Abstract
A number of studies revealed that CCNE1 copy number amplification and overexpression (on mRNA or protein expression level) were associated with prognosis of diverse cancers, however, the results were inconsistent among studies. So we conducted this systematic review and meta-analysis to investigate the prognostic values of CCNE1 amplification and overexpression in cancer patients. PubMed, Cochrane library, Embase, CNKI and WanFang database (last update by February 15, 2018) were searched for literatures. A total of 20 studies were included and 5 survival assessment parameters were measured in this study, which included overall survival (OS), progression free survival (PFS), recurrence free survival (RFS), cancer specific survival (CSS) and distant metastasis free survival (DMFS). Pooled analyses showed that CCNE1 amplification might predict poor OS (HR=1.59, 95% CI: 1.05-2.40, p=0.027) rather than PFS (HR=1.49, 95% CI: 0.83-2.67, p=0.177) and RFS (HR=0.982, 95% CI: 0.2376-4.059, p=0.9801) in various cancers; CCNE1 overexpression significantly correlated with poor OS (HR=1.52, 95% CI: 1.05-2.20, p=0.027), PFS (HR=1.20, 95% CI: 1.07-1.34, p=0.001) and DMFS (HR=1.62, 95% CI: 1.09-2.40, p=0.017) rather than RFS (HR=1.68, 95% CI: 0.81-3.50, p=0.164) and CSS (HR=1.54, 95% CI: 0.74-3.18, p=0.246). On the whole, these results indicated CCNE1 amplification and overexpression were associated with poor survival of patients with cancer, suggesting that CCNE1 might be an effective prognostic signature for cancer patients.
Keywords: cancer, CCNE1, prognosis, systematic review, meta-analysis
Introduction
Cancer is becoming one of the main causes of death and is a hard challenge to the world 1. Early diagnosis and prognosis prediction are helpful for clinical treatment of cancers 2, but traditional detection methods such as biopsy and imaging techniques still have their limitations. Therefore, it is urgently needed to excavate new molecular signatures for the early and precise prognosis prediction of cancers.
Both gene copy number variations and mRNA expression levels (or protein expression levels) in cancer cells could be used as signatures for prognosis and response to therapies. For example, HER2 amplification was an efficient predictor of overall survival in breast cancer patients 3. Amplification, genetic rearrangement and overexpression of cell cycle cyclins genes had been documented in almost all human cancer types and contributed to development and progression of cancers 4. Cyclin E1, encoded by the CCNE1, drives the transition from G1 to S phase by binding and activation of cyclin dependent kinase 2 (CDK2), resulting in the initiation of DNA synthesis 5. Up to now, an increasing number of studies had reported the relations between CCNE1 and prognosis in diverse cancers, but still generated discrepant conclusions. For instance, overexpression and copy number amplification of CCNE1 were related to poor survival in some kinds of malignant tumors such as bladder cancer 6, breast cancer 7, ovarian cancer [8]and so on, while some opposite results existed 9, 10. Therefore, the prognostic value of CCNE1 in cancer patients remained uncertain.
It's worth noting that, in some literatures, Cyclin E1 and Cyclin E2 (encoded by CCNE2) were collectively referred to as Cyclin E 11-13, while Cyclin E1 was formerly called as Cyclin E in some studies 14. Furthermore, Cyclin E2 was also found to be associated with prognosis in several kinds of cancer 15, 16. Therefore, research results derived from articles about Cyclin E without definite indication of Cyclin E1 (or CCNE1) in the full text were not explicit, because we couldn't determine whether these conclusions were the separate effect of Cyclin E1 or the combined effect of Cyclin E1 and Cyclin E2. So in this study, we conducted a rigorous systematic review and meta-analysis for the prognostic values of CCNE1 amplification and overexpression rather than Cyclin E.
Materials and Methods
Literature search
Comprehensive literature search of PubMed, Cochrane library, Embase, CNKI database and WanFang database was performed for articles published prior to February 15, 2018 by using Medical Subject Headings (MeSH) combined with keywords as follows: “cyclin E1, human”, “CCNE1”, “Cyclin E1”, “CCNE”, “Cyclin Es”, “Cyclin Et”, “G1/S-Specific Cyclin-E1”, “Prognosis”, “Survival”, “Prognoses”, “Factor, Prognostic”, “Prognostic Factor”, “Carcinoma”, “Neoplasms”, “Neoplasia”, “Neoplasm”, “Tumor”, “Cancer”, “Malignancy”, “Benign Neoplasm”, “Neoplasm, Benign”, “Malignant Epithelial Neoplasm”, “Epithelioma”, “Carcinomatosis”. Additionally, we conducted the manual search of the references in all included articles to identify potential missing studies that were not found during the preliminary literature searches. Inclusion criteria for the included studies: (1) all articles should be published either in English or Chinese language in the form of full article; (2) observational studies (case-control or cohort studies) that investigated patients with any type of cancer; (3) articles investigating the correlations between CCNE1 expression levels (protein expression level or mRNA expression level) and the prognosis in cancer patients or the correlations of CCNE1 amplification and the prognosis in cancer patients; (4) Hazard ratio (HR) and 95% confidence interval (95% CI) of survival assessment parameters were reported directly, or there was sufficient original data for estimating them in article; (5) when the patient cohorts overlapped among different publications, only the largest or the most recent one was included. Exclusion criteria were as follow: (1) studies without usable or sufficient data; (2) case reports, reviews, letters, comments, conference abstracts and expert opinions; (3) article about Cyclin E in which there was no definite indication of Cyclin E1 (CCNE1); (4) researches about a set of genes rather than CCNE1 alone; (5) results from overlapping patients cohorts; (6) articles based on external datasets. Screening of eligible studies was independently performed by four investigators (HY.Z, JL.W, LS.L, HL.Y), disagreements were resolved by consensus.
Data extraction and quality assessment
Four investigators (HY.Z, JL.W, LS.L, and HL.Y) independently extracted the available data from all included studies. The extracted data was crosschecked and any discrepancy was resolved by discussion. The following information was extracted from each included study: first author name, publication year, study design, cancer type, inclusion period, case and control numbers, research country, region, method for detecting CCNE1 amplification, method for detecting CCNE1 expression level, cut off value, research center, duration of follow-up, HR and 95% CI. If the article only reported Kaplan-Meier curve (K-M curve) without providing HR and 95% CI directly, appropriate data was extracted from the survival curves by using Engauge Digitizer 4.1 software and the logHR and selogHR were calculated according to Tierney et al 17. This method was conducted by three investigators to reduce imprecision. For studies that reported both the univariate and multivariate analysis results, the latter was extracted. Survival assessment parameters with HR and 95% CI available in these included studies were selected in our research, including overall survival (OS), progression free survival (PFS), cancer specific survival (CSS), recurrence free survival (RFS) and distant metastasis free survival (DMFS). The Newcastle-Ottawa Scale (NOS) 18 was used to assess the quality of each included study. The NOS score ranged from 0 to 9, and a study with score >7 was considered as high quality. Three investigators (HY.Z, JL.W, LS.L) independently scored each study, any discrepancy was resolved by discussion.
