Abstract
Notch family plays vital role in carcinogenesis and progression of various cancer, however, its clinical significance and prognostic value in colorectal cancer isn't fully investigated. In present study, we first investigated the NOTCH4 expression in The Cancer Genome Atlas (TCGA) (n=361) and GSE39582 (n=474) database and then validated with our own database (n=248). The transcriptional and protein levels of NOTCH4 were evaluated by RT-PCR and immunohistochemistry study, respectively. Univariate and multivariate survival analyses were performed to explore the relationship between various prognostic factors and survival outcomes. In the univariate analysis, NOTCH3 and NOTCH4 were significantly correlated with prognosis in TCGA and GSE39582 database, respectively (P<0.05). For NOTCH3 has been studied in CRC, we chosen NOTCH4 for further study. NOTCH4 mRNA was higher in liver metastases than their primary colorectal cancer or normal mucosa. Increased NOTCH4 levels significantly correlated with advanced N stage (P= 0.002), M stage (P= 0.002), lymphovascular invasion (P= 0.026), and CEA status (P= 0.030). Patients with high NOTCH4 expression had shorter 5-year disease-free survival (DFS) (HR 6.809; 95% CI 3.334-13.904; P< 0.001) and overall survival (OS) (HR 6.476; 95% CI 3.307-12.689; P<0.001) than those with low NOTCH4 expression. Multivariate survival analysis demonstrated that NOTCH4 was an independent prognostic biomarker for both DFS (HR 7.848; 95% CI 3.777-16.308; P<0.001) and OS (HR 5.323; 95% CI 2.668-10.623; P<0.001).Collectively, NOTCH4 may play critical role in colorectal cancer progression and could serve as a novel biomarker to predict survival after colectomy.
Keywords: NOTCH4, Colorectal cancer, Prognosis
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide resulting in an estimated 50,260 deaths in 2017 in the US alone1. Although gradual improvement in survival with innovative therapeutic strategies, a high proportion of CRC patients still die from metastases and recurrences. The CRC incidence has gradually increased as lifestyles changing in China. Improved understanding of tumor biology and genetics may enhance surgical techniques, chemotherapy methods, and follow-up strategies2. Thus, it is urgent to find new biomarkers for CRC to make accurately predictive for CRC prognosis, which could also enhance the understanding the mechanism of carcinogenesis and progression.
Notch signaling is an evolutionarily conserved pathway that plays crucial role in fetal development, organogenesis, and tissue homeostasis 3, 4. Aberrant activation of Notch signaling was observed in several types of cancer, including colon, breast, pancreatic, cervical, prostate cancer, Large-cell and Hodgkin lymphomas, renal carcinoma and lung cancer4-7. Notch signaling can be an oncogenic or a tumor suppressor, typically depending on the tissue microenvironments, and sometimes each Notch receptor has a paradoxical reaction or effect on clinicopathological features or progression of a special tumor8-11.
The Notch family contains four transmembrane receptors (Notch1-4) and five ligands (Jagged1, Jagged2, and Delta-like 1, 3, and 4) in humans 3.Although there were some previous reports on Notch family in CRC9, 12, 13, but it has not been extensively explored. In this study, we first performed a systematic analysis of the Notch families in The Cancer Genome Atlas (TCGA) and GSE39582 RNA sequence data to find new predictors for prognosis for patients with CRC. As the public available database lacks some vital information, such as the quality of surgery and the adjunctive therapeutic strategies, we then made external validation of the results with patients' samples from our own center.
Materials and Methods
Data selection in TCGA and GSE39582 cohort
The expression of Notch family in human CRC specimens were searched and analyzed in TCGA Illumina RNA-Seq datasets and GSE39582 raw microarray data.
