription is essential for cellular processes, including responses to external and internal signals. At genes, transcription is controlled at multiple, potentially rate-limiting steps (Box 1) including chromatin opening, assembly of the pre-initiation complex (PIC), initiation of transcription, promoter-proximal pausing, elongation, termination and, finally, recycling of the transcription machinery (reviewed in33–34). Global run-on methods, such as Precision Run-On sequencing [G] (PRO-seq), have enabled the identification of the exact positions of transcribing Pol II complexes across the genome35–38. These advances were harnessed to map the transcriptional changes upon heat stress at nucleotide resolution in multiple species. Analyses of the positions of transcribing Pol II complexes in fly9, mouse10 and human11 revealed the rapid repression of thousands of genes, as evidenced by the reduction of Pol II molecules along coding sequences (schematically illustrated in FIG. 2A). While the response in different species is highly conserved, in mammals, Pol II accumulates at the promoter-proximal pause region of heat-repressed genes10,11, whereas in flies, heat stress causes decreased Pol II density across the whole gene9. From fly to human, the rapid activation of hundreds of genes is detected as increased Pol II density both at promoter-proximal regions and along the coding sequences (FIG. 2A).
Box 1. Steps of transcription.
Dynamic coordination of transcription is essential for cellular processes, including responses to external and internal signals. At gene promoters, transcription by RNA polymerase II (Pol II) requires multiple factors and is controlled at several, potentially rate-limiting steps. These include the following (reviewed in33–34):
Promoter opening
Nucleosome remodellers and histone-targeting enzymes increase the accessibility of the regulatory region.
Transcription initiation
A process whereby general transcription factors (GTFs) direct RNA polymerase (Pol) II to promoter or enhancer to form the pre-initiation complex (PIC).
Promoter-proximal pausing
A control step whereby transcriptionally-engaged Pol II pauses 20-50 nucleotides downstream of the transcription start site (TSS). The pause is stabilized against premature termination and elongation by negative elongation factors. Productive elongation is triggered by positive elongation factors, particularly the P-TEFb kinase.
Transcriptional elongation
The progress of Pol II along the DNA template during which nucleotides are added to the nascent RNA chain.
Co-transcriptional processing
The nascent transcript can undergo several modifications as the transcription proceeds:
-
i)
5′-capping adds a methylated guanoside (m7G) to the 5′-end of the nascent RNA as it emerges from the Pol II complex, stabilizing the RNA.
-
ii)
Co-transcriptional splicing can remove introns rapidly after the 5′-end of exon emerges from the transcription bubble.
-
iii)
Cleavage and polyadenylation of the primary transcript adds an untemplated poly(A) stretch to the 3′-end of the RNA, protecting the RNA against degradation and functioning as a signalling moiety.
Transcription termination
Transcription proceeds beyond the cleavage and polyadenylation site; however, the cleavage leaves an unshielded 5′-end to the nascent RNA that Pol II synthetizes, causing its degradation, and termination of transcription.
Recycling
Transcription machinery can be recycled from the 3′-end of the gene to the TSS, for which two non-exclusive models have been presented:
-
i)
Chromatin looping physically brings the 3′-end to the instant vicinity of the TSS.
-
ii)
Chromatin compartmentalization generates an insulated region where transcription machinery is retained in high concentration.
Beyond promoters, distal regulatory elements orchestrate transcription by providing signalling hubs and modulating chromatin compartmentalization (reviewed in80,90).
Figure 2. Heat shock response triggers transcriptional repro o ramming of genes and enhancers across the genome.
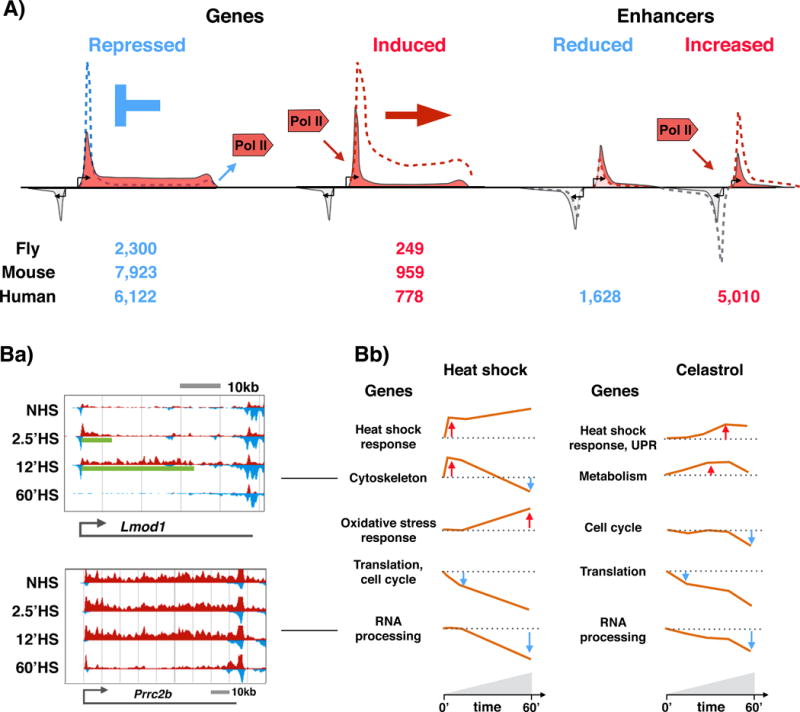
A Schematic representation of RNA polymerase II (Pol II) distribution at heat-responsive genes and enhancers. Heat stress provokes repression of thousands and induction of hundreds of genes in flies9, mice10 and humans11. In mammals, the repression is mediated by inhibiting the release of paused Pol II into elongation, causing accumulation of Pol II at the pause site (blue dashed line. In flies (nuoe.t shown), the heat repression causes decreased Pol II density across the gene. From fly to human, the rapid activation of genes is triggered by releasing promoter-proximal paused Pol II into elongation (red dashed line). Concurrent with reprogramming of genes, the enhancer landscape is re-profiled in eat-stressed cells, provoking educed or increased Pol II density at thousands of enhancers11. Gene transcription is depicted with red; divergent transcription with light grey. At enhancers, both strands (indicated with red and grey) produce short and unstable transcripts. B) Temporal patterns of heat-responsive or celastrol-responsive gene regulation. Ba) Browser-shot examples of rapidly, but transiently, expressed Lmod1 (upper panel) and late repressed Prrc2b (lower panel) genes in wild-type mouse cells under normal conditions (0) and under heat stress of different duration (2.5, 12 and 60 minutes). The transcriptional profile of the plus (red) and minus (blue) strand is depicted. The green line under the transcription profiles (upper panel) indicates the distance that the heat-induced Pol II complexes have travelled at the indicated time point. Bb) Five major temporal patterns appear in heat-shocked (left panels) or celastrol-treated (right panels) cells. The orange line recapitulates the average fold change with respect to the basal transcription level (dotted line) in each category. The main gene ontology terms associated, with each group are indicated to the left of the profile. UPR, unfolded protein response. Panels in Ba and Bb are adapted with permission from10 and 28.
Concurrent with the reprogramming of genes, the transcriptional profiles of enhancers undergo large-scale changes in heat-stressed human cells11, exhibiting reduced or increased Pol II density at thousands of enhancers (FIG. 2A) The changes in gene and enhancer expression are accompanied by changes in chromatin environment at transcribed and untranscribed loci, including nucleosome dynamics29, chromatin sumoylation27, histone acetylation11,29, and trans-activator binding9–11,21,30–32. These changes to chromatin dynamics demonstrate that acute stress triggers massive reshaping of not only gene transcription and the regulatory element landscape but also chromatin structure, which we now discuss below.