Statistical Analysis
We used HR with corresponding 95% CI to calculate pooled data. By convention, HR>1 indicated poor prognosis for the CCNE1 amplification or overexpression group. The heterogeneity among studies was determined by Q test and I2 value 19. When p<0.05 for the Q test or I2>50%, heterogeneity was supposed to be significant, and then the random effect model was applied. On the contrary, fixed effect model was employed (p≥0.05 or I2≤50%). Publication bias was evaluated by Egger's test 20. And when the number of included studies was <10, publication bias analysis was not conducted. Subgroup analysis was performed to find out factors contributing to heterogeneity. One-way sensitivity analysis was performed to assess the stability of results, but when the number of included studies was <3, it was not conducted. P values in all analyses were two sides, and p<0.05 was considered statistically significant. All statistical analyses in this meta-analysis were conducted with STATA 12.0 (Stata Corporation, College Station, TX, USA).
Results
Identification of relevant studies
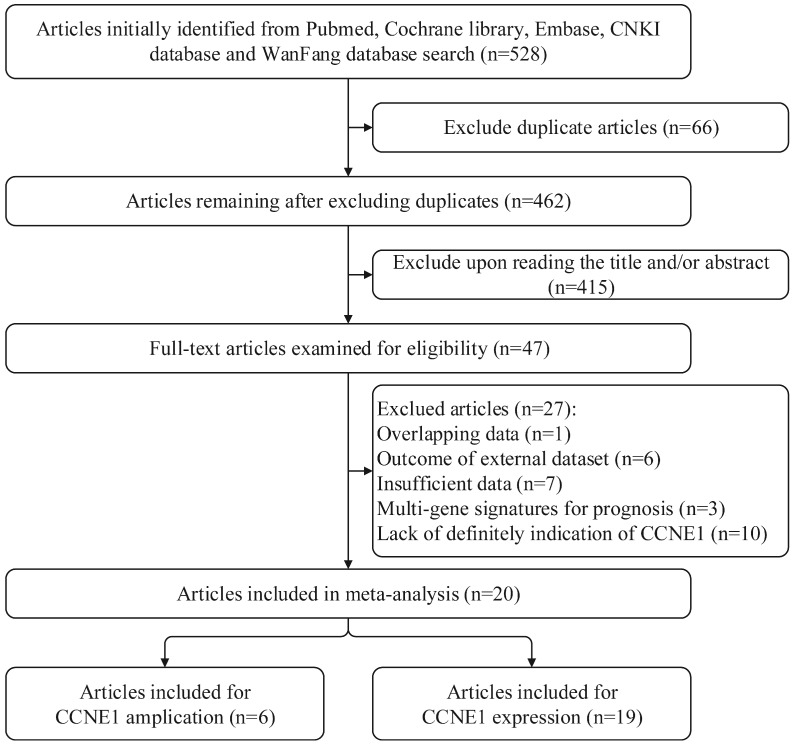
By applying the described searching strategy, we initially collected 528 publications. After exclusion of duplicates and irrelevant studies by reviewing the titles and abstracts, 47 articles remained for further evaluation. We reviewed all of the 47 publications by reading full manuscripts, and excluded 27 articles as follows: 7 21-27 articles lacking sufficient information, 6 28-33 articles based on external datasets, 1 34 article with overlapping data, 3 35-37 articles reporting gene sets including CCNE1 rather than CCNE1 as a single gene signature for prognosis and 10 38-47 articles about Cyclin E in which there were no explicit indication of CCNE1. Finally, 20 studies were qualified and included in our research. Among them, 1 article only exploring the predictive roles of CCNE1 copy number amplification in cancer prognosis 48, 14 articles were only about CCNE1 expression levels (on mRNA or protein expression level) 6, 7, 10, 49-59, and 5 articles examined both CCNE1 amplification and expression levels 8, 9, 60-62. The detailed procedure of literature retrieval is revealed in Figure 1. We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram to design, analyze, and report this meta-analysis 63.
Figure 1.
Flow chart for searching databases and selecting eligible studies.
Study characteristics
The main characteristics of the included 6 studies that explored the prognostic value of CCNE1 amplification in cancers are presented in Table 1A. 1120 patients were enrolled in these researches totally, and the cohort sizes ranged from 83 to 489 cases with the mean of 186. There were 4 articles on ovarian cancer, 1 literature about endometrial carcinomas and 1 research on breast cancer. For detecting the amplification of CCNE1, fluorescence in situ hybridization (FISH) was applied in 3 studies, copy number polymerase chain reaction (PCR) in 1 study, microarray in 1 study and chromogenic in situ hybridization (CISH) in 1 study. CCNE1 amplification proportion varied from 8.33% to 25%, with the mean percentage of 16.54%. HR and 95% CI were directly reported in 5 studies and extracted from K-M curves in 1 study. All studies assessed CCNE1 amplification in tumor tissue.
Table 1A.
Characteristics of the included 6 studies that researching on the predictive value of CCNE1 amplification for cancers prognosis
| Author (Year) | Inclusion period |
Country | Research center |
Tumor Type | Case /Control |
Median/Range of follow-up(Mo) |
Detection method |
Outcome measures |
Amplication percentage |
Variance analysis |
HR source |
Region |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ayhan A(2016) | 1995-2013 | Muti-countrya | multicenter | ovarian cancer | 13/70 | NR | FISH | OS | 15.66 | Univariate | Reported | Mixed |
| Nakayama K(2015) | 1998-2010 | Japan | single | endometrial carcinomas | 9/99 | 52(5-139) | FISH | OS PFS | 8.33 | Multivariate | Reported | Asia |
| Pils D(2014) | 2005-2008 | Muti-countryb | multicenter | ovarian cancer | totalc 167 | 49(1-69) | copy number PCR | OS PFS | 25 | Univariate | Reported | Europe |
| TCGA(2011) | NR | USA | multicenter | ovarian cancer | 106/383 | 30(1-179) | microarray | OS | 21.68 | Univariate | K-M curve | North America |
| Nakayama N(2010) | NR | Japan | single | ovarian cancer | 18/70 | NR | FISH | OS PFS | 20.45 | Multivariate | Reported | Asia |
| Luhtala S(2016) | 2003-2007 | Finland | single | breast cancer | 15/170 | 64.8 | CISH | RFS | 8.11 | Univariate | Reported | Europe |
a: Japan, USA. b: Germany, Belgium, Austria. c: total: the study didn't provide detail number of control and case group.
The main characteristics of the 19 included studies that researched on the predictive roles of CCNE1 expression level in cancers prognosis are shown in Table 1B. The total number of patients included in these articles was 4592, and the sample size ranged from 60 to 857 with the mean value of 242. There were 8 articles on breast cancer, 5 literatures about ovarian cancer, 3 researches on bladder cancer, 1 study about endometrial carcinomas, 1 article about esophageal adenocarcinoma and 1 research on upper tract urothelial carcinoma. For detecting expression levels of CCNE1, immunochemistry (IHC) was applied to evaluate CCNE1 protein expression level in 12 studies, real-time quantitative polymerase chain reaction (RT-qPCR) was performed to detect CCNE1 mRNA expression level in 6 studies, and microarray was conducted to detect CCNE1 mRNA expression level in 1 study. 3 articles treated CCNE1 expression level as continuous type variable, so there were no specific cut-off values in them 9, 56, 57. In the remaining 16 articles, CCNE1 expression level was all classified as high and low group, but the cut-off definitions were various. HR and 95% CI were directly reported in 15 studies and extracted from K-M curves in 4 studies. All studies assessed CCNE1 expression levels in tumor tissue.