Gene-level expression data and their clinical characters of CRC samples were downloaded from TCGA database (https://genome-cancer.ucsc.edu/) and GSE39582 database (https://www.ncbi.nlm.nih.gov/geo/). The inclusion criteria were, (1) pathological diagnosed with adenocarcinoma; (2) have no neoadjunctive therapy; (3) with intact survival information (OS for TCGA database and RFS for GSE39582 database). Patients who recurred or died with CRC at last follow-up were treated as the clinical endpoint for recurrence or tumor specific survival. Follow-up was completed on Apr 27, 2016 in TCGA database on Feb 24, 2017 in GSE39582 database.
Patients for external validation
A total of 248 patients who had radical surgery for CRC between January 2005 and December 2010 were included for this study. The group composed of 155 men and 130 women with a mean age of 65 (range, 21-92) years. The tissue were made from paired tumour and non-tumour tissue from each patient. All patient-derived specimens were collected and archived under protocols approved by the Institutional Review Boards of Taizhou Hospital. The diagnoses were pathological confirmed by at least two pathologists and restaged according to the 8th American Joint Committee on Cancer (AJCC) guidelines.
Immunohistochemistry (IHC) study
Construction of the tissue microarray and IHC staining was performed as previously described 14, 15. Briefly, sections were incubated with 1:100 dilution of anti-NOTCH4 (Q99466, Signalway Antibody, Maryland, USA.) overnight at 4C, and then incubated with goat anti-rabbit Envision System Plus-HRP (Dako Cytomation) for 30 min at room temperature. After rinsing three times in PBS for 10 min each, the sections were incubated with DAB for 1 min, counterstained with Mayer hematoxylin, dehydrated, and mounted. Phosphate-buffered saline was used as a negative control.
Data were assessed by two independent single-blinded pathologists. A semiquantitative scoring system 16 was used to evaluate both staining intensity (0, no staining; 1+, weak staining; 2+, moderate staining; 3+, strong staining) and the percentage of stained cells (0, <5%; 1, 5%-25%; 2, 26%-50%; 3, 51%-75%; and 4, >75%). The scores for staining intensity and percentage of positive cells were then multiplied to generate the immunoreactivity score for each case 17, 18. All cases were sorted into two groups according to the immunoreactivity score. High expression of NOTCH4 was defined as detectable immunoreactions in the nucleus with an immunoreactivity score of ≥4.
Real-time PCR
Fifteen paired liver metastases, primary cancer tissues, and their normal control were collected immediately stored in RNA-later at -80°C after they resected from synchronous colorectal cancer liver metastases patients. NOTCH4 mRNA levels were analyzed using a real-time PCR assay as previously described18. The total RNA from tissues was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions, and reversely transcripted to cDNA with PrimeScript™ RT Master Mix (Perfect Real Time) kit (RR036A, Takara) based on the manufacturer's instruction. RT-PCR was performed using the SYBR Green Master Mix (Roche, Mannheim, Germany) on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) in triplicate, and non-template controls were run for each assay under the same conditions. Primers were as follows: GAPDH- F, 5'- GCACCGTCAAGGCTGAGAAC-3', GAPDH- R, 5'-TGGTGAAGACGCCAGTGGA-3'; NOTCH4-F, 5'- TGTGAACGTGATGTCAACGAG -3', NOTCH4-R, 5'- ACAGTCTGGGCCTATGAAACC -3'.
The cycling conditions were as follows: initial denaturation for 10 min at 95℃ followed by 35 cycles of denaturation (15s at 95℃), annealing and elongation (30s at 60℃).The relative expression of NOTCH4 was calculated and normalized using the RQ value method relative to GAPDH.
Preoperative CEA examination
Preoperative blood samples were drawn in the morning from all patients. Plasma was separated from the blood cells by centrifuging the blood sample at 1000 g for 10 minutes. Magnetic particle enzyme immunoassay using the UniCel DxI 800 Access immunoassay system (Beckman Coulter Inc., Fullerton, CA, USA) was performed to detect CEA levels. According to the manufacturer's instructions, 5.2 ng/mL was chosen as cut-off value for serum CEA.