Reprogramming of gene expression
Temporal patterns of gene expression changes
Many studies have analysed heat-induced changes in mRNA or protein expression using microarray, mRNA-seq, mass spectrometry, or antibody based methods21,31–32,39–42 (reviewed in1,43). Collectively, these studies have identified similar classes of heat-induced or heat-repressed genes across species, revealing core processes that need to be adjusted in stressed cells (reviewed in1,43). Heat-induced genes encode chaperones and proteases, metabolic regulators, DNA and RNA repair machinery, signalling molecules and transport proteins (reviewed in1,43). Heat-repressed genes, by contrast, have gained less focus than the heat-induced genes, primarily due to limitations in inferring transcriptional changes, particularly repression, from steady-state RNA and protein levels. However, the heat-repressed genes have been reported to comprise translation machinery, cell growth regulators and RNA processors39–40 (reviewed in43).
The limitations for quantitative and temporal analyses of transcription were recently overcome by studies that used PRO-seq to map profiles of nascent transcription9–11,28. These studies largely confirmed the results of steady-state RNA analyses, additionally discovering a massive downregulation of gene expression upon treatment with either heat (FIG. 2A, 2B) or the small molecular compound celastrol [G] (FIG. 2Bb). Moreover, the temporal analyses of Pol II progression along the genes revealed several different patterns of gene expression9–11,28 (FIG 2B). From fly to human, components of the translation machinery are transcriptionally repressed within 10 minutes of stress exposure, whereas RNA-processing complexes show a more delayed decline (FIG. 2B). Furthermore, the heat-induced progression of Pol II along activated genes can be detected within 2.5 minutes of heat stress (FIG. 2B), indicating instant responsiveness of genes that encode chaperone machineries and, in mouse fibroblasts, cytoskeletal components10. Intriguingly, cytoskeletal genes are transiently induced, producing a burst of expression, after which their transcription diminishes below basal expression10 (FIG. 2B). This sequential activation and inactivation during heat stress demonstrates that the diverse responses of gene classes to stress require sophisticated control mechanisms.
The analyses of nascent transcription upon heat9–11,27 and celastrol28 enable a direct comparison of transcriptional changes to distinct stresses. Somewhat surprisingly, heat and celastrol induce strikingly similar patterns of transcription10–11,28 (FIG. 2B): In essence, celastrol-triggered transcriptional changes follow slower kinetics and cause smaller fold enrichments than heat shock, but also trigger synthesis of chaperone complexes and repress production of translation machinery and RNA-processing proteins28. An intriguing difference in celastrol-exposed cells compared with heat-treated cells is the selective induction of components of the unfolded protein response [G] of the endoplasmic reticulum (UPRER) (FIG. 2B). These comparisons indicate that heat and celastrol stresses activate the same pathways to increase expression of cytosolic chaperones, but differ in their effect on cellular compartments, such as ER. Measuring transcription dynamics from distinct stresses is particularly informative for elucidating the mechanisms that prepare and execute genome- wide changes in gene expression, and for identification of compartment-specific pathways. Given that patho-physiological conditions are associated with a variety of stressors, understanding the molecular details of each response is critical in clarifying causes of stress-related pathological conditions, and particularly, for targeted prevention of disease states.
Regulation at stress-activated genes
The rapid and robust induction of HSP gene expression upon heat stress has served as a model for transcriptional stress responses, fostering many groundbreaking insights into the regulatory steps of gene expression (Box 1; reviewed in6,8,12). An important part of these analyses has been the real-time in vivo imaging of the sequential recruitment of transcription factors, chromatin remodellers, and Pol II to the activated Hsp70 promoter in polytene chromosomes of Drosophila melanogaster44–47. Importantly, genome-wide assays that measure nascent transcription9–11, nucleosome localization29,48 and positioning of chromatin-bound regulators9–11,49–50 have shown that the mechanisms that govern the regulation of HSP genes are largely generalizable to other heat-activated genes, and likely also to inducible transcriptional changes in general, such as detected during development (reviewed in34). These regulatory events include the action of chromatin remodellers at the promoter and along the gene29,48, the recruitment dynamics of transcription factors9–11,21,30–32, establishment and maintenance of promoter-proximal Pol II pausing9,51, and the release of paused Pol II into productive RNA elongation9–11,27–28.
Promoter opening and pre-establishment of directionality at heat-activated genes
Transcription requires accessible chromatin and, accordingly, the majority of heat-induced genes have established an open promoter state prior to the stress exposure9,11,31–32. In Drosophila melanogaster, GAGA-associated factor (GAF), which recognizes GAGA-rich sequences, is essential for maintenance of the nucleosome-free chromatin state51, as well as pausing of Pol II at a number of heat-responsive promoters9 (FIG. 3a). GAF has been shown to interact with chromatin remodelers, including NUcleosome Remodeling Factor (NURF)52, which catalyses the sliding of nucleosomes and is required for proper expression of hundreds of genes53–54. In mammals, Replication Protein A (RPA) and FACilitates Transcription (FACT) create nucleosome-free regions at heat-responsive promoters55 (FIG. 3b).
Figure 3. Promoter opening, establishment of directionality and rapid release of paused RNA polymerase II.

Before heat shock, GAGA-associated Factor (GAF) and nucleosome remodelling factor (NURF) create nucleosome-free promoters at heat-induced genes in Drosophila melanogaster, where strong promoter elements direct the pre-initiation initiation complex (PIC), and hence RNA polymerase II (Pol II), to the coding strand (part a). In mammals, replication protein A (RPA) and the FACT (facilitates chromatin transcription) complex contribute to promoter opening (part b). Preferential binding of the PIC to the core promoter of the coding strand creates directionality by orienting Pol II towards the gene. The highly paused and highly directional genes are rapidly activated by releasing paused Pol II into productive elongation in flies (part c) and mammals (part d). The pause-release is triggered by heat-activated transactivators, such as heat shock factor 1 (HSF1) (parts c and d) and possibly serum response factor (SRF) (part d), through (direct or indirect) recruitment of positive transcription elongation factor b (P-TEFb) and chromatin modifiers. Nucleosome remodelling by FACT and acetyltransferases allows efficient Pol II progression along the gene. Sumoylation (S) and poly(ADP-ribose) polymerase (PARP1)-mediated parylation can modify transcriptional regulators and facilitate chromatin compartmentalization. The arrows indicate recruitment of Pol II (grey), transactivators HSF1 and SRF (red) and P-TEFb (purple). The shaded grey areas demonstrate Pol II density. Ac, acetylation; DSIF, DRB sensitivity-inducing inducing factor; NELF, negative elongation factor; P, phosphorylation.