Table 1B.
Characteristics of the included 19 studies that researching on the predictive value of CCNE1 overexpression for cancers prognosis.
| Author (Year) | Inclusion period |
Country | Research center |
Tumor Type |
Case/ Control |
Median/ Range of follow-up(Mo) |
Detection method |
Cut-off | Outcome measures |
High expression percentage |
Variance analysis |
HR source |
Region |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ayhan A(2016) | 1995-2013 | Muti-countrya | multicenter | ovarian cancer | 20/63 | NR | IHC | 80%b | OS | 24.1 | Multivariate | Reported | Mixed |
| Nakayama K(2015) | 1998-2010 | Japan | single | EC | 54/54 | 52(5-139) | IHC | medianc | OS PFS | 50 | Univariate | Reported | Asia |
| Zhou Z(2014) | 2002-2008 | USA | single | EAC | totald 112 | 39(0.3-142) | IHC | 10%e | OS | NR | Univariate | K-M curve | North America |
| Wu S(2014) | NR | China | single | UTUC | 59/44 | 39(1-98) | IHC | 5f | OS | 57.28 | Multivariate | Reported | Asia |
| Nakayama N(2010) | NR | Japan | single | ovarian cancer | 44/44 | NR | IHC | median | OS PFS | 50 | Univariate | Reported | Asia |
| Farley J(2003) | NR | USA | multicenter | ovarian cancer | 62/77 | NR | IHC | meang | OS | 44.6 | Multivariate | Reported | North America |
| Shariat SF(2006) | 1987-2002 | USA | single | bladder cancer | 125/99 | NR | IHC | 30%h | CSS RFS | 55.31 | Univariate | K-M curve | North America |
| Chappuis PO(2005) | 1980-1995 | Canada | single | breast cancer | 67/186 | 96 | IHC | 50%i | CSS | 26.48 | Univariate | K-M curve | North America |
| Lotan Y(2013) | 2002-2012 | USA | single | bladder cancer | 183/216 | NR | IHC | 30%j | CSS RFS | 84.72 | Univariate | Reported | North America |
| Lundgren C(2015) | 1993-2004 | Sweden | multicenter | breast cancer | 186/186 | NR | IHC | NR | CSS DMFS | 48.45 | Univariate | Reported | Europe |
| Luhtala S(2016) | 2003-2007 | Finland | single | breast cancer | 74/128 | 64.8 | IHC | 50%k | RFS | 36.63 | Univariate | Reported | Europe |
| Matsushita R(2015) | 2003-2013 | Japan | single | bladder cancer | 30/30 | NR | RT-qPCR | median | OS | 50 | Univariate | K-M curve | Asia |
| Pils D(2014) | 2005-2008 | Muti-countryl | multicenter | ovarian cancer | totald 167 | 49(1-69) | RT-qPCR | none | OS PFS | none | OS:Multivariate PFS:Univariate |
Reported | Europe |
| Marchini S(2008) | 1992-2003 | Italy | single | ovarian cancer | OS:9/59; PFS:10/57 | NR | RT-qPCR | OS(0.124); PFS(0.34) |
OS PFS | OS:13.23 PFS:14.93 |
Multivariate | Reported | Europe |
| Sieuwerts AM(2006) | 1979-1995 | Netherland | multicenter | breast cancer | 317/318 | 95(11-202) | RT-qPCR | median | OS DMFS | 49.92 | Multivariate | Reported | Europe |
| Jansen MP(2011) | 1981-1996 | Netherland | single | breast cancer | totald 226 | 89(10-165) | RT-qPCR | none | PFS | none | Multivariate | Reported | Europe |
| Kreike B(2010) | 1984-1995 | Netherland | single | breast cancer | totald 291 | 80.4(0.6-237.9) | microarray | none | RFS | none | Univariate | Reported | Europe |
| Desmedt C(2006) | NR | UK | single | breast cancer | totald 205 | 97.32 | RT-qPCR | median | RFS | 50 | Univariate | Reported | Europe |
| Fredholm H(2017) | 1992-2005 | Swedish | multicenter | breast cancer | 435/422 | 115.2 | IHC | 10%m | DMFS | 50.76 | Univariate | Reported | Europe |
a: Japan, USA. b: diffuse and intense immunoreactivity in 80% of tumor cells were considered highly expressed. c: median value. d: total: the study didn't provide detail number of control and case group. e: 10% or more of cells stained with a moderate to strong intensity was considered highly expressed. f: staining index score = 5. g: mean value. h: CCNE1 immunoreactivity was considered high when samples demonstrated more than 30% nuclear reactivity. i: CCNE1 immunoreactivity was considered high when samples demonstrated more than 50% nuclear reactivity. j: CCNE1 immunoreactivity was considered high when samples demonstrated more than 30% nuclear reactivity. k: CCNE1 immunoreactivity was considered high when samples demonstrated more than 50% nuclear reactivity. l: Germany, Belgium, Austria. m: CCNE1 immunoreactivity was considered high when samples demonstrated more than 10% nuclear reactivity.
As shown in Table S1, global quality assessment scores of the 20 included studies in this study were generally high with 14 (70%) articles being more than 7 points, ranged from 6 to 8.
Survival analysis for CCNE1 amplification carriers with cancers
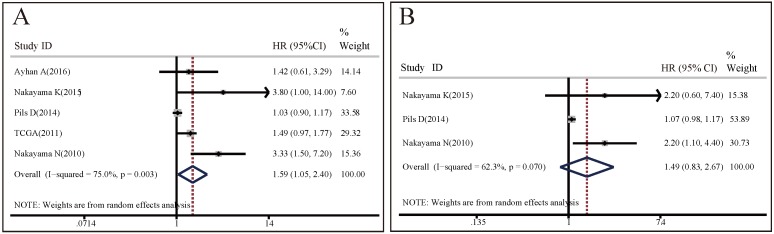
OS analysis: 5 studies with totally 935 patients investigated the association between CCNE1 amplification and OS. A random effect model was used to estimate the pooled HR and 95% CI, due to a significant heterogeneity among these 5 studies (I2=75%, p=0.003). As shown in Figure 2A, the results demonstrated that CCNE1 amplification carriers were associated with worse OS than non-carriers, with a pooled HR of 1.59 (95% CI: 1.05-2.40, p=0.027). The results of subgroup analyses were presented in Table S2A. It revealed that analysis method, detection method, region, and research center might contribute to the heterogeneity. It was noteworthy that, of these 5 included studies, 4 studies focused on ovarian cancer, but CCNE1 amplification had no significant relationship with OS in ovarian cancer patients (HR=1.46, 95% CI: 0.98-2.18, p=0.064). In studies adopting multivariate analysis rather than univariate analysis, CCNE1 amplification carriers had statistically significantly worse OS (multivariate analysis: HR=3.45, 95% CI: 1.76-6.77, p<0.001; univariate analysis: HR=1.22, 95% CI: 0.90-1.65, p=0.193).
