Highlights
-
•
Dairy processing wastewater biotreatment using IASBR technology is proposed.
-
•
Minor (0.2 LPM) aeration rate variations heavily influenced efficiency and ecology.
-
•
Optimal nutrient removal efficiencies (≥92%) achieved under 0.6LPM aeration rate.
-
•
Comamonadaceae stably enriched in reactor biomass (>80% relative abundance).
-
•
Comamonadaceae top contributor of nitrogen and phosphorus assimilation genes.
Keywords: Biotreatment, Dairy wastewater, Sludge biomass, Intermittently aerated sequencing batch reactor (IASBR), Biological nutrient removal (BNR), Comamonadaceae
Abstract
Dairy processing generates large volumes of wastewater that require extensive nutrient remediation prior to discharge. Significant commercial opportunities exist therefore for cost-effective biotechnologies capable of achieving this requirement. In this study the authors evaluated the use of intermittently aerated sequencing batch reactors, (IASBRs), as a single-tank biotreatment system for co-removal of COD, nitrogen and phosphorus from synthetic dairy processing wastewater. Variation of the IASBR aeration rates, (0.8, 0.6 and 0.4 L/min), had significant impacts on the respective nutrient removal efficiencies and underlying microbial diversity profiles. Aeration at 0.6 L/min was most effective and resulted in >90% co-removal of orthophosphate and ammonium. 16S rRNA based pyrosequencing of biomass DNA samples revealed the family Comamonadaceae was notably enriched (>80% relative abundance) under these conditions. In silico predictive metabolic modelling also identified Comamonadaceae as the major contributor of several known genes for nitrogen and phosphorus assimilation (nirK, nosZ, norB, ppK, ppX and phbC).
1. Introduction
The European dairy industry is experiencing a period of rapid growth following the abolition of European milk quotas in 2015, with a 50% increase in Irish milk production predicted by 2020. In Ireland, dairy processors consume an average of 2.3 L of water per litre of milk processed [1] but can produce up to 10 L of effluent depending on the end product [2]. Such effluents are considered an important source of potential water pollution due to their high nutrient composition. Total Kjeldahl nitrogen (TKN) concentrations up to 1462 mg L−1 and total phosphorus (TP) concentrations of 640 mg L−1 have been reported in dairy processing wastewater [3]. Dairy processing plant effluent discharges into the environment must not impair the quality of the receiving water bodies and ensure that Environmental Quality Standards (EQS) are not exceeded. Licensed discharge limits can depend on the sensitivity of the receiving water body but typical dairy processing sector limits are currently: 5–25 mg L-1 total nitrogen (TN), 2–5 mg L-1 TP and 10 mg L-1 total ammonia (NH4-N) [4]. Thus there are numerous drivers for sustainable waste management strategies in the dairy processing sector.
Dairy wastewaters are highly biodegradable and therefore amenable to biological secondary treatments that consist of aerobic and anaerobic processes, typically in combination. Such biological nutrient removal (BNR) systems offer a cost-effective alternative to chemical treatments for the removal of nitrogen and phosphorus [5]. In such systems, conventional nitrogen removal is achieved in a two-stage process composed of aerobic nitrification and anoxic denitrification [6]. Phosphorus remediation also involves aerobic/anaerobic cycling conditions in a process referred to as enhanced biological phosphorus removal (EBPR) [7]. Technologies focused on achieving nutrient removal in parallel with improved sustainability have begun to emerge in recent decades. These novel processes include: completely autotrophic nitrogen removal over nitrite (CANON) [8], anaerobic ammonium oxidation (ANAMMOX) [9], single reactor system for high activity ammonium removal over nitrite (SHARON) [10], oxygen-limited autotrophic nitrification–denitrification (OLAND) [11], partial nitrification-denitrification [12] and, simultaneous nitrification-denitrification (SND) and phosphorus removal [13]. The capacity of these systems to improve sustainability is reflected in advantages such as reduced energy/chemical additive inputs and reduced volumes of sludge biomass and/or chemical precipitants requiring downstream treatment/disposal [6]. For example, coupled partial nitrification and denitrification systems have been shown to reduce aeration costs by 25%, biomass generation by 30% [14,15] and process CO2 emissions by 20% [12,16].
Intermittently aerated sequencing batch reactors (IASBRs) represent one such BNR process with the capacity for co-remediation of nitrogen and phosphorus within a single bioreactor [17]. Each IASBR operational cycle incorporates multiple, alternating anaerobic and aerobic periods, potentially reducing operational costs and sludge production volumes. The intermittent aeration process has been shown to achieve long-term, stable partial nitrification resulting in a reduced oxygen demand for ammonia conversion and a reduced organic substrate requirement for subsequent denitrification [18]. Nutrient removal performances using IASBR technology have previously been assessed for domestic and slaughterhouse wastewater [19,20]. Pan et al. [20] compared SBR and IASBR system efficiencies for the removal of nitrogen and phosphorus in synthetic domestic wastewater. Total nitrogen (TN) and phosphorus (TP) removal efficiencies of 79% and 63% in the SBR system increased to 90% and 74% with the application of the IASBR approach, respectively. In addition, SND efficiencies of 90.4% and 79% were reported in the IASBR and SBR systems, respectively. Li et al. [19] reported average TN and TP removal efficiencies of 96% and 99%, respectively, from slaughterhouse influents treated in IASBRs.
Characterisation of microbial diversity and ecosystem function are essential to understanding and optimising biological wastewater treatment processes [21]. Previous studies have demonstrated the influence of operational conditions and influent compositions on the microbial ecology of bioreactor systems and associated key metabolic activities of nitrification, denitrification and phosphorus accumulation [[22], [23], [24]]. To date, the microbial characterisation of IASBR systems has been limited to a single fluorescence in situ hybridization study to determine the relative, spatial abundance of ammonium (12%) and nitrite oxidizing (7%) bacteria within the general (EUB) bacterial community [25]. The present study investigated the application of an IASBR to the remediation of synthetic dairy processing wastewater with a focus on the impacts of differing aeration rates, (0.4, 0.6, 0.8 L/min) and characterisation of the associated microbial communities based on pyrosequencing of 16S rRNA gene V5–V9 hypervariable regions.