Once chromatin is accessible, general transcription factors (GTFs) can bind to the DNA and position Pol II at the transcription start site (TSS). In flies, strong promoter elements, such as TATA box and initiator (Inr), are localized close to the TSS of the coding strand36, positioning the pre-initiation complex (PIC) upstream of the gene (FIG. 3a,c). A very small amount of divergent transcription [G] occurs in fly promoters36–37; however, it is particularly evident in mammals35,56, where two core promoters [G] initiate transcription in opposing directions, one towards the gene (sense) and the other upstream (FIG. 3b,d). The two coupled core promoters harbour a similar number and order of DNA elements36,57, and consequently, mammalian promoters need to establish direction-specific signals to efficiently initiate stable transcription of the gene, while upstream transcription of the anti-sense strand produces short and unstable transcripts35. High-resolution chromatin immunoprecipitation assays, such as ChIP-exo58 and ChIP-nexus59, have provided valuable insights into positioning of transcriptional complexes within the regulatory element. Examining this high-resolution promoter architecture [G] with respect to the gene’s activity — or ability to fire upon heat stress — revealed that positioning of PIC to the core promoter of the coding strand mediates transcriptional directionality at divergent promoters11 (FIG. 3b). The highly directional transcription at divergent promoters of both highly transcribed and heat-induced genes demonstrates that the cell can distinguish between core promoters of a single regulatory element to efficiently direct Pol II towards the gene.
Promoter-proximal pausing enables rapid and synchronous gene activation
One of the most striking features of the transcription of the Drosophila model gene Hsp70 is its promoter-proximal pause and the regulated release of Pol II into elongation60–61, now known to be a necessary step at the vast majority of genes in flies and mammals9–11,27–28,35–38,62–64. Indeed, Pol II pausing has emerged as a major regulatory step that enables rapid and synchronous activation (reviewed in34), as well as global repression10–11,28 of genes. Besides nascent RNA sequencing, high-resolution ChIP assays58–59 and permanganate mapping of transcription bubble (PIP-seq)49–50 indicate that the majority of Pol II molecules at the 5′-region of the gene reside at the promoter-proximal pause site. In fact, a relatively large fraction of the total transcriptionally engaged Pol II on the genome resides at the promoter-proximal pause site36–37. Together, a variety of genome-wide assays have established that the coordinated release of promoter-proximal paused Pol II into productive elongation and the subsequent recruitment of new Pol II molecules to the promoter region serve as the major regulatory events of transcription, determining the repertoire of active (or inducible) genes, as well as the level of their mRNA biogenesis.
At the promoter-proximal pause site, 20-50 nucleotides (nt) downstream of the TSS, the Pol II complex is stabilized by Negative ELongation Factor (NELF) and unphosphorylated DRB Sensitivity-Inducing Factor (DSIF), which prevent premature termination by increasing the residence time of Pol II at the pause site (reviewed in33–34). Despite slight differences in the molecular machineries that execute promoter opening and create directionality in fly and mammals, the result of these actions at heat-responsive gene promoters is conserved, that is, the establishment of highly-directional, highly-paused genes that are prewired for rapid and synchronous activation (FIG. 3a, 3b).
HSF1-dependent and HSF1-independent gene activation
The activator of HSP genes in eukaryotes is Heat Shock Factor protein 1 (HSF1), an evolutionarily conserved transcription factor that is essential for production of heat-induced chaperones65–67 (reviewed in6,8). Beyond stress, HSF1 is activated in a variety of conditions, such as development, cancer and metabolic processes, which positions HSF1 at the crossroads of sensing and responding to cellular states (FIG. 1). HSF1 is held in an inactive monomeric state through interactions with chaperones (reviewed in6,8,12). These interactions are relieved when protein-damaging conditions sequester chaperones and release HSF1 to its active trimeric state (FIG. 1). As a trimer, HSF1 binds tightly to DNA, and recruits chromatin remodelers and factors such as Positive Elongation Factor b (P-TEFb) (reviewed in6,8,12). Of note, HSF1 drives distinct gene expression programmes in different physiological conditions (reviewed in8), indicating that the genome-regulatory mechanisms of HSF1 extent well beyond stress responses. Indeed, chromatin structure, composition of factors co-localizing at HSF1-binding sites11,32, developmental stage, and the cell cycle phase31,68–69 each create unique environments that affect the binding and trans-activation capacity of HSF1.
The instant responsiveness of chaperone genes to heat stress is mediated via the open chromatin state and pre-established paused Pol II (FIG. 3a, 3b). HSF1 has been reported to participate in the establishment and maintenance of the open chromatin state via direct interactions with the SWI/SNF complex70, FACT55,71, and Tip6071, and through indirect mechanisms that promote acetylation of histone H472–73 and H2A71. The primary function of HSF1 upon stress, however, seems to be to release paused Pol II into productive elongation9–10 (FIG. 3c, 3d).
The importance of HSF1 for trans-activating genes that are highly induced upon heat stress has become evident from analyses of mRNA expression (reviewed in8,12) and nascent transcription9–11. The transcriptional response to stress, however, extends beyond HSF1-activated genes, as transcription of the majority of heat-induced genes does not depend on HSF19–10,74. Furthermore, global genome-wide repression of transcription occurs independently of HSF19–10, underscoring the importance of multiple pathways and executor molecules for the global coordination of transcriptional stress responses. In mouse fibroblasts, Serum Response Factor (SRF) was recently shown to mediate the rapid induction of cytoskeletal genes, exemplifying an HSF1-independent trans-activator that triggers reprogramming of RNA synthesis in heat-stressed cells (FIG. 3d). Intriguingly, SRF binding upon heat shock is transient10, which implies extremely dynamic mechanisms that control both activation and inactivation of SRF during heat stress.
Release of paused Pol II and recruitment of new Pol II
At the Hsp70 gene in Drosophila melanogaster, the heat-induced release of paused Pol II into elongation requires HSF1-dependent recruitment of P-TEFb75. Since P-TEFb-mediated phosphorylation of NELF and the C-terminal domain (CTD) of Pol II are essential for gene expression in mammals (reviewed in76), it is tempting to speculate that increased P-TEFb concentration at the heat-induced genes could overcome the global transcriptional repression. The entry of new Pol II molecules to the TSS is limited by the rate of release of Pol II into productive elongation77, as might be expected from the position of the pause site and the size of the Pol II complex, which causes steric hindrance for new initiation78. Coupling P-TEFb-mediated pause-release to the entry of new Pol II78 molecules might be an important mechanism that drives high transcriptional activity, such as that triggered by heat shock.
Regulation at stress-repressed genes
The genome-wide transcriptional repression in heat- and celastrol-stressed cells occurs via inhibiting the release of promoter-proximal Pol II into elongation (FIG. 2)10–11,28. The reduced entry into elongation, subsequently, causes transcription machineries to clear off from the gene bodies of thousands of repressed genes (FIG. 2)9–11,28. In mammals, which harbour long genes, this clearance is likely to cause a profound increase in the pool of unengaged Pol II, (FIG. 4a) which could be readily exploited to rapidly fill freed pause sites at stress-induced genes (FIG. 4b). In addition to Pol II accumulation at induced genes, its density increases also at the pause sites of repressed genes (FIG. 4c), where the presence of PIC is directing Pol II molecules to the TSS, but inhibited release from the pause site causes Pol II to accumulate10–11,28 (FIG. 2A and 4c). This increased pausing of repressed genes could be, in essence, an efficient mechanism to ensure their rapid re-activation once the stress is ameliorated. In flies, where the genes in general are short, the clearance of Pol II from heat-repressed genes frees a more limited pool of unengaged Pol II9, which is likely needed in its entirety at heat-induced genes. As a consequence, the global redistribution of Pol II is less prominent in flies, despite the largely similar mechanism that coordinate the Pol II molecules genome-wide. The molecular events that inhibit Pol II pause-release at thousands of genes upon stress remain to be shown, but are likely to involve sequestering and/or limited activity of P-TEFb or some other general component of the transcriptional regulatory machinery.
Figure 4. Redistribution of transcription machinery upon heat shock.