Figure 2.
Forest plots of associations between CCNE1 amplification and survival in cancer patients. (A) Effect of CCNE1 amplification on overall survival. (B) Effect of CCNE1 amplification on overall survival progression free survival.
PFS analysis: 3 studies of 363 patients assessed the predictive value of CCNE1 amplification for PFS. Significant heterogeneity was found across these included studies (I2=62.3%, p=0.07), so the pooled HR was calculated based upon a random effects model. Meta-analysis results demonstrated no significant association between CCNE1 amplification and PFS (HR=1.49, 95% CI: 0.83-2.67, p=0.177) (Figure 2B). As shown in Table S2B, the results of subgroup analyses revealed that analysis method, detection method, region, and research center might contribute to the heterogeneity. We also found that there was no statistical difference in PFS between CCNE1 amplification carriers and non-carriers, regardless of tumor type or study quality. However, it was worth noting that, in studies adopting multivariate analysis, CCNE1 amplification carriers showed significantly worse PFS (HR=2.20, 95% CI: 1.20-4.04, p=0.011).
RFS analysis: only 1 study enrolling 185 patients reported the predictive values of CCNE1 amplification for RFS in breast cancer, with the HR of 0.982 (95% CI: 0.2376-4.059, p=0.9801), indicating no significant relationship between them in breast cancer patients.
Survival analysis for CCNE1 overexpression carriers with cancers
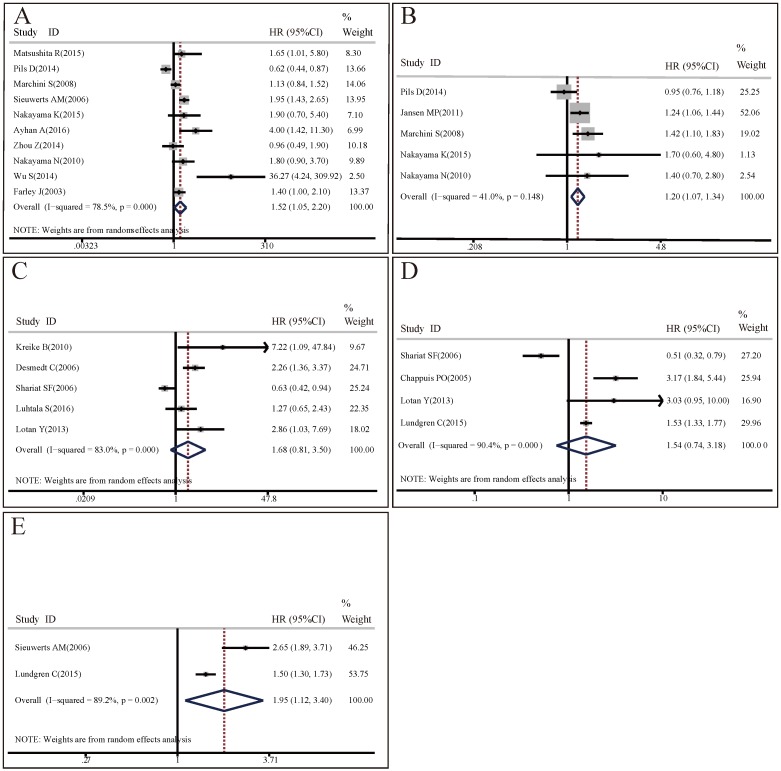
OS analysis: 10 studies with 1562 patients researched on the association between CCNE1 expression levels and OS. The random effect model was applied to estimate the pooled HR with corresponding 95% CI. As shown in Figure 3A, there was significant heterogeneity among these included studies (I2=78.5%, p<0.001), meta-analysis results demonstrated that there was significant difference in OS between high CCNE1 expression level group and low CCNE1 expression level group. CCNE1 overexpression carriers were associated with worse OS than non-carriers, with a pooled HR of 1.52 (95% CI: 1.05-2.20, p=0.027). Subgroup analyses revealed that CCNE1 overexpression carriers had statistically significantly shorter OS compared to non-carriers in studies as followed: on breast cancer or upper tract urothelial carcinoma; detecting by IHC method; about protein expression level; conducted in Asia or mixed region; with HR and 95% CI reported directly or with high quality. Noteworthily, of these 10 included studies, 5 studies were about ovarian cancer, but CCNE1 overexpression had no significant relationship with OS in ovarian cancer patients (HR=1.28, 95% CI: 0.81-2.03, p=0.283) (Table S3A).
Figure 3.
Forest plots of associations between CCNE1 overexpression and survival in cancer patients. (A) Effect of CCNE1 overexpression on overall survival. (B) Effect of CCNE1 overexpression on progression free survival. (C) Effect of CCNE1 overexpression on recurrence free survival. (D) Effect of CCNE1 overexpression on cancer specific survival. (E) Effect of CCNE1 overexpression on distant metastasis free survival.
PFS analysis: Since there was no significant heterogeneity across 5 included studies involving 666 patients (I2=41%, p=0.148), the fixed effect model was applied. Compared with low CCNE1 expression group, CCNE1 overexpression group had a significantly worse PFS (HR=1.20, 95% CI: 1.07-1.34, p=0.001) (Figure 3B). The results of subgroup analyses were presented in Table S3B. It revealed that analysis method might contribute to the heterogeneity. In studies adopting multivariate analysis, CCNE1 overexpression predicted statistically significantly poorer PFS (multivariate analysis: HR=1.29, 95% CI: 1.13-1.47, p<0.001; univariate analysis: HR=1.01, 95% CI: 0.82-1.24, p=0.958). However, of these 5 included studies, 3 studies were about ovarian cancer, but CCNE1 overexpression didn't show prognostic prediction ability for PFS in ovarian cancer patients (HR=1.14, 95% CI: 0.97-1.34, p=0.109).
RFS analysis: 5 studies encompassing 1140 patients investigated the predictive values of CCNE1 overexpression for RFS in cancers. As the result of meta-analysis exhibited obvious heterogeneity (I2=83.0%, p<0.001), the random effect model was used. As shown in Figure 3C, meta-analysis results revealed no association between CCNE1 overexpression and RFS, with a pooled HR of 1.68 (95% CI: 0.81-3.50, p=0.164). Subgroup analyses results showed that the source of HR might contribute to the heterogeneity. It was notable that, of these 5 included studies, there were 4 studies with HR and 95% CI from literatures reported directly, and CCNE1 overexpression were associated with poorer RFS (HR=2.1, 95% CI: 1.31-3.36, p=0.002) in them (Table S3C).
CSS analysis: 4 studies enrolling 881 patients assessed the association between CCNE1 expression levels and CSS. The p and I2 values of heterogeneity test were <0.001 and 90.4% respectively. After using random effect model, the pooled HR was 1.54 (95% CI: 0.74-3.18, p=0.246), indicating CCNE1 expression level didn't have significant correlation with CSS in cancer patients (Figure 3D). The results of subgroup analyses were presented in Table S3D.