2. Material and methods
2.1. Dairy synthetic wastewater
Six Irish dairy processing plants with on-site wastewater treatment facilities were sampled to determine effluent organic matter, nitrogen and phosphorus levels. The average compositions were as follows: chemical oxygen demand (COD) 3513 mg L−1, soluble COD 3307 mg L−1, TN 122.2 mg L−1, TP 51.9 mg L−1, ammonia (NH4-N) 48.9 mg L−1, orthophosphate (PO4-P) 25.4 mg L−1. These characteristics were used to model the synthetic wastewater, incorporating a formulation previously reported by Henry [26]. The final composition contained NaOAc 2929 mg L−1, yeast extract 218 mg L−1, dried milk powder 872 mg L−1, NH4CL 167.3 mg L−1, urea 129.9 mg L−1, Na2HPO4 126 mg L−1, KHCO3 50 mg L−1, NaHCO3 130 mg L−1, MgSO4·7H2O 50 mg L−1, FeSO4 ·7H2O 10 mg L−1, MnSO4·H2O 2 mg L-1 and CaCl2·6H2O mg L−1. The pH of the synthetic wastewater was 7.9.
2.2. Laboratory-scale IASBR system set up and operation
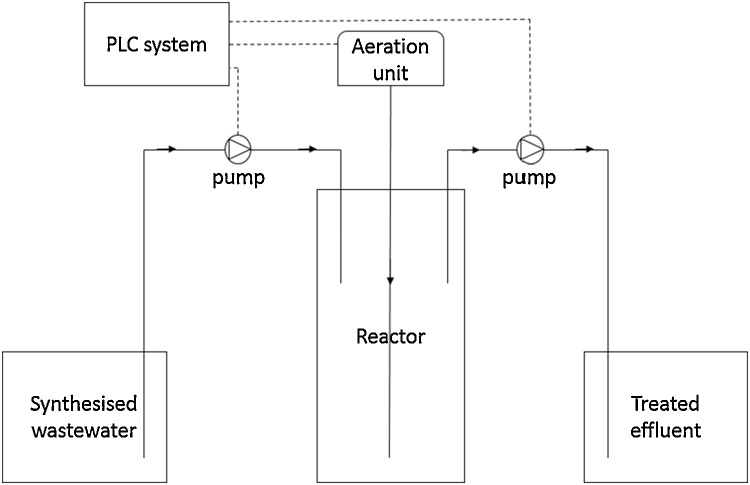
Three laboratory-scale IASBR systems were operated at the Environmental Engineering laboratory in the Department of Civil Engineering, National University of Ireland, Galway. Three identical reactors were operated in triplicate, each bioreactor having an eight litre working volume (Fig. 1). The reactors were located in a temperature controlled environment at approximately 11 °C, in order to replicate average annual temperatures in Ireland. The system was initially seeded with return sludge from a municipal wastewater treatment plant, located in Tuam, Co. Galway (Ireland). The seed sludge contained 8000 mg L−1 total suspended solids (TSS) and 6200 mg L-1 volatile suspended solids (VSS) respectively, with a 5 l volume being used to inoculate reactors. The operational conditions of the IASBR are summarised in Table 1.
Fig. 1.
Schematic of the laboratory-scale IASBR system.
Table 1.
IASBR operational conditions.
| Bioreactor volume (L) | 8 |
| Hydraulic retention time (days) | 4 |
| Solid retention time (days) | 20 |
| Temperature (°C) | 11 |
| Aeration rate (litres/min) | 0.8-0.6-0.4 |
| Operation cycle | |
| Aeration period (minutes) | 60 |
| Non-aeration period (minutes) | 100 |
The IASBR system was operated in 12 h cycles. At the beginning of each cycle synthetic wastewater was pumped into the system (1 L), followed by four repeat periods of alternating non-aeration (100 min) and aeration (60 min) under continuous mixing. A final 80 min period without aeration or mixing was incorporated to facilitate sludge settling and effluent decanting (800 ml), before the next cycle commenced. A single 400 ml volume of mixed liquor was removed from the reactor once each day as sludge waste, resulting in a 20 day solids retention time (SRT). Samples for metagenomic analyses were collected twice weekly between days 50–201. Three different aerations rates were applied during this period: 0.8, 0.6 and 0.4 L/min. The aeration rates were determined according to preliminary tests of the oxygen concentration profiles in the reactor and from previous work described by Pan et al. [27], and Li et al. [19]. At day 55 the initial aeration rate of 1 L/min was reduced to 0.8 L/min and sustained for 20 days. Between days 76–161, aeration was further reduced to 0.6 L/min. On day 161 the aeration rate underwent a final reduction to 0.4 L/min, which was maintained until the conclusion of the trial on day 201.
2.3. Physicochemical profile of the IASBR
Standardized analytical procedures [28] were performed to test influent/effluent suspended solids (SS), dissolved oxygen (DO), chemical oxygen demand (COD) and biological oxygen demand (BOD5). TN, TP and total organic carbon (TOC) were tested using a Biotector TOC, TN, TP Analyser. In addition, TN and TP were also measured using the HACH TNT methods: 100062, 10,127 and 8190, respectively. Quantification of ammonium (NH4-N), nitrite (NO2-N), total oxidized nitrogen (TON), orthophosphate (PO4-P) and calcium carbonate (CaCO3) for alkalinity were analysed using a Konelab 20 Nutrient Analyser (Thermo Scientific), in accordance with the manufacturer’s instructions. Parameters were analysed on a daily basis.
2.4. Biomass collection and metagenomic DNA extraction
Mixed liquor samples were routinely collected during the third aeration period within the IASBR cycle. A subset of these samples were selected for metagenomic analyses and comprised representatives of each SRT, varying nutrient removal performances and the different aeration rates between days 50–201, respectively (Table 2). Samples were collected in sterile bottles and immediately placed at −20 °C until microbial diversity studies were performed at University College Cork, Ireland.
Table 2.
IASBR biomass sampling schedule.
| Sample ID | T1a | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 |
|---|---|---|---|---|---|---|---|---|---|
| Day since starting | 39 | 62 | 82 | 108 | 131 | 150 | 168 | 182 | 201 |
| Aeration (litres/min) | 1 | 0.8 | 0.6 | 0.6 | 0.6 | 0.6 | 0.4 | 0.4 | 0.4 |
T1 reference sample represents aeration rate applied during bioreactor stabilisation.
To ensure sufficient biomass for optimal nucleic acid extraction, 6 ml of sludge was centrifuged for 15 min at 5000 r.p.m, before re-suspending pellets in 1 ml of supernatant. A 300 μl volume of the concentrated biomass was then processed using a PowerSoil DNA Isolation Kit (MOBIO Laboratories) for DNA extraction, according to the manufacturer’s instructions. Extractions were quantified via spectrophotometry using a NanoDrop (ND-1000, Thermo-Fisher, DE, USA) and visualized via 1% agarose gel electrophoresis, SafeView (NBS Biologicals) staining and UV trans-illumination.