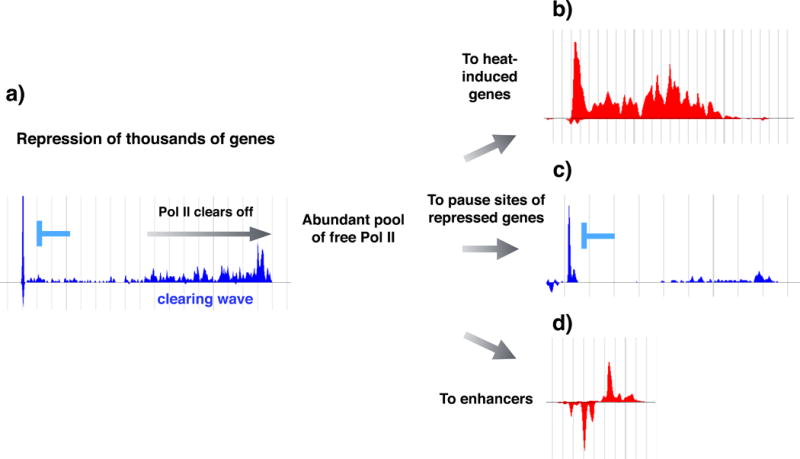
Inhibition of pause-release causes RNA polymerase II (Pol II) to clear off from thousands of gene bodies (part a). In mammals, where genes are generally long, the clearing of Pol II causes an abundant pool of unengaged Pol II in the cell. The increased availability of Pol II can be utilized for rapid filling of the pause sites at heat-induced genes where the rate of pause-release profoundly increases (part b), as well as the filling of the available pause sites at heat-repressed genes where inhibition of pause-release causes high promoter-proximal pause signal (part c). The increased availability of Pol II is likely to be also contributing to the tuning up of the enhancer repertoire, as an increased number and transcription of enhancers are detected upon heat11 and celastrol28 stresses (part d). The Pol II profile at these example genes and enhancer is obtained from the raw data presented in ref.11, where human K562 cells were subjected to a 30-minute heat stress at 42 °C. Transcriptionally repressed genes (parts a and c) are indicated in blue, and a transcriptionally induced gene (part b) and enhancer (part d) are depicted in red.
Genome-wide changes in the enhancer landscape
Enhancers are regulatory elements that can activate gene transcription over large distances, and they have emerged as important definers of cell-type specific patterns of transcription79, (reviewed in80–81). With the advent of high-throughput sequencing, enhancers have been shown to produce short and unstable transcripts termed enhancer RNAs (eRNAs)82–83, and to share many structural features with promoters35,84–85. Especially in mammals35, but also in flies36, eRNAs are in general produced from divergent regulatory elements, which harbour two core initiation regions [G] that launch transcription in opposing directions. The exact coordination of enhancer transcription and its gene regulatory activity is currently elusive, however, transcribed enhancers in general produce unstable and short transcripts to both directions35 (FIG. 2A). Interestingly, enhancers harbour a similar composition, yet lower levels, of trans-activators and transcription machinery than promoters35,11, suggesting comparable regulatory events at genes and enhancers. Furthermore, enhancer transcription is coordinated by P-TEFb and DSIF-mediated pause-release84, emphasizing the generalities of transcriptional mechanisms between transcribed regulatory elements.
Remodelling of the enhancer repertoire and eRNA production
With PRO-seq, the genome-wide enhancer transcription was measured at a nucleotide resolution in non-stressed and stressed human cells11,28. This direct quantification of eRNA synthesis uncovered the immediate establishment of a distinct repertoire of transcribed enhancers under acute stress11, indicating genome-wide changes in enhancer activity. Concurrent with the transcriptional reprogramming of genes, heat stress or celastrol treatment cause an overall increase in Pol II density at enhancers11,28. Since enhancer transcription, in general, correlates with its enhancing capacity84–85, the increased transcription at many enhancers likely indicates their increased activity11,28. However, the collectively higher transcription at enhancers upon heat or celastrol treatment may reflect the increased availability of Pol II in the cell (discussed above), contributing to the rapid changes in enhancer transcription (FIG. 2A, FIG. 4). Beyond the enhancers that show either induced or reduced Pol II density along the regulatory element length (FIG. 2A), a subset of enhancers show restricted progression of Pol II upon heat stress11. This changed pattern of Pol II elongation at enhancers could indicate that regulation of Pol II progression acts as an indicator, or possibly a rate-limiting step, in establishing enhancer activity. Indeed, given the recently identified P-TEFb-mediated pause-release at enhancers in Drosophila melanogaster84, it seems plausible that enhancer transcription is also globally coordinated at both initiation and pausing. However, like the functional relevance of eRNAs, the consequences of Pol II pause-release for enhancer activity remains to be shown.
Lineage-specific factors mark enhancers
While promoters are marked for rapid activation by abundant levels of GTFs and highly paused Pol II, heat-activated enhancers harbour much lower levels of transcription machinery under normal conditions11. Instead, heat-induced enhancers contain high levels of lineage-specific factors, such as GATA-binding proteins (GATA1 and GATA2) and TAL BHLH Transcription Factor 1 (TAL1) in non-stressed human K562 cells11. Lineage-specific factors are important determinants of the enhancer repertoire during development81, and their preexistence at stress-responsive enhancers indicates similar mechanisms that establish enhancer responsiveness during stress and development. As waves of enhancer transcription precede or co-occur with changes in gene expression program during development86 and stress11,28, the general mechanisms of lineage-specific factors likely involve defining condition-specific gene expression programs by preparing the repertoire of inducible enhancers.
HSF1 activates enhancer transcription
Genome-wide profiling of HSF1 target sites has shown that the majority of HSF1-bound regions lie distal to gene promoters10,21,30–32. By comparing HSF1 target sites to the coordinates of transcribed regulatory regions, HSF1 was found to be recruited upon sensing of heat to hundreds of heat-activated enhancers11. Whether the role of HSF1 at enhancers is to induce eRNA production, reach to paused Pol II at the promoter-proximal region, or to increase the local concentration of transcriptional regulators in the connected promoter–enhancer hubs, remains to be shown. However, previous results have demonstrated that Pol II pause-release can be triggered from enhancers87–89, and that HSF1 contributes to the transcriptional induction as well as to the recruitment of transcription machinery and chromatin modifiers47,70–73,75. All of these features of HSF1 activity could be exploited at enhancers. Since enhancers define the cell-type specific patterns of transcription, it is likely that at least a subset of the enhancer-mediated transcriptional reprogramming upon stress contributes to cell-type-specific stress responses. In comparison, the ubiquitously heat-induced HSP genes across eukaryote species gain HSF1 binding to the promoters, suggesting that the conserved, promoter-driven response to heat stress could be complemented with enhancer-dependent specificity. The mechanisms by which HSF1 drives transcriptional programs via promoters and enhancers are likely to extend from stress to other genome-regulatory responses, such as detected during development68 and cancer progression21.
Chromatin remodelling upon stress
Genomes are three-dimensionally organized structures where the accessibility of DNA elements, and the proximity of distal regions are coordinated in space and time (reviewed in90). The connectivity network of the genome is composed of both short and long-range interactions, which provide the organizational platform for transcriptional regulation and enhancer-promoter interactions90. Transcriptional reprogramming upon stress co-occurs with global changes in chromatin landscape at genes, enhancers and untranscribed regions11,27,29. Moreover, heat-induced changes in connectivity and organization of the genome have been reported in yeast91 and fly92.