DMFS analysis: A random effect model was used to estimate the pooled HR with corresponding 95% CI, due to a significant heterogeneity across 3 included studies with 1678 patients (I2=86.6%, p=0.001). As shown in Figure 3E, overexpression of CCNE1 might predict poorer DMFS in cancer patients, with a pooled HR of 1.62 (95% CI: 1.09-2.40, p=0.017). The results of subgroup analyses were presented in Table S3E.
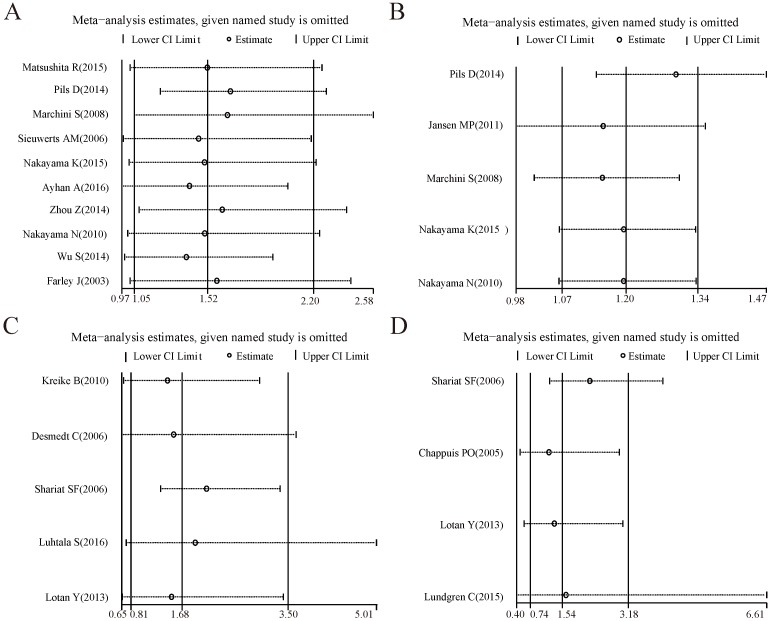
Sensitivity analysis
For the analysis groups about prognostic values of CCNE1 amplification on OS and PFS or about prognostic values of CCNE1 overexpression on OS, PFS, CSS, RFS and DMFS in cancer patients, sensitivity analyses were all performed by removing each study in turn from the pooled analysis. As showed in Figure 4-5, removing any of the included studies had no significant influence on the overall results of these groups mentioned above.
Figure 4.
Forest plots of sensitivity analysis for the included studies on CCNE1 amplification. Pooled hazard ratios and 95% confidence intervals by omitting each study. (A) For overall survival group. (B) For progression free survival group.
Figure 5.
Forest plots of sensitivity analysis for the included studies on CCNE1 overexpression. Pooled hazard ratios and 95% confidence intervals by omitting each study. (A) For overall survival group. (B) For progression free survival group. (C) For recurrence free survival group. (D) For cancer specific survival group. (E) For distant metastasis free survival group.
Publication bias
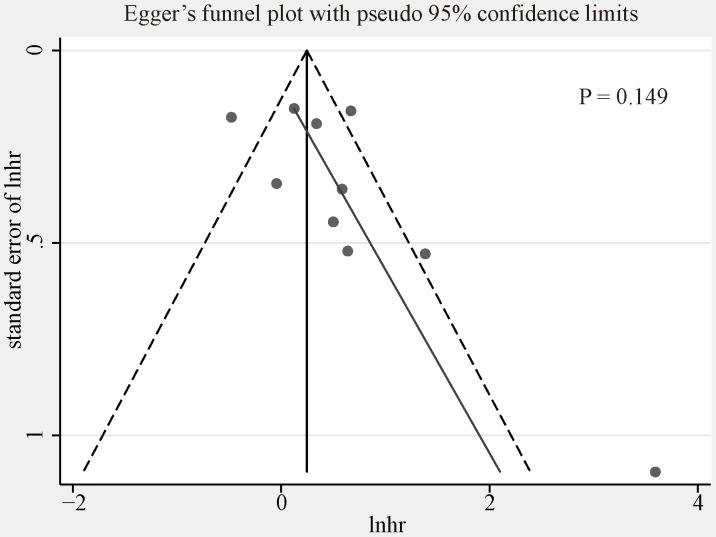
We used funnel plot and Egger's tests to evaluate the publication bias of included studies researching on the association between CCNE1 overexpression and OS (Figure 5, Egger's test, p=0.149). No evidence of significant publication bias was found. We did not analyze the publication bias of meta-analyses for the other groups mentioned above as the quantity of included studies was too small (<10) to make a valid statistical test.
Discussion
In cancer development, deregulation of cell cycle regulators was considered to be a prerequisite. CCNE1 located on the chromosomal band 19q12, was first isolated in 1991 64. Cyclin E1 functioning as a regulatory subunit of CDK2, played significant roles in chromosomal instability and apoptosis in cancer cells 5. In addition, researchers recently discovered that Cyclin E1 also had kinase-independent functions in cell-cycle progression 13. Overexpression and amplification of CCNE1 were reported to be associated with poor prognosis in a variety of cancers such as bladder cancer 6, ovarian cancer 8 and so on, while some opposite results existed 9, 10. Even in subtypes of a single type cancer, CCNE1 had different prognosis roles. For instance, CCNE1 overexpression was related to a significantly increased risk of death and relapse in basal-like and triple receptor-negative breast cancers, while there was a lack of such association in hormone receptor-positive and luminal breast cancers 65. So the prognostic value of CCNE1 in cancer patients remained unclear.
CCNE2 was located on the chromosomal band 8q22.1 66. Although Cyclin E1 and E2 were often coordinately regulated, and exhibited high homology within their important functional regions (47% between the overall sequences and 70% identity between the cyclin box) 67, which appeared to be functionally redundant, recently, more and more evidence suggested that Cyclin E1 and E2 were frequently expressed independently of one another in cancers, which had unique relationship with cancer prognosis. For example, it had been reported that Cyclin E2 overexpression was an independent and better prognostic signature for HER2 and luminal B breast cancer, when compared with Cyclin E1 12. Besides, both CCNE2 overexpression and amplification were related to endocrine therapy resistance in breast cancer 12, 68. However, in many studies, Cyclin E1 and Cyclin E2 were collectively referred to as Cyclin E, while in other articles, Cyclin E1 was called as Cyclin E. This inconsistency led us to conduct this meta-analysis for the prognosis value of Cyclin E1 in cancers rather than Cyclin E. We excluded articles about Cyclin E without definite indication of Cyclin E1 (or CCNE1) to avoid the possible confounding affect caused by Cyclin E2.