Statistical Analysis
The χ2 test or Fisher's exact test was used appropriately to analyze the relationship between NOTCH4 expression and clinicopathological variables. Multiple comparisons between groups were determined using Bonferroni's correction with type I error adjustment. For OS analysis, patients who died at the last follow-up were defined as clinical endpoints. For analysis of DFS, tumor progression after surgical resection was the clinical endpoint, documented as either tumor recurrence or metastasis. Follow-up data were recorded by phone or medical records. The survival rates were calculated by the Kaplan-Meier method and the differences between the survival curves were examined by the log-rank test. Univariate Cox proportional hazards regressions were applied to estimate the individual hazard ratio (HR) for the DFS and OS. The significant variables in the univariate analyses (P<0.05) were then put into the multivariate analysis. The HR with 95% confidence interval (CI) was measured to estimate the hazard risk of individual factors. A two-sided P-value of <0.05 was considered to indicate statistical significance. Analyses were performed using the SPSS statistical software program version 17.0 (SPSS Inc., Chicago, IL).
Results
Notch family expression in TCGA database and GSE39582 database
Although previous studies have shown that some Notch family genes play crucial roles in CRC, whether other Notch gene families that might play critical roles in CRC remains unknown. Therefore, we first systematically studied the Notch family in the TCGA database to search for new biomarkers for CRC. We identified a total of 361 patients with CRC in TCGA database, including 201 male and 160 female (Table 1). The median follow-up time was 734 days. In the univariate survival analysis, NOTCH3 (hazard ratio [HR], 1.312; 95% confidence interval [CI], 1.065-1.617; P=0.011), NOTCH4 (HR, 1.378; 95% CI, 1.057-1.796; P=0.018), and DLL4 (HR, 1.570; 95% CI, 1.061-2.324; P=0.024) were significantly correlated with OS (Table 2).
Table 1.
Clinical characteristics of patients with colorectal cancer in the TCGA and GSE39582 database
| Variable | TCGA | GSE39582 | |||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Sex | Male | 201 | 55.7 | 213 | 44.9 |
| Female | 160 | 44.3 | 261 | 55.1 | |
| Age | 64 | 31-90 | 67 | 22-97 | |
| T stage | T1/T2 | 68 | 18.8 | 52 | 11.0 |
| T3/T4 | 291 | 80.6 | 418 | 88.2 | |
| TX | 2 | 0.6 | 4 | 0.8 | |
| N stage | N0 | 198 | 54.8 | 283 | 59.7 |
| N1 | 98 | 27.1 | 108 | 22.8 | |
| N2 | 63 | 17.5 | 78 | 16.5 | |
| Nx | 2 | 0.6 | 5 | 1.1 | |
| M stage | M0 | 299 | 82.9 | 474 | 100 |
| M1 | 50 | 13.9 | / | / | |
| Mx | 12 | 3.3 | / | / | |
| Lymphovascular invasion | Negative | 203 | 56.2 | / | / |
| Positive | 112 | 31.0 | / | / | |
| Unknown | 46 | 12.7 | / | / | |
Table 2.
Univariate Cox proportional hazards analysis of NOTCH gene expression and overall survival (TCGA) or relapse free survival (GSE39582) for patients with colorectal cancer.