Histone modifications at transcribed and untranscribed loci
Rapid stress-induced gene activation is accompanied by the dynamic remodelling of nucleosomes, not only at promoters but also along coding regions29,47–48,71. The rapid regulation of chromatin state across the transcribed genome has been globally demonstrated during the UPRER; upon its activation the majority of promoters of active genes and their gene bodies show increased accessibility29. The increased nucleosome accessibility at genes involves histone acetylation11,29, which loosens the DNA-nucleosome interactions, subsequently facilitating transcription factor binding and the progression of Pol II (reviewed in93). Importantly, histone acetylation occurs also at transcriptionally activated enhancers and at HSF1-bound genomic regions that do not initiate detectable levels of transcription11. Whether the untranscribed islands of histone H4 acetylation are enhancers without transcriptional activity, architectural regions for genomic interactions, or platforms for transcription factor binding, are currently unknown. However, the instant reprogramming of enhancers and the appearance of untranscribed islands of histone H4 acetylation indicate reshaping of the regulatory element landscape in stressed cells, and highlights the dynamic interplay of genes, distal regulatory elements, and the chromatin state.
Chromatin compartmentalization
Genome architecture is mediated via protein-mediated chromatin looping that creates domains in which regulatory processes can be coordinated and molecules or histone modifications concentrated or depleted90. Such looping has been reported to occur via CCCTC-binding Factor (CTCF)94, cohesin95, and Yin-Yang factor 1 (YY1)96. Beyond the interactions between protein bound DNA elements, chemical compartmentalization, such as parylation [G], sumoylation [G], and phase separation [G], can create insulated regions (reviewed in97–99), which we discuss here below.
The ordered recruitment of trans-activators, remodelling complexes and Pol II at the Hsp70 gene in D. melanogaster occurs concurrently with compartmentalization of the locus44,47,71. This heat-induced compartmentalization is mediated by polyADP-Ribose Polymerase (PARP), which generates linear or branched polymers of ADP-Ribose (PAR) (FIG. 3c), and is associated with chromatin decondensation100–101. The produced network of PAR molecules (FIG. 3c) can retain transcription machinery in the locus, thereby enabling efficient recycling of Pol II and high levels of transcription47. Parylation at the HSP70 locus has been demonstrated also in human cells102–103, and HSF1-dependent PARP1 recruitment was recently shown to occur during DNA-damage response104. At heat-activated genes, the increased levels and spreading of PARP1 is thought to create an insulated region for transcription47,71, whereas during DNA-damage response, HSF1-dependent PARP1-recruitment to heat shock element enables efficient repair at near-by loci104
Besides parylation, conjugation of Small Ubiquitin-like MOdifier (SUMO) to chromatin-bound proteins has been suggested to contribute to formation of chromatin domains and insulate regions during stress (reviewed in98,105). Sumoylation of the proteome is, indeed, a hallmark of heat, osmotic, ethanol, metabolic and genotoxic stresses105–109, and a large-scale, selective chromatin sumoylation occurs upon heat shock, particularly at active genes and Pol II pause sites27. As a number of chromatin bound proteins from trans-activators to chromatin modifiers and histones become sumoylated during stress108,110, the chromatin sumoylation likely targets a multitude of individual proteins. Intriguingly, parylation at the chromatin has been coupled to sumoylation of PARP1102–103, indicating potential crosstalk between chemical moieties, perhaps adding regulatory layers to condition-specific chromatin compartmentalization.
The model on transcriptional compartments is particularly important in terms of enhancer-promoter connectivity, as transcriptional hubs, constituting of genes and their regulatory elements, are proposed to create phase-separated domains where transcriptional components are both concentrated and effectively retained111. Indeed, one of the mechanisms of enhancer-mediated transcriptional control involves increasing the local concentration of the transcription machinery, which could be coordinated by compartments that retain, or restrict, the access of transcriptional regulators112. The dynamic formation of chromatin domains likely involves protein networks including architectural proteins90, transcription machinery and factor binding113, as well as chemical moieties at the chromatin98,105.
Co-transcriptional processing
Retention of introns versus co-transcriptional splicing
The discovery of co-transcriptional splicing has profoundly increased our understanding of how transcription is intricately intertwined with RNA processing (reviewed in114). Upon heat shock, both splicing and polyadenylation have been shown to be globally reduced115–117. Co-transcriptional splicing was recently measured for individual genes in mouse cells, showing that severe 44°C heat treatment causes retention of introns at 27% of transcripts that have at least two introns and adequate levels of expression for quantification by RNA-seq117. In general, transcripts that retained introns remained in the nucleus during stress exposure (FIG. 5), and their encoded proteins are known to be involved in various processes of gene expression, such as tRNA synthetases and components of spliceosome117. The genes encoding these transcripts showed high mRNA levels prior to the heat exposure117; and therefore, their adequate production and storage as unspliced transcripts during stress might ensure rapid recovery. In contrast, heat-induced genes were co-transcriptionally spliced and translated to proteins117, indicating highly selective mechanisms that recognize genes for rapid heat-induced synthesis of mature mRNA (FIG. 5).
Figure 5. Co-transcriptional processing in heat-stressed cells.
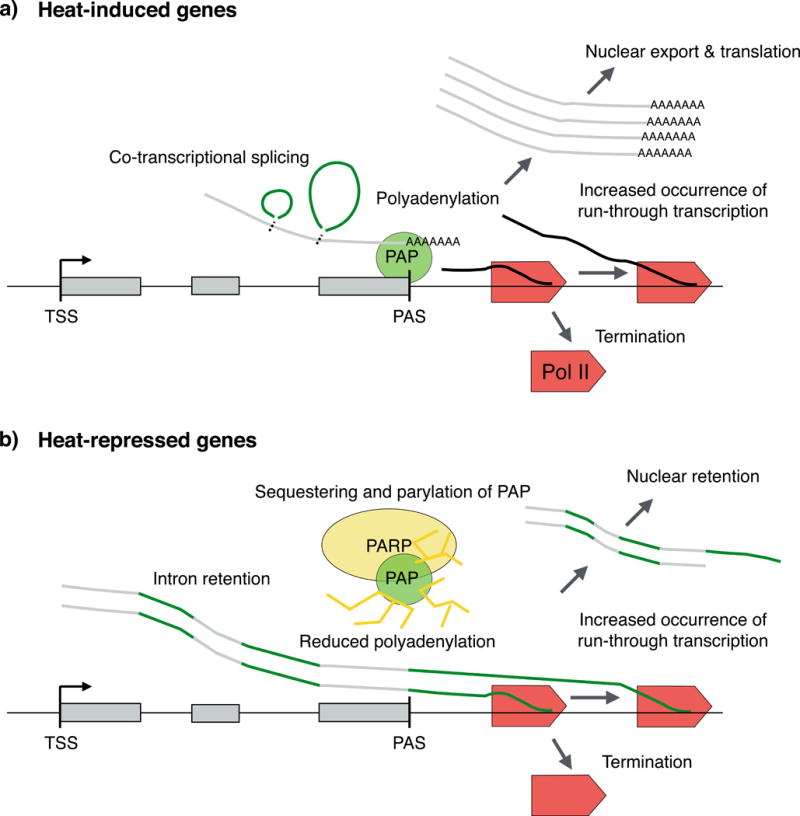
Transcripts of heat-induced genes are co-transcriptionally spliced, efficiently polyadenylated and exported to the cytosol (part a). By contrast, transcripts of a number of uninduced genes retain introns, show reduced polyadenylation and increased readthrough transcription, and are retained in the nucleus during heat exposure (part b). Retention of intron-containing transcripts could be a mechanism to store pre-mRNA until the stress is ameliorated and cellular processes are restored. PAP, poly(A) polymerase; PARP1, poly(ADP-ribose) polymerase 1; PAS, polyadenylation and cleavage site; Pol II, RNA polymerase II; TSS, transcription start site.