In this meta-analysis, a total of 20 studies were included and 5 survival assessment parameters (OS, PFS, RFS, CSS and DMFS) were measured. The results of meta-analysis indicated that CCNE1 amplification predicted worse OS rather than PFS and RFS in cancer patients. However, it was noteworthy that the number of the included studies about CCNE1 amplification for PFS or RFS was relatively small. And in studies adopting multivariate analysis, CCNE1 amplification carriers showed significantly worse PFS. CCNE1 overexpression was associated with worse OS, PFS and DMFS rather than RFS and CSS. Nevertheless, of 5 included studies for the relationship between CCNE1 overexpression and RFS, there were 4 studies with HR and 95% CI reported directly from literatures, and CCNE1 overexpression predicted poorer RFS in these 4 studies. Therefore, generally, CCNE1 amplification and overexpression indicated worse prognosis of patients with cancer. Heterogeneity was found to be significant for each analysis group in this meta-analysis, except for the group researching on the correlation of CCNE1 overexpression with PFS (I2=41%, p=0.148). So we carried out detailed subgroup analysis to examine the potential sources of heterogeneity under a broader range of study level circumstances, which showed that for different analysis groups, the causes of heterogeneity were diversified. Contrary to the pooled result of all the involving cancers in each analysis group, in ovarian cancer patients, CCNE1 amplification had no prognosis value on OS, and CCNE1 overexpression also didn't predict worse OS or PFS. It was quite unexpected, as researches about ovarian cancer accounted for the largest share in the included studies of these analysis groups. However, some studies had reported that CCNE1 amplification or overexpression could select ovarian cancer patients more likely to benefit from CDK2 targeted therapies 69, 70. And then, because the prognostic value of CCNE1 was reported to be affected by germline or somatic mutations of BRCA1/2 in ovarian cancer 48, so we suspected that the mutation rate of BRCA1/2 in different studies affected the assessment of the prognostic value of CCNE1 in ovarian cancer. In the future, we believed that the research for the role of CCNE1 in the prognosis of ovarian cancer needed to be carried adjusting for BRCA1/2. The sensitivity analysis showed that no individual study could significantly influence the conclusion in each analysis group of our research, indicating the credibility and stability of our meta-analysis results.
There were several limitations of this meta-analysis, which should be addressed. First, there were a variety of survival assessment parameters, while only 20 articles were included, therefore studies specific to some analysis groups were relatively insufficient, limited data availability for these groups reduced the credibility of our results. Second, we excluded a number of articles for Cyclin E without indication of Cyclin E1 (or CCNE1). Although this strategy avoided the possible confounding affect from Cyclin E2, it may inevitably delete some researches exactly about CCNE1. Third, despite no significant publication bias was observed in studies researching on the association between CCNE1 overexpression and OS, publication bias may still exist. Because we did not perform publication bias analysis for the other analysis groups as the quantity of included studies in them was too little; most articles reported positive results, whereas those with negative findings generally were less likely to be published; despite we tried to collect all relevant researches, but inevitably, some studies may still be missing. Fourth, the cut-off definition for CCNE1 overexpression was not unified among the included studies. Finally, HR and 95% CI in some studies were calculated based on the data extracted from survival curve, which may be less reliable than that reported directly in articles.
Despite the limitations described above, to our knowledge, this was the first meta-analysis about the predictive roles of CCNE1 amplification and overexpression in the prognosis of various cancers. Our systematic review and meta-analysis suggested that, on the whole, CCNE1 amplification and overexpression were associated with poor prognosis in cancer patients, which meant that CCNE1 could be hopefully applied to clinical work as a prognostic signature for cancers. Additionally, CCNE1 overexpression showed a more important role in prognosis than CCNE1 amplification in cancer patients. Unexpectedly, both amplification and overexpression of CCNE1 had no prognostic prediction ability in ovarian cancer patients. At last, the total number of our included studies was relatively small, more clinical investigations with larger sample size, multicenter, higher quality and prospective design were strongly needed to further validate the prognostic role of CCNE1 in cancer patients.
Figure 6.
Egger's funnel plot for publication bias among the included studies exploring prognostic values of CCNE1 overexpression for overall survival.
Acknowledgments
This work was generously supported by the National Natural Science Foundation for the Youth of China (No.81601992), the Natural Science Foundation of Jiangsu Province (No.BK20160343), Jiangsu Provincial Clinical Orthopedic Center.
Abbreviations
- NR
not reported
- EC
endometrial carcinomas
- EAC
esophageal adenocarcinoma
- UTUC
upper tract urothelial carcinoma
- OS
overall survival
- PFS
progression free survival, RFS: recurrence free survival, CSS: cancer specific survival
- DMFS
distant metastasis free survival
- CDK2
cyclin dependent kinase 2
- MeSH
Medical Subject Headings
- HR
Hazard ratio
- 95% CI
95% confidence interval
- K-M curve
Kaplan-Meier curve
- NOS
Newcastle-Ottawa Scale
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- FISH
fluorescence in situ hybridization
- PCR
polymerase chain reaction
- CISH
chromogenic in situ hybridization
- IHC
immunochemistry
- RT-qPCR
real-time quantitative polymerase chain reaction.
Supplementary Material
Supplementary tables.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Sequeiros T, Rigau M, Chiva C. et al. Targeted proteomics in urinary extracellular vesicles identifies biomarkers for diagnosis and prognosis of prostate cancer. Oncotarget. 2017;8:4960–76. doi: 10.18632/oncotarget.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slamon DJ, Clark GM, Wong SG. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 4.Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–92. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–86. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 6.Matsushita R, Seki N, Chiyomaru T. et al. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br J Cancer. 2015;113:282–9. doi: 10.1038/bjc.2015.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sieuwerts AM, Look MP, Meijer-van Gelder ME. et al. Which cyclin E prevails as prognostic marker for breast cancer? Results from a retrospective study involving 635 lymph node-negative breast cancer patients. Clin Cancer Res. 2006;12:3319–28. doi: 10.1158/1078-0432.CCR-06-0225. [DOI] [PubMed] [Google Scholar]
- 8.Nakayama N, Nakayama K, Shamima Y. et al. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer. 2010;116:2621–34. doi: 10.1002/cncr.24987. [DOI] [PubMed] [Google Scholar]