2.5. Pyrosequencing and processing of 16S rRNA sequence data
Universal primers U905 F (5′-TGAAACTYAAAGGAATTG-3′) and U1492R (5′- GGTTACCTTGTTACGACTT-3′) with 10 nt unique barcodes (Table S1) were used to amplify the V5-V9 regions of bacterial and archaeal 16S rRNA genes from the extracted DNA [29,30]. Each sample was amplified in triplicate to ensure representative sampling. PCR cycling parameters were as follows: initial denaturation at 98 °C × 5 min and 30 cycles of 94 °C × 40 s, 55 °C × 40 s and 72 °C × 50 s with a final extension at 72 °C for 5 min. PCR products were purified using a QIAquick PCR Purification Kit (QIAGEN) and quantified on a Qubit 3.0 Fluorometer (ThermoFisher). The purified products were pooled in equimolar quantities and forwarded to an external service provider for emulsion PCR and 454 GS FLX + pyrosequencing, MACROGEN (Seoul).
Pyrosequenced amplicon data were corrected using Acacia [31] and subsequent analyses were carried out using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline [32]. Chimeras were filtered out and removed using USEARCH v6.1 [33]. Quality-filtered sequences (minimum length 200 bp, with no ambiguous bases and homopolymers of 6 bp as maximum) were aligned via PynAST against the SILVA 123 reference database [34]. Taxonomy was assigned to each OTU using the RDP classifier at a 0.97 threshold. The filtered alignments were clustered into OTUs at the 97% identity level in an open-reference OTU picking process implemented in QIIME.
To compute the diversity analysis, singletons were filtered out from the OTU table before normalizing to ensure that the observed differences were caused by biological origin and not due to random variations in relative sequencing depths [35]. The technique used for normalization was cumulative sum scaling (CSS) [36]. Alpha diversity within each sample was calculated following QIIME pipeline procedures.
2.6. Predictive functional metabolic modelling
Based on the 16S rRNA sequences, the functional potential of the microbial communities in the bioreactor was predicted using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) approach [37]. The recommended parameters according to the PICRUSt manual were applied for closed reference OTU picking using the GreenGenes 13_5 reference dataset in QIIME. The OTU table was then filtered for singletons and normalized using the CSS method in QIIME. Using PICRUSt in the web-based Galaxy platform (http://huttenhower.sph.harvard.edu/galaxy), the CSS normalized OTU table was then normalized by known/predicted 16S copy number abundance. Based on the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database [38], the metagenome functional prediction was performed and categorized by the KEGG Orthology (KO) hierarchical levels 1, 3, and 4. In accordance with the PICRUSt guidelines, the Nearest Sequenced Taxon Index (NSTI) cut-off of < 0.15 was used for quality control of the predictions from the samples. Metagenome contributions were computed in PICRUSt for the prediction of the top contributors for target genes of interest. Principal component analysis (PCA) for the functional predictions from the different samples was performed with the vegan package in R, using the RStudio integrated development environment [39]. The plots were generated using R built-in functions combined with the ggplot2 package [40].
2.7. Sequence data accession number
Raw sequence data were submitted to the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena) under accession no. PRJEB23305.
3. Results
3.1. Nutrient removal performance in the IASBR
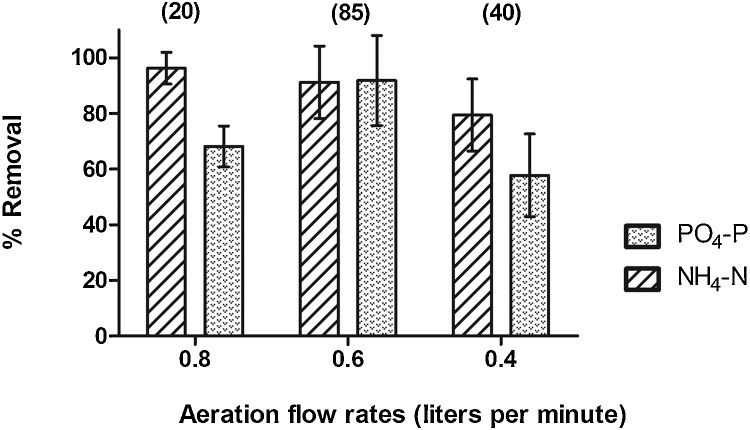
Orthophosphate (PO4-P) and ammonia (NH4-N) percent removal efficiencies were found to vary depending on the IASBR aeration rate applied, (0.8, 0.6 or 0.4 L/min), as shown in Fig. 2. Sustained nitrogen removal of ∼96% was observed for the 0.8 L/min rate with a concomitant 68% removal of PO4-P. Under 0.6 L/min aeration, removal efficiencies of approximately 92% were achieved for both PO4-P and NH4-N. The IASBR performance deteriorated following a shift to 0.4 L/min aeration, with average removal efficiencies of 79% and 57% observed for NH4-N and PO4-P, respectively. The capacity of the system to treat synthetic dairy processing wastewater correlated well with previously reported NH4-N and PO4-P removal from slaughterhouse effluents subjected to IASBR treatment [19,25,27].
Fig. 2.
Average % removal of PO4-P and NH4-N at varying aeration rates. In brackets, the duration of each aeration period in days.
3.2. System community richness
A total of 82,176 high quality reads were detected after bioinformatics quality control analyses. The coverage index for each sample was ≥0.8, suggesting that the relative number of species were well represented among samples, (Table 3). With respect to alpha diversity metrics, the library size of each sample was normalized due to varying depths of coverage across the samples. The species richness, calculated by Chao 1 index and the observed OTUs at a 3% cut off level, is summarized in Table 3.
Table 3.
Diversity and species richness within the metagenomic dataset.