Read-through transcription and polyadenylation in heat-stressed cells
Transcription terminates when Pol II proceeds beyond the polyadenylation and cleavage site (PAS) where the transcript is cleaved, and Poly(A) Polymerase (PAP) synthesizes an untemplated adenosine stretch to the 3′-end of the RNA (reviewed in118). After cleavage, Pol II continues transcription, but is ‘chased down’ by exonucleases that target the subsequently un-capped nascent RNA. Upon osmotic, oxidative or heat stress, transcriptional patterns of thousands of genes show prolonged Pol II progression, extending several kilobases further downstream of the PAS than observed in non-stress conditions119. Reduced poly(A)-processing has been coupled to read-through transcription120–122, and RNA-seq studies indicate that read-through transcription, indeed, produces long RNA molecules that mostly remain in the nucleus119 (FIG. 5).
Reduced polyadenylation in heat-stressed cells involves PARP1-mediated parylation and subsequent removal of PAP from the 3′-ends of non-induced genes116. How the heat-induced genes evade parylation of PAP and its release from the RNA is not currently known. However, PARP1 levels have been shown to increase at the 5′-, and reduce at the 3′-end of heat-induced genes upon stress, suggesting tightly regulated PARP1 localization along the gene116. The heat-induced repression of splicing could be coupled to increased read-through transcription and reduced polyadenylation, producing long intron-containing transcripts that are retained in the nucleus (FIG. 5), perhaps until the stress conditions are reversed and the cell restores its basal functions, including splicing and translation. The heat-induced transcription, by comparison, is coupled to co-transcriptional splicing, efficient polyadenylation, translocation into the cytosol and abundant translation, demonstrating how coupled transcription and RNA-processing can be streamlined for swift protein synthesis.
Conclusions and perspectives
Rapidly provoked stress responses have paved the way for elucidating key cellular and biological processes. Over the past several years, genome-wide characterizations of nascent transcription, chromatin state and co-transcriptional processes have fuelled a change from gene-centric analyses to genome-wide measuring of transcriptional control. The major transcriptional discoveries from stress responses include the ordered regulatory events that orchestrate heat-induced transcription, as well as the identification of promoter-proximal pausing of Pol II as the major step at which this rapid and global reprogramming is defined. Moreover, with the ability to map the exact locations of transcription complexes, the rules that create transcriptional directionality are becoming evident, and identification of enhancers and measures of their eRNA production can be conducted at nucleotide resolution.
Despite recent advances, there are a number of questions that remain to be answered. While pre-established chromatin architecture and trans-activator binding triggers transcription upon stress9–11, the molecular mechanisms that simultaneously repress thousands of genes are unknown. On a similar note, although recent studies have identified how the enhancer repertoire is re-profiled within minutes of stress11,28, the correlation of enhancer transcription and chromatin state with the capacity to activate connected genes remain to be elucidated. Another major question to be answered in the field of cell biology is whether every cell in a population responds to a stress signal. The individual cells in Drosophila melanogaster salivary glands elicit a strikingly synchronous stress response with highly similar recruitment kinetics, execution rates and compartmentalization dynamics at Hsp70 loci44–47. In Caenorhabditis elegans, two sensory neurons are essential for the heat response of the animal16, indicating communication between tissues and organismal control over cells. Moreover, previous experience of the animal can prime HSF1-mediated responses123, and the metabolic state of the organism is integrated via neuroendocrine networks and transcellular chaperones16–17 to the cells’ ability to provoke transcriptional changes (FIG. 1). Single-cell sequencing methods hold great promises for the elucidation of the global transcriptional response and the fate of each cell in a culture, tissue or organism. In the future, when targeted RNA or DNA-capture from single cells becomes highly efficient, methods such as ChIP–seq and nascent RNA-seq will provide insights into the mechanisms of regulation at the single-cell level. Nonetheless, single-cell measurements of mRNA expression have already enabled the dissection of transcriptional responses for each signalling pathway of the UPRER, revealing that stress responses can be executed in markedly different ways within distinct cells of an apparently homogeneous cell population124. Expanding mechanistic analyses on gene and enhancer coordination to single-cell studies and to the physiological models of tissues and organisms will profoundly increase our knowledge of how cellular processes are coordinated within organisms, and how neurodegenerative diseases or cancer progression is intertwined with the ability to maintain homeostasis. Taken together, the future holds great promise for uncovering how transcriptional networks of genes and enhancers are coordinated in single cells and across the organism in health and disease.
Acknowledgments
We apologize to our many colleagues whose important work was only indirectly cited. This work was financially supported by The Sigrid Jusélius Foundation (A.V.) and NIH grant RO1-GM25232 (J.T.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Glossary terms
- Chaperones
Proteins that assist in folding, unfolding, assembly and disassembly of macromolecular structures
- Celastrol
Chemical compound (pentacyclic triterpenoid) that induces heat shock response and unfolded protein response, and exhibits anti-inflammatory, anti-cancer and antioxidant activities
- Precision Run-On sequencing (PRO-seq)
A method that maps the exact locations, orientation, and amounts of transcribing RNA polymerases across the genome
- Unfolded protein response
Cellular response to accumulation of misfolded proteins
- Promoter architecture
Positioning and dynamics of nucleosomes, chromatin remodelers, transcriptional regulators, pre-initiation complex, and transcriptionally engaged Pol II at the promoter
- Divergent transcription
A widespread phenomenon in various species, in which active genes and enhancers are transcribed in both directions. At genes, the coding strand (sense direction) encodes a stable mRNA, whereas the non-protein-coding antisense transcripts are short and unstable. At enhancers, the transcripts, called enhancer RNAs (eRNAs), in both directions are short and unstable
- Core promoters
Short (~50 nt) regions surrounding the TSS that provide a binding platform for general transcription factors (GTFs) and direct RNA polymerase (Pol) II to initiation sites
- Promoters
Regions including the core promoter and upstream sequences (usually 1 kb or less) that contain binding sites for transcription factors, which coordinate the expression of the downstream gene. At genes with divergent transcription, the promoter includes the region between the two core promoters
- Enhancers
Regions distal to gene promoters that have the potential to activate one or several genes
- Core initiation regions
Similar to core promoters, these regions are the sites of Pol II assembly and transcription initiation at promoters or enhancers
- Parylation
Post-translational modification of a single molecule, or chains of poly ADP-ribose that are covalently attached to the catalytic enzyme (PARP) itself or other proteins
- Sumoylation
Post-translational modification, whereby small ubiquitin-like modifier (SUMO) is covalently attached
- Phase-separation
Formation of multimolecular, membrane-less assemblies that can compartmentalize biochemical reactions
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Toivola DM, Strnad P, Habtezion A, Omary MB. Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 2010;20:79–91. doi: 10.1016/j.tcb.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 4.Quirós PM, Langer T, López-Otín C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 5.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:4–19. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gidalevitz T, Prahlad V, Morimoto RI. The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a009704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vihervaara A, Sistonen L. HSF1 at a glance. J Cell Sci. 2014;127:261–266. doi: 10.1242/jcs.132605. [DOI] [PubMed] [Google Scholar]