- 9.Pils D, Bachmayr-Heyda A, Auer K. et al. Cyclin E1 (CCNE1) as independent positive prognostic factor in advanced stage serous ovarian cancer patients - a study of the OVCAD consortium. Eur J Cancer. 2014;50:99–110. doi: 10.1016/j.ejca.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Shariat SF, Ashfaq R, Sagalowsky AI. et al. Correlation of cyclin D1 and E1 expression with bladder cancer presence, invasion, progression, and metastasis. Hum Pathol. 2006;37:1568–76. doi: 10.1016/j.humpath.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Caldon CE, Sergio CM, Sutherland RL. et al. Differences in degradation lead to asynchronous expression of cyclin E1 and cyclin E2 in cancer cells. Cell Cycle. 2013;12:596–605. doi: 10.4161/cc.23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caldon CE, Sergio CM, Kang J. et al. Cyclin E2 overexpression is associated with endocrine resistance but not insensitivity to CDK2 inhibition in human breast cancer cells. Mol Cancer Ther. 2012;11:1488–99. doi: 10.1158/1535-7163.MCT-11-0963. [DOI] [PubMed] [Google Scholar]
- 13.Geng Y, Lee YM, Welcker M. et al. Kinase-independent function of cyclin E. Mol Cell. 2007;25:127–39. doi: 10.1016/j.molcel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 14.Geng Y, Yu Q, Whoriskey W. et al. Expression of cyclins E1 and E2 during mouse development and in neoplasia. Proc Natl Acad Sci U S A. 2001;98:13138–43. doi: 10.1073/pnas.231487798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosco EE, Wang Y, Xu H. et al. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest. 2007;117:218–28. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Zhao S, Frasor JM. et al. An integrated bioinformatics approach identifies elevated cyclin E2 expression and E2F activity as distinct features of tamoxifen resistant breast tumors. PLoS One. 2011;6:e22274. doi: 10.1371/journal.pone.0022274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tierney JF, Stewart LA, Ghersi D. et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells G, Shea B, O'connell D, The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses. Department of Epidemiology and Community Medicine, University of Ottawa, Canada. www ohr i ca/programs/clinical_ epidemiology/oxford htm; 2012. [Google Scholar]
- 19.Higgins JP, Thompson SG, Deeks JJ. et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egger M, Davey Smith G, Schneider M. et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapoznik S, Aviel-Ronen S, Bahar-Shany K. et al. CCNE1 expression in high grade serous carcinoma does not correlate with chemoresistance. Oncotarget. 2017;8:62240–7. doi: 10.18632/oncotarget.19272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Passoni N, Gayed B, Kapur P. et al. Cell-cycle markers do not improve discrimination of EORTC and CUETO risk models in predicting recurrence and progression of non-muscle-invasive high-grade bladder cancer. Urol Oncol. 2016;34:485.e7–e14. doi: 10.1016/j.urolonc.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto T, Yanaihara N, Okamoto A. et al. Cyclin D1 predicts the prognosis of advanced serous ovarian cancer. Exp Ther Med. 2011;2:213–9. doi: 10.3892/etm.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callagy G, Pharoah P, Chin SF. et al. Identification and validation of prognostic markers in breast cancer with the complementary use of array-CGH and tissue microarrays. J Pathol. 2005;205:388–96. doi: 10.1002/path.1694. [DOI] [PubMed] [Google Scholar]
- 25.Etemadmoghadam D, deFazio A, Beroukhim R. et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15:1417–27. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayr D, Kanitz V, Anderegg B. et al. Analysis of gene amplification and prognostic markers in ovarian cancer using comparative genomic hybridization for microarrays and immunohistochemical analysis for tissue microarrays. Am J Clin Pathol. 2006;126:101–9. doi: 10.1309/n6x5mb24bp42kp20. [DOI] [PubMed] [Google Scholar]
- 27.Etemadmoghadam D, George J, Cowin PA. et al. Amplicon-dependent CCNE1 expression is critical for clonogenic survival after cisplatin treatment and is correlated with 20q11 gain in ovarian cancer. PLoS One. 2010;5:e15498. doi: 10.1371/journal.pone.0015498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vermeulen MA, Doebar S, van Deurzen C, Copy number profiling of oncogenes in ductal carcinoma in situ of the male breast. Endocr Relat Cancer; 2017. [DOI] [PubMed] [Google Scholar]
- 29.Yang H, Zhang X, Cai XY. et al. From big data to diagnosis and prognosis: gene expression signatures in liver hepatocellular carcinoma. PeerJ. 2017;5:e3089. doi: 10.7717/peerj.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song BN, Kim SK, Chu IS. Bioinformatic identification of prognostic signature defined by copy number alteration and expression of CCNE1 in non-muscle invasive bladder cancer. Exp Mol Med. 2017;49:e282. doi: 10.1038/emm.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao L, Ding J, Perdue A. et al. Cyclin E1 is a common target of BMI1 and MYCN and a prognostic marker for neuroblastoma progression. Oncogene. 2012;31:3785–95. doi: 10.1038/onc.2011.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Etemadmoghadam D, Au-Yeung G, Wall M. et al. Resistance to CDK2 inhibitors is associated with selection of polyploid cells in CCNE1-amplified ovarian cancer. Clin Cancer Res. 2013;19:5960–71. doi: 10.1158/1078-0432.CCR-13-1337. [DOI] [PubMed] [Google Scholar]
- 33.Wiedemeyer WR, Beach JA, Karlan BY. Reversing Platinum Resistance in High-Grade Serous Ovarian Carcinoma: Targeting BRCA and the Homologous Recombination System. Front Oncol. 2014;4:34. doi: 10.3389/fonc.2014.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shariat SF, Ashfaq R, Sagalowsky AI. et al. Association of cyclin D1 and E1 expression with disease progression and biomarkers in patients with nonmuscle-invasive urothelial cell carcinoma of the bladder. Urol Oncol. 2007;25:468–75. doi: 10.1016/j.urolonc.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Pluciennik E, Krol M, Nowakowska M. et al. Breast cancer relapse prediction based on multi-gene RT-PCR algorithm. Med Sci Monit. 2010;16:Cr132–6. [PubMed] [Google Scholar]
- 36.Jensen LB, Bartlett JM, Witton CJ. et al. Frequent amplifications and deletions of G1/S-phase transition genes, CCND1 and MYC in early breast cancers: a potential role in G1/S escape. Cancer Biomark. 2009;5:41–9. doi: 10.3233/CBM-2009-0570. [DOI] [PubMed] [Google Scholar]
- 37.Bieche I, Tozlu S, Girault I. et al. Identification of a three-gene expression signature of poor-prognosis breast carcinoma. Mol Cancer. 2004;3:37. doi: 10.1186/1476-4598-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, Zhang G, Wang HL. et al. Analysis of expression of cyclin E, p27kip1 and Ki67 protein in colorectal cancer tissues and its value for diagnosis, treatment and prognosis of disease. Eur Rev Med Pharmacol Sci. 2016;20:4874–9. [PubMed] [Google Scholar]
- 39.Alexander A, Karakas C, Chen X. et al. Cyclin E overexpression as a biomarker for combination treatment strategies in inflammatory breast cancer. Oncotarget. 2017;8:14897–911. doi: 10.18632/oncotarget.14689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melincovici CS, Mihu CM, Marginean M. et al. The prognostic significance of p53, Bax, Bcl-2 and cyclin E protein overexpression in colon cancer - an immunohistochemical study using the tissue microarray technique. Rom J Morphol Embryol. 2016;57:81–9. [PubMed] [Google Scholar]