| Sample ID | T1 | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 |
|---|---|---|---|---|---|---|---|---|---|
| High quality reads | 11675 | 13220 | 5568 | 9994 | 13770 | 10182 | 4936 | 5026 | 7805 |
| Normalized reads | 1914 | 2047 | 1495 | 1703 | 1290 | 1199 | 1072 | 1144 | 1462 |
| Observed OTUs | 674 | 676 | 493 | 551 | 366 | 355 | 314 | 329 | 416 |
| Chao 1 index | 740.5 | 789 | 719.8 | 776.5 | 366 | 355 | 314 | 329 | 416 |
| Coverage index | 0.9 | 0.9 | 0.8 | 0.8 | 1 | 1 | 1 | 1 | 1 |
3.3. Microbial ecology profiling of the bioreactor
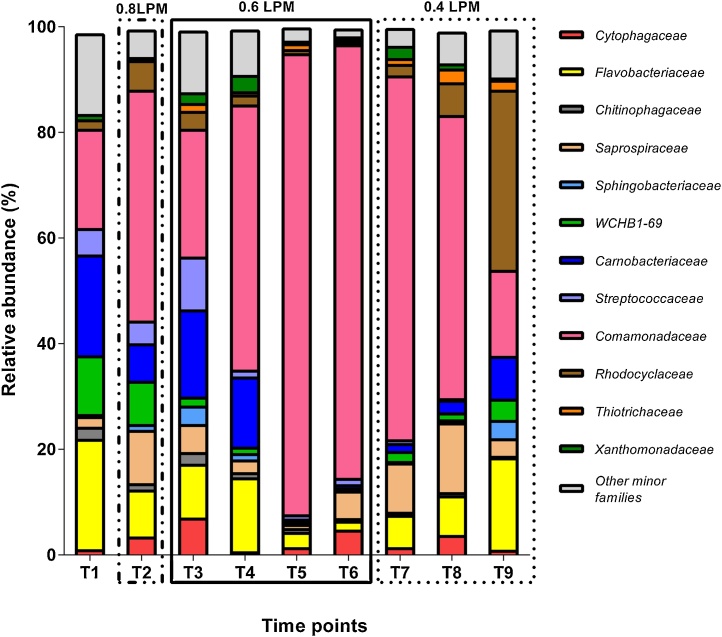
Bacterial community compositions in the IASBR system were determined using the high-throughput pyrosequencing method which targeted the V5-V9 region. Only sequences with OTU assignment similarities of ≥ 97% were included in the analyses. Ecological profiles at family level are shown in Fig. 3. In summary, 12 dominant families, (defined as having ≥1% total relative abundance), were identified. The “Other minor families” category represents grouped families with total relative abundance values lower than 1%. A notable observation was the dominance of the Comamonadaceae family within the IASBR community profile and the impact of the relative aeration rates on their overall levels. In the reference sample (T1), the abundance of Comamonadaceae was 18.8% and increased up to a maximum of 43.7% within the first aeration rate investigated (0.8 L/min). In the subsequent shift to reactor operation at 0.6 L/min over 4 SRTs, (T3–T6), Comamonadaceae relative abundance steadily increased to sustained maxima of 87% (T5) and 82.1% (T6), respectively. The final reduction in reactor aeration to 0.4 L/min correlated with a gradual decrease in Comamonadaceae from days 168 (T7) to 201 (T9), where levels dropped from 68.9% to comparable reference sample values of 16.3%. While Comamonadaceae dominated the majority of profiled samples, other families previously reported to be involved in nitrogen and phosphorus remediation processes were also observed e.g. Flavobacteriaceae and Rhodocyclaceae [[41], [42], [43]]. However, their low, respective relative abundances of 1.7% and 0.4% during optimal performance under 0.6 L/min, (see T5 and T6 in Fig. 3), appears to suggest a limited role in the system.
Fig. 3.
Overview of IASBR bacterial community structure at family level. T1 to T9 represents differing sample time-points (see Table 2).
3.4. Functional potential of the microbial communities
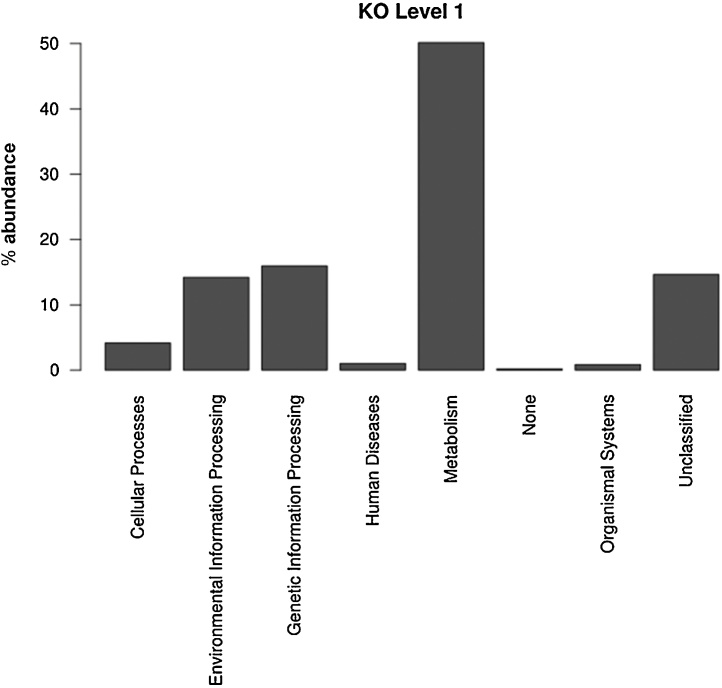
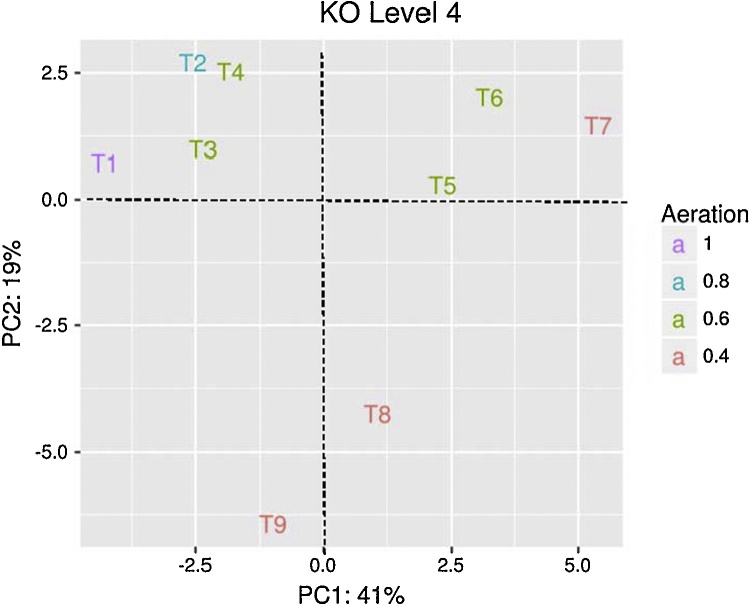
PICRUSt predicted metabolic functionality from the metagenomic profiling of microbial communities in the IASBR at KO hierarchy level 1 is shown in Fig. 4. Among predicted KEGG pathways, “Metabolism” (50.14%) was the most abundant category followed by “Genetic Information Processing” (15.98%), “Unclassified” (14.66%), and “Environmental Information Processing” (14.23%). Principal Component Analyses (PCA) were computed to investigate potential correlations between bacterial community metabolic profiles and the varying aeration rates applied to the IASBR system. As shown in Fig. 5, three distinct clusters emerged which indicated a shift in the functional/metabolic profiles of the microbial communities in response to the varied aeration conditions. The plots also revealed the time dependent nature of these shifts, e.g. T3–T4 versus T5–T6 during 0.6 L/min aeration conditions, which correlate with the observed bacterial diversity profiles shown in Fig. 3. The KO database also facilitated analysis of the metagenomic data set for relative abundances of genes known to contribute to nitrogen and phosphorus remediation. Key genes associated with denitrification (nitrite reductase (nirK), nitric oxide reductase (norB) and N2O reductase (nosZ), and EBPR processes (polyphosphate kinase (ppk), exopolyphosphatase (ppx) and polyhydroxyalkanoate synthase (phaC) were identified. OTUs contributing the genes of interest described above were then computed using PICRUSt. In order to select the top contributors to the genes of interest, OTUs for the metagenome prediction were merged up to the family level. Taxa that did not contribute ≥ 1% of the total relative abundance for one or more of the genes of interest were excluded. As shown in Tables 4 and S1, Comamonadaceae represented the top contributor for the described genes, which correlated with their observed taxonomic dominance in the IASBR system (Fig. 3). However, it was also noted that some of the less well represented taxa, such as for example Xanthomonadaceae (2.2% relative abundance), had a significant contribution to the predicted functional profile of the microbial communities (Table S1).