- 9.Duarte FM, Fuda NJ, Mahat DB, Core LJ, Guertin MJ, Lis JT. Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev. 2016;30:1731–1746. doi: 10.1101/gad.284430.116. Demonstrates that pausing is an indispensable step for heat-induced transcription, and maps the global transcriptional changes in heat-stressed Drosophila cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. Maps detailed kinetics of nascent transcription in heat-stressed mouse cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vihervaara A, et al. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun. 2017;8:255. doi: 10.1038/s41467-017-00151-0. Quantifies the transcriptional change at nucleotide resolution across genes and enhancers in human cells, identifying mechanisms that establish directionality and prewire trans-activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guertin MJ, Petesch SJ, Zobeck KL, Min IM, Lis JT. Drosophila heat shock system as a general model to investigate transcriptional regulation. Cold Spring Harb Symp Quant Biol. 2010;75:1–9. doi: 10.1101/sqb.2010.75.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prostko CR, Brostrom MA, Brostrom CO. Reversible phosphorylation of eukaryotic initiation factor 2 alpha in response to endoplasmic reticular signaling. Mol Cell Biochem. 1993;127–128:255–265. doi: 10.1007/BF01076776. [DOI] [PubMed] [Google Scholar]
- 14.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 15.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. Identifies organismal control over cellular stress responses in C elegans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Oosten-Hawle P, Porter RS, Morimoto RI. Regulation of organismal proteostasis by transcellular chaperone signaling. Cell. 2013;153:1366–1378. doi: 10.1016/j.cell.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama T, et al. Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat Cell Biol. 1999;1:479–485. doi: 10.1038/70265. [DOI] [PubMed] [Google Scholar]
- 19.Yoo BC, Kim SH, Cairns N, Fountoulakis M, Lubec G. Deranged expression of molecular chaperones in brains of patients with Alzheimer’s disease. Biochem Biophys Res Commun. 2001;280:249–258. doi: 10.1006/bbrc.2000.4109. [DOI] [PubMed] [Google Scholar]
- 20.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendillo ML, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez-Pastor R, et al. Abnormal degradation of the neuronal stress-protective transcription factor HSF1 in Huntington’s disease. Nat Commun. 2017;8:14405. doi: 10.1038/ncomms14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sannino S, Brodsky JL. Targeting protein quality control pathways in breast cancer. BMC Biol. 2017;15:109. doi: 10.1186/s12915-017-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritossa FM. A new puffing pattern induced by a temperature shock and DNP in Drosophila. Experientia. 1962;18:571–573. [Google Scholar]
- 25.Tissières A, Mitchell HK, Tracy UM. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol. 1974;84:389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- 26.DiDomenico BJ, Bugaisky GE, Lindquist S. Heat shock and recovery are mediated by different translational mechanisms. Proc Natl Acad Sci USA. 1982;79:6181–6185. doi: 10.1073/pnas.79.20.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niskanen EA, Malinen M, Sutinen P, Toropainen S, Paakinaho V, Vihervaara A, Joutsen J, Kaikkonen MU, Sistonen L, Palvimo JJ. Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol. 2015;16:153. doi: 10.1186/s13059-015-0717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dukler N, Booth GT, Huang YF, Tippens N, Danko CG, Lis JT, Siepel A. Nascent RNA sequencing reveals a dynamic global transcriptional response at genes and enhancers to the natural medicinal compound celastrol. Genome Res. 2017;27:1816–1829. doi: 10.1101/gr.222935.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller B, et al. Widespread changes in nucleosome accessibility without changes in nucleosome occupancy during a rapid transcriptional induction. Genes Dev. 2017;31:451–462. doi: 10.1101/gad.293118.116. Globally shows the increased chromatin accessibility and nucleosome remodeling at stress-induced genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahn JS, Hu Z, Thiele DJ, Iyer VR. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol. 2004;24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vihervaara A, et al. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc Natl Acad Sci USA. 2013;110:E3388–3397. doi: 10.1073/pnas.1305275110. Maps HSF1 and HSF2 target loci in freely cycling and mitotic human cells, demonstrating how HSF2 marks genes for post-mitotic transcription while HSF1 is largely excluded from the dividing chromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guertin MJ, Lis JT. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 2010;6:e1001114. doi: 10.1371/journal.pgen.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. Identifies enhancers across the genome by their divergent pattern of transcription that produces unstable transcripts to both directions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Core LJ, et al. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. Development of global run-on sequencing methodology and identification of divergent transcription from genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. Refines global run-on sequencing to nucleotide resolution and identifies mechanisms that coordinate initiation and pausing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray JI, et al. Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell. 2004;15:2361–2374. doi: 10.1091/mbc.E03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell. 2004;15:1254–1261. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorensen JG, Nielsen MM, Kruhoffer M, Justesen J, Loeschcke V. Full genome gene expression analysis of the heat stress response in Drosophila melanogaster. Cell Stress Chaperones. 2005;10:312–328. doi: 10.1379/CSC-128R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 44.Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- 45.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 46.Yao J, Zobeck KL, Lis JT, Webb WW. Imaging transcription dynamics at endogenous genes in living Drosophila tissues. Methods. 2008;45:233–241. doi: 10.1016/j.ymeth.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zobeck KL, Buckley MS, Zipfel WR, Lis JT. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell. 2010;40:965–975. doi: 10.1016/j.molcel.2010.11.022. Detailed identification of the kinetic events that mediate trans-activation of the Drosophila Hsp70 gene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teves SS, Henikoff S. Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes Dev. 2011;25:2387–2397. doi: 10.1101/gad.177675.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, et al. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol Cell. 2013;50:711–722. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai WK, Pugh BF. Genome-wide uniformity of human ‘open’ pre-initiation complexes. Genome Res. 2017;27:15–26. doi: 10.1101/gr.210955.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuda NJ, et al. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet. 2015;11:e1005108. doi: 10.1371/journal.pgen.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao H, et al. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 53.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 54.Badenhorst P, et al. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujimoto M, et al. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell. 2012;48:182–194. doi: 10.1016/j.molcel.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 56.Andersson R, et al. Human gene promoters are intrinsically bidirectional. Mol Cell. 2015;60:346–347. doi: 10.1016/j.molcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scruggs BS, et al. Bidirectional Transcription Arises from Two Distinct Hubs of Transcription Factor Binding and Active Chromatin. Mol Cell. 2015;58:1101–1112. doi: 10.1016/j.molcel.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pugh BF, Venters BJ. Genomic Organization of Human Transcription Initiation Complexes. PLoS One. 2016;11:0149339. doi: 10.1371/journal.pone.0149339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Q, Johnston J, Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol. 2015;33:395–401. doi: 10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. Identifies promoter-proximal pausing of Pol II by showing that the Pol II complex that resides 20-50nt downstream of TSS is transcriptionally engaged. [DOI] [PubMed] [Google Scholar]
- 61.Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci USA. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilchrist DA, Santos dos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen F, Gao X, Shilatifard A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev. 2015;29:39–47. doi: 10.1101/gad.246173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Topol J, Ruden DM, Parker CS. Sequences required for in vitro transcriptional activation of a Drosophila hsp70 gene. Cell. 1985;42:527–537. doi: 10.1016/0092-8674(85)90110-2. [DOI] [PubMed] [Google Scholar]
- 66.Wiederrecht G, Shuey DJ, Kibbe WA, Parker CS. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987;48:507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- 67.Wu C, et al. Purification and properties of Drosophila heat shock activator protein. Science. 1987;238:1247–1253. doi: 10.1126/science.3685975. [DOI] [PubMed] [Google Scholar]