- 41.Tzankov A, Gschwendtner A, Augustin F. et al. Diffuse large B-cell lymphoma with overexpression of cyclin e substantiates poor standard treatment response and inferior outcome. Clin Cancer Res. 2006;12:2125–32. doi: 10.1158/1078-0432.CCR-05-2135. [DOI] [PubMed] [Google Scholar]
- 42.Frei E, Visco C, Xu-Monette ZY. et al. Addition of rituximab to chemotherapy overcomes the negative prognostic impact of cyclin E expression in diffuse large B-cell lymphoma. J Clin Pathol. 2013;66:956–61. doi: 10.1136/jclinpath-2013-201619. [DOI] [PubMed] [Google Scholar]
- 43.Richter J, Wagner U, Kononen J. et al. High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am J Pathol. 2000;157:787–94. doi: 10.1016/s0002-9440(10)64592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heeran MC, Hogdall CK, Kjaer SK. et al. Limited prognostic value of tissue protein expression levels of cyclin E in Danish ovarian cancer patients: from the Danish 'MALOVA' ovarian cancer study. Apmis. 2012;120:846–54. doi: 10.1111/j.1600-0463.2012.02913.x. [DOI] [PubMed] [Google Scholar]
- 45.Grabowski P, Schrader J, Wagner J. et al. Loss of nuclear p27 expression and its prognostic role in relation to cyclin E and p53 mutation in gastroenteropancreatic neuroendocrine tumors. Clin Cancer Res. 2008;14:7378–84. doi: 10.1158/1078-0432.CCR-08-0698. [DOI] [PubMed] [Google Scholar]
- 46.Potemski P, Kusinska R, Pasz-Walczak G. et al. Prognostic relevance of cyclin E expression in operable breast cancer. Med Sci Monit. 2009;15:Mt34–40. [PubMed] [Google Scholar]
- 47.Takano Y, Kato Y, van Diest PJ. et al. Cyclin D2 overexpression and lack of p27 correlate positively and cyclin E inversely with a poor prognosis in gastric cancer cases. Am J Pathol. 2000;156:585–94. doi: 10.1016/S0002-9440(10)64763-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Z, Bandla S, Ye J. et al. Cyclin E involved in early stage carcinogenesis of esophageal adenocarcinoma by SNP DNA microarray and immunohistochemical studies. BMC Gastroenterol. 2014;14:78. doi: 10.1186/1471-230X-14-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu S, Chen J, Dong P. et al. Global gene expression profiling identifies ALDH2, CCNE1 and SMAD3 as potential prognostic markers in upper tract urothelial carcinoma. BMC Cancer. 2014;14:836. doi: 10.1186/1471-2407-14-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farley J, Smith LM, Darcy KM. et al. Cyclin E expression is a significant predictor of survival in advanced, suboptimally debulked ovarian epithelial cancers: a Gynecologic Oncology Group study. Cancer Res. 2003;63:1235–41. [PubMed] [Google Scholar]
- 52.Chappuis PO, Donato E, Goffin JR. et al. Cyclin E expression in breast cancer: predicting germline BRCA1 mutations, prognosis and response to treatment. Ann Oncol. 2005;16:735–42. doi: 10.1093/annonc/mdi149. [DOI] [PubMed] [Google Scholar]
- 53.Lotan Y, Bagrodia A, Passoni N. et al. Prospective evaluation of a molecular marker panel for prediction of recurrence and cancer-specific survival after radical cystectomy. Eur Urol. 2013;64:465–71. doi: 10.1016/j.eururo.2013.03.043. [DOI] [PubMed] [Google Scholar]
- 54.Lundgren C, Ahlin C, Holmberg L. et al. Cyclin E1 is a strong prognostic marker for death from lymph node negative breast cancer. A population-based case-control study. Acta Oncol. 2015;54:538–44. doi: 10.3109/0284186X.2014.965274. [DOI] [PubMed] [Google Scholar]
- 55.Marchini S, Mariani P, Chiorino G. et al. Analysis of gene expression in early-stage ovarian cancer. Clin Cancer Res. 2008;14:7850–60. doi: 10.1158/1078-0432.CCR-08-0523. [DOI] [PubMed] [Google Scholar]
- 56.Jansen MP, Reijm EA, Sieuwerts AM. et al. High miR-26a and low CDC2 levels associate with decreased EZH2 expression and with favorable outcome on tamoxifen in metastatic breast cancer. Breast Cancer Res Treat. 2012;133:937–47. doi: 10.1007/s10549-011-1877-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kreike B, Hart G, Bartelink H. et al. Analysis of breast cancer related gene expression using natural splines and the Cox proportional hazard model to identify prognostic associations. Breast Cancer Res Treat. 2010;122:711–20. doi: 10.1007/s10549-009-0588-6. [DOI] [PubMed] [Google Scholar]
- 58.Desmedt C, Ouriaghli FE, Durbecq V. et al. Impact of cyclins E, neutrophil elastase and proteinase 3 expression levels on clinical outcome in primary breast cancer patients. Int J Cancer. 2006;119:2539–45. doi: 10.1002/ijc.22149. [DOI] [PubMed] [Google Scholar]
- 59.Fredholm H, Magnusson K, Lindstrom LS. et al. Breast cancer in young women and prognosis: How important are proliferation markers? Eur J Cancer. 2017;84:278–89. doi: 10.1016/j.ejca.2017.07.044. [DOI] [PubMed] [Google Scholar]
- 60.Ayhan A, Kuhn E, Wu RC. et al. CCNE1 copy-number gain and overexpression identify ovarian clear cell carcinoma with a poor prognosis. Mod Pathol. 2017;30:297–303. doi: 10.1038/modpathol.2016.160. [DOI] [PubMed] [Google Scholar]
- 61.Nakayama K, Rahman MT, Rahman M. et al. CCNE1 amplification is associated with aggressive potential in endometrioid endometrial carcinomas. Int J Oncol. 2016;48:506–16. doi: 10.3892/ijo.2015.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luhtala S, Staff S, Tanner M. et al. Cyclin E amplification, over-expression, and relapse-free survival in HER-2-positive primary breast cancer. Tumour Biol. 2016;37:9813–23. doi: 10.1007/s13277-016-4870-z. [DOI] [PubMed] [Google Scholar]
- 63.Moher D, Liberati A, Tetzlaff J. et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–9. doi: 10.7326/0003-4819-151-4-200908180-00135. W64. [DOI] [PubMed] [Google Scholar]
- 64.Koff A, Cross F, Fisher A. et al. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell. 1991;66:1217–28. doi: 10.1016/0092-8674(91)90044-y. [DOI] [PubMed] [Google Scholar]
- 65.Agarwal R, Gonzalez-Angulo AM, Myhre S. et al. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin Cancer Res. 2009;15:3654–62. doi: 10.1158/1078-0432.CCR-08-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zariwala M, Liu J, Xiong Y. Cyclin E2, a novel human G1 cyclin and activating partner of CDK2 and CDK3, is induced by viral oncoproteins. Oncogene. 1998;17:2787–98. doi: 10.1038/sj.onc.1202505. [DOI] [PubMed] [Google Scholar]
- 67.Martinerie L, Manterola M, Chung SS. et al. Mammalian E-type cyclins control chromosome pairing, telomere stability and CDK2 localization in male meiosis. PLoS Genet. 2014;10:e1004165. doi: 10.1371/journal.pgen.1004165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Neut M, Shum A, Cuevas BD. et al. Stimulation of hERG1 channel activity promotes a calcium-dependent degradation of cyclin E2, but not cyclin E1, in breast cancer cells. Oncotarget. 2015;6:1631–9. doi: 10.18632/oncotarget.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Noske A, Henricksen LA, LaFleur B. et al. Characterization of the 19q12 amplification including CCNE1 and URI in different epithelial ovarian cancer subtypes. Exp Mol Pathol. 2015;98:47–54. doi: 10.1016/j.yexmp.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 70.Yang L, Fang D, Chen H. et al. Cyclin-dependent kinase 2 is an ideal target for ovary tumors with elevated cyclin E1 expression. Oncotarget. 2015;6:20801–12. doi: 10.18632/oncotarget.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.