Fig. 4.
Functional predictions of bacterial diveristy of IASBR treating dariy synthetic wastewater. KEGG metagenome functional predictions of OTUs at KO.
Fig. 5.
Principal Components Analyses (PCA) at gene level (KO level 4) considering the aeration rates and the time-points.
Table 4.
Correlation of taxonomy (up to family level) and relative contributions to genes of interest: ppk, ppx, phaC, nirK, norB and nosZa.
| Total relative abundance (%) |
Taxonomy | Relative gene contributions (%) |
|||||
|---|---|---|---|---|---|---|---|
| ppk | ppx | phaC | nirK | norB | nosZ | ||
| 38.2 | Comamonadaceae | 30.90 | 30.36 | 58.33 | 5.10 | 50.12 | 31.13 |
| 10.4 | Order SC-I-84 | 0.88 | 0.88 | 1.71 | – | 1.75 | – |
| 8.2 | Rhodocyclaceae | 6.19 | 6.71 | 13.96 | 17.33 | 12.28 | 22.77 |
| 8 | Flavobacteriaceae | 11.61 | 10.70 | – | 20.70 | 8.03 | 8.27 |
| 4.1 | Thiotrichaceae | 3.16 | 3.14 | 6.11 | – | – | – |
| 3.6 | Saprospiraceae | 6.68 | 3.15 | – | – | – | 16.93 |
| 2.4 | Cytophagaceae | 2.27 | 4.31 | – | – | – | 0.51 |
| 2.2 | Xanthomonadaceae | 10.37 | 10.17 | 9.90 | 34.63 | 15.89 | 0.71 |
| 1.5 | Weeksellaceae | 2.57 | 2.55 | – | 9.48 | 4.04 | 0.92 |
| 0.9 | Chitinophagaceae | 4.44 | 2.96 | – | – | – | 7.73 |
| 0.6 | Sphingobacteriaceae | 4.44 | 4.12 | – | – | – | – |
| 0.4 | Porphyromonadaceae | 1.03 | 0.71 | – | – | – | – |
| 0.4 | Peptococcaceae | – | 1.30 | 1.27 | – | – | – |
| 0.4 | Rhodobacteraceae | 1.45 | 2.88 | 1.41 | 6.85 | 2.65 | 6.53 |
| 0.3 | Order Bacteroidales | 1.29 | 1.14 | – | – | – | – |
| 0.3 | SB-1 | 1.11 | 1.10 | – | – | – | – |
| 0.3 | Cryomorphaceae | 0.91 | 1.16 | – | – | – | – |
| 0.1 | Lachnospiraceae | 1.24 | 0.31 | – | – | – | – |
| 0.1 | Sinobacteraceae | 0.94 | 0.94 | 1.83 | – | – | – |
“- “= no detected contribution of gene of interest.
1% cut-off was applied.
4. Discussion
4.1. Impact of the aeration rates in nutrient removal performance
The dairy industry forms a key pillar of the agri-food sector in numerous countries with projected 2020 global milk production outputs estimated to reach approximately 830 million tonnes [44]. A significant percentage of liquid milk undergoes processing into a range of consumer products such as whey protein, butter, cheese and milk powder, which can consume 2–6m3 of water per tonne of milk processed [45]. The resulting high volumes of wastewater can present a considerable remediation challenge due to high nutrient loadings ranging from 3 to 70 kg/m3 COD, 0.05–1.4 kg/m3 total nitrogen and 0.01–0.7 kg/m3 total phosphorous, respectively [46]. The potential of IASBR technology for high efficiency nutrient removal from industrial as well as domestic wastewaters has previously been reported [47,20,25,27]. In the current study the scope of IASBR application has been expanded to include the potential remediation of dairy processing wastewater. In summary, optimum PO4-P and NH4-N co-remediation efficiencies (>90%) were achieved with synthetic dairy wastewater at 0.6 L/min, but the IASBR performance was found to be significantly affected at aeration rates above or below this value (i.e. 0.8 or 0.4 L/min, respectively). When the aeration rate was 0.4 L/min, the DO concentrations in the reactors were low, potentially reducing the ammonium oxidation rate by autotrophic nitrifiers. It may also explain lower phosphorous uptake by phosphorus accumulating organisms (PAOs) wherein polyphosphate accumulation occurs under aerobic conditions in conjunction with intracellular polyhydroxyalkanoate degradation. Overall results appear to suggest that under optimal aeration, IASBR could offer an effective treatment option for dairy processing wastewaters, coupled with reduced energy and infrastructural demands when compared with traditional BNR multistage systems.