- 68.Åkerfelt M, et al. Heat shock transcription factor 1 localizes to sex chromatin during meiotic repression. J Biol Chem. 2010;285:34469–34476. doi: 10.1074/jbc.M110.157552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elsing AN, et al. Expression of HSF2 decreases in mitosis to enable stress-inducible transcription and cell survival. J Cell Biol. 2014;206:735–749. doi: 10.1083/jcb.201402002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol. 2001;21:5826–37. doi: 10.1128/MCB.21.17.5826-5837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petesch SJ, Lis JT. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell. 2012;45:64–74. doi: 10.1016/j.molcel.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corey LL, Weirich CS, Benjamin IJ, Kingston RE. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 2003;17:1392–1401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomson S, Hollis A, Hazzalin CA, Mahadevan LC. Distinct stimulus-specific histone modifications at hsp70 chromatin targeted by the transcription factor heat shock factor-1. Mol Cell. 2004;15:585–594. doi: 10.1016/j.molcel.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 74.Solís EJ, et al. Defining the Essential Function of Yeast Hsf1 Reveals a Compact Transcriptional Program for Maintaining Eukaryotic Proteostasis. Mol Cell. 2016;63:60–71. doi: 10.1016/j.molcel.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 76.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shao W, Zeitlinger J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet. 2017;49:1045–1051. doi: 10.1038/ng.3867. [DOI] [PubMed] [Google Scholar]
- 78.Gressel S, et al. CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife. 2017;6:e29736. doi: 10.7554/eLife.29736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 80.Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012;28:276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Henriques T, et al. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. doi: 10.1101/gad.309351.117. Published in Advance January 29, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mikhaylichenko O, et al. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. doi: 10.1101/gad.308619.117. Published in Advance January 29, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arner E, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schaukowitch K, et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bradner JE, Hnisz D, Young RA. Transcriptional Addiction in Cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen FX, et al. PAF1 regulation of promoter-proximal pause release via enhancer activation. Science. 2017;357:1294–1298. doi: 10.1126/science.aan3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dekker J, Misteli T. Long-Range Chromatin Interactions. Cold Spring Harb Perspect Biol. 2015;7:a019356. doi: 10.1101/cshperspect.a019356. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chowdhary S, Kainth AS, Gross DS. Heat Shock Protein Genes Undergo Dynamic Alteration in Their Three-Dimensional Structure and Genome Organization in Response to Thermal Stress. Mol Cell Biol. 2017;37:e00292–17. doi: 10.1128/MCB.00292-17. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li L, et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol Cell. 2015;58:216–31. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galvani A, Thiriet C. Nucleosome dancing at the tempo of histone tail acetylation. Genes. 2015;6:607–621. doi: 10.3390/genes6030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo Y. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci USA. 2012;109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weintraub AS, et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell. 2017;171:1573–1588. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bai P. Biology of Poly(ADP-Ribose) Polymerases: The Factotums of Cell Maintenance. Mol Cell. 2015;58:947–958. doi: 10.1016/j.molcel.2015.01.034. [DOI] [PubMed] [Google Scholar]
- 98.Niskanen EA, Palvimo JJ. Chromatin SUMOylation in heat stress: To protect, pause and organise?: SUMO stress response on chromatin. Bioessays. 2017;39 doi: 10.1002/bies.201600263. [DOI] [PubMed] [Google Scholar]
- 99.Haddad N, Jost D, Vaillant C. Perspectives: using polymer modeling to understand the formation and function of nuclear compartments. Chromosome Res. 2017;25:35–50. doi: 10.1007/s10577-016-9548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 101.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 102.Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, Zhang XD, Bischof O, Seeler JS, Dejean A. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J. 2009;28:3534–3548. doi: 10.1038/emboj.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ouararhni K, et al. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 2006;20:3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fujimoto M, et al. The HSF1-PARP13-PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis. Nat Commun. 2017;8:1638. doi: 10.1038/s41467-017-01807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guo C, Henley JM. Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life. 2014;66:71–77. doi: 10.1002/iub.1244. [DOI] [PubMed] [Google Scholar]
- 107.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 108.Blomster HA, et al. Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol Cell Proteomics. 2009;8:1382–1390. doi: 10.1074/mcp.M800551-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pellegrino S, Altmeyer M. Interplay between Ubiquitin, SUMO, and Poly(ADP-Ribose) in the Cellular Response to Genotoxic Stress. Front Genet. 2016;7:63. doi: 10.3389/fgene.2016.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fukuto A, et al. SUMO modification system facilitates the exchange of histone variant H2A.Z-2 at DNA damage sites. Nucleus. 2018;9:87–94. doi: 10.1080/19491034.2017.1395543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Strom AR, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tippens ND, Vihervaara A, Lis JT. Enhancer transcription: what, where, when, and why? Genes Dev. doi: 10.1101/gad.311605.118. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Herzel L, Ottoz DSM, Alpert T, Neugebauer KM. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol. 2017;18:637–650. doi: 10.1038/nrm.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 116.Di Giammartino DC, Shi Y, Manley JL. PARP1 represses PAP and inhibits polyadenylation during heat shock. Mol Cell. 2013;49:7–17. doi: 10.1016/j.molcel.2012.11.005. Identifies parylation of PAP, and shows how its regulation defines polyadenylation in heat-stressed cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shalgi R, Hurt JA, Lindquist S, Burge CB. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep. 2014;7:1362–1370. doi: 10.1016/j.celrep.2014.04.044. Measures co-transcriptional splicing across the transcriptome in stressed and non-stressed mouse cells. [DOI] [PubMed] [Google Scholar]
- 118.Proudfoot NJ. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016;352:aad9926–aad9926. doi: 10.1126/science.aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vilborg A. Comparative analysis reveals genomic features of stress-induced transcriptional readthrough. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1711120114. [Epub ahead of print] Describes global run-through transcription upon osmotic, oxidative or heat stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grosso AR, et al. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. eLife. 2015;4:e09214. doi: 10.7554/eLife.09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rutkowski AJ, et al. Widespread disruption of host transcription termination in HSV-1 infection. Nat Commun. 2015;6:7126. doi: 10.1038/ncomms8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA. Widespread inducible transcription downstream of human genes. Mol Cell. 2015;59:449–461. doi: 10.1016/j.molcel.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ooi FK, Prahlad V. Olfactory experience primes the heat shock transcription factor HSF-1 to enhance the expression of molecular chaperones in C. elegans. Sci Signal. 2017;10:eaan4893. doi: 10.1126/scisignal.aan4893. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Adamson B, et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882. doi: 10.1016/j.cell.2016.11.048. Identifies transcriptional responses mediated by the three UPRER signaling pathways, and demonstrates that cellular stress responses can be executed differently in distinct cells of a seemingly homogeneous population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu B, Chen P, Xi D, Zhu H, Gao Y. ATF4 regulates CCL2 expression to promote endometrial cancer growth by controlling macrophage infiltration. Exp Cell Res. 2017;360:105–112. doi: 10.1016/j.yexcr.2017.08.031. [DOI] [PubMed] [Google Scholar]
- 126.Acosta-Alvear D, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 127.Fiorese CJ, et al. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol. 2016;26:2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao Q, et al. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]