4.2. IASBR microbial community profiling
It is generally accepted that microbial communities within activated sludge are affected by changes in bioreactor configurations, which can exert influences over system stability and robustness [48,49]. However, IASBR systems are not well characterised in this regard. In an effort to establish some insight into IASBR community structure the authors applied next generation sequencing (NGS) to 16S profiling of multiple samples collected under varying operational aeration rates. Subtle changes in the aeration rates appeared to significantly impact on the observed community structure within the reactor (Fig. 3). The most notable observation was the dominance of the family Comamonadaceae within the biomass, (82–87% relative abundance), at 0.6 L/min aeration; which coincided with optimal nutrient removal performance within the reactor (Fig. 2). It is possible that a threshold oxygen concentration provides a selective pressure for Comamonadaceae specific metabolism which becomes optimal under 0.6 L/min. A partial enrichment appears to operate under 0.8 L/min. However, the competitive advantage appears to dissipate at 0.4 L/min and, rather than drop off sharply, Comamonadaceae gradually decline toward reference sample levels over a 60 day period. Xin and co-workers recently demonstrated that varying aeration pressures, (0.2–0.6 MPa), significantly impacted on the relative abundance of Comamonadaceae in a sequencing batch reactor kettle (SBRK) system treating municipal wastewater [50]. In an earlier study, Sadaie and colleagues reported the gradual dominance of Comamonadaceae (52.3%) following reduced air supply (<1 mg/L) to a conventional activated sludge process treating food processing waste (180 m3, BOD5 = 1000 mg/L) [51]. The disparity between the compositions of municipal, food processing and dairy wastewaters suggests the influent is unlikely to be the selective pressure in Comamonadaceae enrichment, but rather reduced dissolved oxygen. Several Comamonadaceae species, belonging to at least 12 different genera, have been isolated from activated sludge and linked with nutrient removal from wastewaters [50,52,53]. Evidence from the literature suggests a positive correlation between several members of the Comamonadaceae and denitrification processes [53,54]. Recently, Ge and colleagues reported a novel clade within Comamonadaceae linked with high capacity phosphorus uptake from abattoir waste streams [55]. The authors achieved >90% orthophosphate removal, (influent load 24 mg L-1), in an SBR system operated at a solid retention time of <4 days. Fluorescent in situ hybridisation (FISH) and intracellular poly-phosphate granule staining with 4′,6′-diamidino-2-phenylindole confirmed Comamonadaceae representatives as key contributors to orthophosphate uptake within the system. Collectively, these recent studies suggest that Comamonadaceae members may well play a number of important roles in biological nutrient removal processes where they constitute a sizeable fraction of the microbial biomass.
4.3. Predictive metagenomic profiling of the IASBR microbial community metabolome
In order to gain a fuller understanding or describe the microbial ecology of a system, functional correlations are required. In an effort to fully mine the ngs data for potential correlations between taxanomic abundance and possible contributors to nutrient removal efficiencies, a predictive modelling approach, PICRUSt, was applied. Ahmed et al. [56] previously employed this approach to model the diversity and abundance of antibiotic resistance genes in raw versus secondary effluents from four Australian municipal treatment facilities. In a separate study, Gao et al. also employed PICRUSt analyses to suggest that the removal of pathogenic microorganisms from sewage sludge via anaerobic digestion did not significantly reduce the genetic capacity within the sludge to contribute “human disease” [57]. Our study represents the first application of PICRUSt modelling on an IASBR system.
IASBR systems have been reported to involve partial nitrification to remove nitrogen via nitrite intermediates [25,58]. Such processes require aerobic denitrifiers and the associated nirK, norB and nosZ genes [59]. PICRUSt analysis indicated that Comamonadaceae potentially contribute >50% of the norB and >30% of the nosZ genes within the community. With respect to phosphorus removal genes, ppk and ppx, were selected regarding their roles in poly-P synthesis and degradation, respectively [60,61]. Poly-hydroxy-alkanoate (PHA) metabolism has also been linked with EBPR, and involves a phaC encoded synthase [53,62]. In our analyses Comamonadaceae was also predicted to be the top contributor of ppk, ppx and phaC genes within the community (Tables 4 and S1). The authors did note that a strict relationship between relative taxonomic abundance and metagenomic contribution was not observed. Rhodobacteraceae for example, a known denitrifying proteobacteria [63,64] accounted for only 0.4% relative taxonomic abundance within the dataset, however its predicted functional contribution of denitrification genes was over 6% for nirK and nosZ genes (Table 4 and S1). Further, comprehensive analytical investigation of the IASBR system will however be required (e.g. FISH, biopolymer specific staining and gene expression analyses) to establish the functional significance of the modelled outputs and to provide further insights into our understanding of the microbial ecology underpinning successful IASBR application.
5. Conclusions
With the introduction of legislation such as the EU Water Framework Directive (Directive 2000/60/EC) and more stringent licensing requirement, cost efficient, sustainable treatment of wastewater is becoming increasingly important. IASBRs have the potential to provide a high efficiency treatment approach for dairy processing wastewater; reducing the need/costs for high level aeration and chemical precipitant addition while decreasing the volume of sludge produced. The single reactor IASBR system also offers a reduced infrastructural footprint when compared with traditional anoxic/oxic multistage systems. In conclusion, IASBR application to dairy processing wastewater remediation is a promising technological approach. However, optimisation is critically dependent on operational aeration rates, which greatly influence the ecological shifts within the system. Metagenomic based metabolic profiling suggests members of the Comamonadaceae family may contribute significantly to nitrogen and phosphate remediation processes. Currently, the authors are investigating functional correlations between the IASBR performance and ecological profiles reported here, in addition to determining the impacts of real-time dairy processing wastewater inputs to the system.
Conflict of interest statement
The authors have no conflict of interest to declare.
Acknowledgements
The authors would like to thank the Department of Agriculture, Food and the Marine (Ref.: 13‐F‐507) for funding the Dairy Water Project. For additional details: www.dairywater.ie.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2018.e00263.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Finnegan W., Goggins J., Clifford E., Fitzhenry K., Zhan X. World Water Congress. 2015. Water consumption and direct energy use in the Irish dairy processing industry.http://iwra.org/congress/resource/3027663.pdf [Google Scholar]
- 2.Lateef A., Chaudhry M.N., Ilyas S. Biological treatment of dairy wastewater using activated sludge. Sci. Asia. 2013;39:179–185. [Google Scholar]
- 3.Britz T.J., van Schalkwyk C., Hung Y.T. Waste Treatment in the Food Processing Industry. 2006. Treatment of dairy processing wastewaters; pp. 1–28. [Google Scholar]
- 4.European Commission . 2008. BAT Guidance Note on Best Available Techniques for the Dairy Processing Sector. [Google Scholar]
- 5.EPA . 2007. Biological Nutrient Removal Processes and Costs. [Google Scholar]
- 6.Breisha G.Z., Winter J. Bio-removal of nitrogen from wastewaters–a review. J. Am. Sci. 2010;6:508–528. [Google Scholar]
- 7.Seviour R.J., Mino T., Onuki M. The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol. Rev. 2003;27:99–127. doi: 10.1016/S0168-6445(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 8.Sliekers A.O., Derwort N., Gomez J.C., Strous M., Kuenen J.G., Jetten M.S. Completely autotrophic nitrogen removal over nitrite in one single reactor. Water Res. 2002;36:2475–2482. doi: 10.1016/s0043-1354(01)00476-6. [DOI] [PubMed] [Google Scholar]
- 9.Jetten M.S., Strous M., van de Pas-Schoonen K.T., Schalk J., van Dongen U.G., van de Graaf A.A., Logemann S., Muyzer G., van Loosdrecht M.C. The anaerobic oxidation of ammonium. FEMS Microbiol. Rev. 1998;22:421–437. doi: 10.1111/j.1574-6976.1998.tb00379.x. [DOI] [PubMed] [Google Scholar]
- 10.Dongen L.G., Jetten M.S., van Loosdrecht M.C., Kujawa-Roeleveld . IWA Publishing; 2001. The Combined Sharon/Anammox Process, A Sustainable Method for N-Removal from Sludge Water. [Google Scholar]
- 11.Pynaert K., Smets B.F., Beheydt D., Verstraete W. Start-up of autotrophic nitrogen removal reactors via sequential biocatalyst addition. Environ. Sci. Technol. 2004;38:1228–1235. doi: 10.1021/es030081+. [DOI] [PubMed] [Google Scholar]
- 12.Kornaros M., Dokianakis S.N., Lyberatos G. Partial nitrification/denitrification can be attributed to the slow response of nitrite oxidizing bacteria to periodic anoxic disturbances. Environ. Sci. Technol. 2010;44:7245–7253. doi: 10.1021/es100564j. [DOI] [PubMed] [Google Scholar]
- 13.Tsuneda S., Ohno T., Soejima K., Hirata A. Simultaneous nitrogen and phosphorus removal using denitrifying phosphate-accumulating organisms in a sequencing batch reactor. Biochem. Eng. J. 2006;27:191–196. [Google Scholar]
- 14.Gut L., Płaza E., Hultman B. Assessment of a two-step partial nitritation/Anammox system with implementation of multivariate data analysis. Chemom. Intell. Lab. Syst. 2007;86:26–34. [Google Scholar]
- 15.Rodriguez-Sanchez A., Gonzalez-Martinez A., Martinez-Toledo M.V., Garcia-Ruiz M.J., Osorio F., Gonzalez-Lopez J. The effect of influent characteristics and operational conditions over the performance and microbial community structure of partial nitritation reactors. Water. 2014;6:1905–1924. [Google Scholar]
- 16.Shalini S.S., Joseph K. Nitrogen management in landfill leachate: application of SHARON, ANAMMOX and combined SHARON-ANAMMOX process. Waste Manag. 2012;32:2385–2400. doi: 10.1016/j.wasman.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Orhon D., Karahan O., Zengin G.E., Olsson O., Bauer M. IWA Publishing; 2005. Mechanism and Design of Sequencing Batch Reactors for Nutrient Removal. [Google Scholar]
- 18.Li J., Elliott D., Nielsen M., Healy M.G., Zhan X. Long-term partial nitrification in an intermittently aerated sequencing batch reactor (SBR) treating ammonium-rich wastewater under controlled oxygen-limited conditions. Biochem. Eng. J. 2011;55:215–222. [Google Scholar]
- 19.Li J.P., Healy M.G., Zhan X., Norton D., Rodgers M. Effect of aeration rate on nutrient removal from slaughterhouse wastewater in intermittently aerated sequencing batch reactors. Water Air Soil Pollut. 2008;192:251–261. [Google Scholar]
- 20.Pan M., Chen T., Hu Z., Zhan X. Assessment of nitrogen and phosphorus removal in an intermittently aerated sequencing batch reactor (IASBR) and a sequencing batch reactor (SBR) Water Sci. Technol. 2013;68:400–405. doi: 10.2166/wst.2013.190. [DOI] [PubMed] [Google Scholar]
- 21.Sanz J.L., Köchling T. Molecular biology techniques used in wastewater treatment: an overview. Process. Biochem. 2007;42:119–133. [Google Scholar]
- 22.Valentín-Vargas A., Toro-Labrador G., Massol-Deyá A.A. Bacterial community dynamics in full-scale activated sludge bioreactors: operational and ecological factors driving community assembly and performance. PLoS One. 2012;8 doi: 10.1371/journal.pone.0042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S.H., Kang H.J., Park H.D. Influence of influent wastewater communities on temporal variation of activated sludge communities. Water Res. 2015;73:132–144. doi: 10.1016/j.watres.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Martinez A., Rodriguez-Sanchez A., Garcia-Ruiz M.J., Muñoz-Palazon B., Cortes-Lorenzo C., Osorio F., Vahala R. Performance and bacterial community dynamics of a CANON bioreactor acclimated from high to low operational temperatures. Chem. Eng. J. 2016;287:557–567. [Google Scholar]
- 25.Pan M., Henry L.G., Liu R., Huang X., Zhan X. Nitrogen removal from slaughterhouse wastewater through partial nitrification followed by denitrification in intermittlently aerated sequencing batch reactors at 11 °C. Environ. Technol. 2013;35:470–477. doi: 10.1080/09593330.2013.832336. [DOI] [PubMed] [Google Scholar]
- 26.Henry L.G. NUI Galway; Ireland: 2014. Title Comparison of Intermittently Aerated Sequencing Batch Reactors (IASBRs) and Conventional Sequencing Batch Reactors (Csbrs) in Wastewater Treatment. PhD Disseration.https://aran.library.nuigalway.ie/handle/10379/4249 [Google Scholar]
- 27.Pan M., Hu Z., Liu R., Zhan X. Effects of loading rate and aeration on nitrogen removal and N2O emissions in intermittently aerated sequencing batch reactors treating slaughterhouse wastewater at 11 °C. Bioprocess Biosyst. Eng. 2015;38:681–689. doi: 10.1007/s00449-014-1307-1. [DOI] [PubMed] [Google Scholar]
- 28.Federation WE & American Public Health Association (APHA) 21st edition. 2005. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- 29.Wang Y., Qian P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Z.M., Wang Y., Tian R.M., Lee O.O., Wong Y.H., Batang Z.B., Al-Suwailem A., Lafi F.F., Bajic V.B., Qian P.Y. Pyrosequencing revealed shifts of prokaryotic communities between healthy and disease-like tissues of the Red Sea sponge Crella cyathophora. PeerJ. 2015;3:e890. doi: 10.7717/peerj.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bragg L., Stone G., Imelfort M., Hugenholtz P., Tyson G.W. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat. Methods. 2012;9:425–426. doi: 10.1038/nmeth.1990. [DOI] [PubMed] [Google Scholar]
- 32.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 34.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMurdie P.J., Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014 doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulson J.N., Stine O.C., Bravo H.C., Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Vega Thurber R.L., Knight R., Beiko R.G., Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Team R . RStudio, Inc.; Boston (MA): 2015. RStudio: Integrated Development for R. [Google Scholar]
- 40.Wickham H. ggplot2: elegant graphics for data analysis. J. Stat. Softw. 2009;35:65–68. [Google Scholar]
- 41.Guo Y., Peng Y., Wang B., Li B., Zhao M. Achieving simultaneous nitrogen removal of low C/N wastewater and external sludge reutilization in a sequencing batch reactor. Chem. Eng. J. 2016;306:925–932. [Google Scholar]
- 42.Kamika I., Coetzee M., Mamba B.B., Msagati T., Momba M.N.B. The impact of microbial ecology and chemical profile on the enhanced biological phosphorus removal (EBPR) process: a case study of northern wastewater treatment works. Johannesbg. Int. J. Environ. Res. Public Health. 2014;11:2876–2898. doi: 10.3390/ijerph110302876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong Y., Xia Y., Nielsen J.L., Nielsen P.H. Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiol. 2007;153(12):4061–4073. doi: 10.1099/mic.0.2007/007245-0. [DOI] [PubMed] [Google Scholar]
- 44.Bojnec S., Ferto I. Export competitiveness of dairy products on global markets: the case of the European Union countries. J. Dairy Sci. 2014;97:6151–6163. doi: 10.3168/jds.2013-7711. [DOI] [PubMed] [Google Scholar]
- 45.Demirel B., Yenigun O., Onay T.T. Anaerobic treatment of dairy wastewaters: a review. Process Biochem. 2005;40:2583–2595. [Google Scholar]
- 46.Minescu A., Wall D., Murphy J., Dobson A.D.W., O’ Leary N.D. Biopolymer production from Irish dairy industry wastewaters. EPA Rep. 2016;190:1–50. ISBN 978-1-84095-670-2. [Google Scholar]
- 47.Li J.P., Sorenson K., Zhan X.M., Healy M.G., Norton D., Rodgers M. Nutrient removal from slaughterhouse wastewater in an intermittently aerated sequencing batch reactor. Bioresour. Technol. 2008;99(16):7644–7650. doi: 10.1016/j.biortech.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Wagner M., Loy A. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 2002;13:218–227. doi: 10.1016/s0958-1669(02)00315-4. [DOI] [PubMed] [Google Scholar]
- 49.Werner J.J., Knights D., Garcia M.L., Scalfone N.B., Smith S., Yarasheski K., Cummings T.A., Beers A.R., Knight R., Angenent L.T. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4158–4163. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xin X., He J., Wang Y., Feng J., Qiu W. Role of aeration intensity on performance and microbial community profiles in a sequencing batch reaction kettle (SBRK) for wastewater nutrients rapid removal. Bioresour. Technol. 2016;201:140–147. doi: 10.1016/j.biortech.2015.11.053. [DOI] [PubMed] [Google Scholar]
- 51.Sadaie T., Sadaie A., Takada M., Hamano K., Ohnishi J., Ohta N., Matsumoto K., Sadaie Y. Reducing sludge production and the domination of comamonadaceae by reducing the oxygen supply in the wastewater treatment procedure of a food-processing factory. Biosci. Biotechnol. Biochem. 2007;71(3):791–799. doi: 10.1271/bbb.60632. [DOI] [PubMed] [Google Scholar]
- 52.Weissbrodt D.G., Shani N., Holliger C. Linking bacterial population dynamics and nutrient removal in the granular sludge biofilm ecosystem engineered for wastewater treatment. FEMS Microbiol. Ecol. 2014;88:579–595. doi: 10.1111/1574-6941.12326. [DOI] [PubMed] [Google Scholar]
- 53.Willems A. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. 4th ed. Springer; Reference: 2014. The family Comamonadaceae; pp. 777–851. [Google Scholar]
- 54.Calderer M., Martí V., De Pablo J., Guivernau M., Prenafeta-Boldú F.X., Viñas M. Effects of enhanced denitrification on hydrodynamics and microbial community structure in a soil column system. Chemosphere. 2014;111:112–119. doi: 10.1016/j.chemosphere.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 55.Ge H., Batstone D.J., Keller J. Biological phosphorus removal from abattoir wastewater at very short sludge ages mediated bynovel PAO clade Comamonadaceae. Water Res. 2015;69:173–182. doi: 10.1016/j.watres.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 56.Ahmed W., Staley C., Sidhu J., Sadowsky M., Toze S. Amplicon-based profiling of bacteria in raw and secondary treated wastewater from treatment plants across Australia. Appl. Microbiol. Biotechnol. 2017;101:1253–1266. doi: 10.1007/s00253-016-7959-9. [DOI] [PubMed] [Google Scholar]
- 57.Gao J., Liu G., Li H., Xu L., Du L., Yang B. Predictive functional profiling using marker gene sequences and community diversity analyses of microbes in full-scale anaerobic sludge digesters. Bioprocess. Biosyst. Eng. 2016;39:1115–1127. doi: 10.1007/s00449-016-1588-7. [DOI] [PubMed] [Google Scholar]
- 58.Mota C., Head M.A., Ridenoure J.A., Cheng J.J., Francis L. Effects of aeration cycles on nitrifying bacterial populations and nitrogen removal in intermittently aerated reactors. Appl. Environ. Microbiol. 2005;71(12):8565–8572. doi: 10.1128/AEM.71.12.8565-8572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan C., Yang X., Lee D.J., Du M., Wan F., Chen C. Aerobic denitrification by novel isolated strain using as nitrogen source. Bioresour. Technol. 2011;102:7244–7248. doi: 10.1016/j.biortech.2011.04.101. [DOI] [PubMed] [Google Scholar]
- 60.Zheng X., Wu R., Chen Y. Effects of ZnO nanoparticles on wastewater biological nitrogen and phosphorus removal. Environ. Sci. Technol. 2011;45:2826–2832. doi: 10.1021/es2000744. [DOI] [PubMed] [Google Scholar]
- 61.Chen H., Wang D., Li X., Yang Q., Luo K., Zeng G., Tang M. Effects of Cd(II) on wastewater biological nitrogen and phosphorus removal. Chemosphere. 2014;117:27–32. doi: 10.1016/j.chemosphere.2014.05.057. [DOI] [PubMed] [Google Scholar]
- 62.Sakai K., Miyake S., Iwama K., Inoue D., Soda S., Ike M. Polyhydroxyalkanoate (PHA) accumulation potential and PHA-accumulating microbial communities in various activated sludge processes of municipal wastewater treatment plants. J. Appl. Microbiol. 2015;118:255–266. doi: 10.1111/jam.12683. [DOI] [PubMed] [Google Scholar]
- 63.Motlagh A.M., Goel R. Water Reclamation and Sustainability. Elsevier Science Ltd.; Waltham, MA: 2014. Sustainability of activated sludge processes. [Google Scholar]
- 64.Heylen K., Vanparys B., Wittebolle L., Verstraete W., Boon N., De Vos P. Cultivation of denitrifying bacteria: optimization of isolation conditions and diversity study. Appl. Environ. Microbiol. 2006;72:2637–2643. doi: 10.1128/AEM.72.4.2637-2643.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.