| TCGA | GSE39582 | |||
|---|---|---|---|---|
| Factor | HR (95% CI) | P | HR (95% CI) | P |
| NOTCH1 | 0.922(0.638-1.332) | 0.667 | 0.960(0.622-1.483) | 0.856 |
| NOTCH2 | 0.979(0.692-1.385) | 0.907 | 1.471(0.976-2.217) | 0.065 |
| NOTCH3 | 1.312(1.065-1.617) | 0.011 | 1.982(1.435-2.737) | <0.001 |
| NOTCH4 | 1.378(1.057-1.796) | 0.018 | 2.995(1.650-5.435) | <0.001 |
| JAG1 | 1.102(0.865-1.403) | 0.431 | 0.847(0.639-1.122) | 0.248 |
| JAG2 | 1.050(0.781-1.412) | 0.748 | 0.832(0.656-1.055) | 0.129 |
| DLL1 | 1.106(0.867-1.413) | 0.417 | 0.968(0.668-1.402) | 0.865 |
| DLL3 | 1.065(0.921-1.231) | 0.397 | 2.044(1.012-4.130) | 0.046 |
| DLL4 | 1.570(1.061-2.324) | 0.024 | 1.648(0.990-2.742) | 0.055 |
Abbreviation: CI, confidence interval; HR, hazard ratio
Bold type indicates statistical significance
In GSE39582 cohort, there was a total of 474 eligible patients were identified, including 213 male and 261 female (Table 1). All patients had no distant metastases. The primary study point was recurrence free survival (RFS). In the univariate survival analysis, we found that NOTCH3 (HR, 1.982; 95% CI, 1.435-2.737, P<0.001), NOTCH4 (HR, 2.995; 95% CI, 1.650-5.435; P<0.001), and DLL3 (HR, 2.044; 95% CI, 1.012-4.130, P=0.046) were the four predictors for RFS in patients with colon cancer (Table 2).
As NOTCH3 and NOTCH4 were upregulated in both TCGA and GSE39582 database, and NOTCH3 has been studied in CRC 13, 19, we then chosen NOTCH4 for further study.
Expression patterns of NOTCH4 in CRC
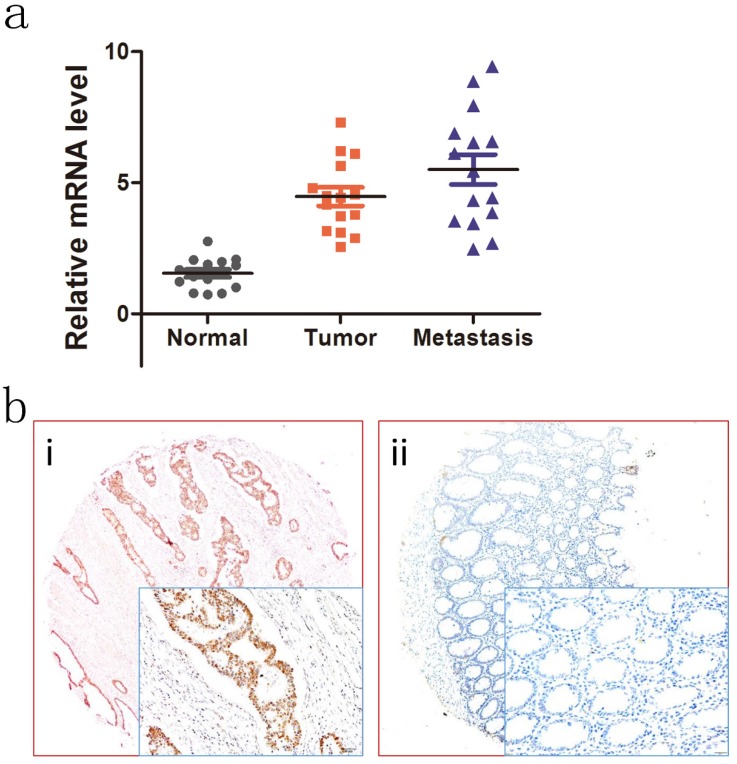
To investigate the expression patterns of NOTCH4 in CRC, we first explored NOTCH4 transcriptional expressions in 15 paired liver metastases, primary colon cancer, and adjacent normal colon tissues by RT-PCR analysis. NOTCH4 mRNA expression was found to be different among the groups. The most obviously difference was between adjacent normal controls and liver metastases (Fig.1a). The result indicated that NOTCH4 may affect the progression and metastasis of the CRC.
Figure 1.
Expression of NOTCH4 in colon cancer tumors and adjacent normal mucosa. (a) Relative NOTCH4 mRNA levels in 15 matched liver metastases, primary tumors and adjacent normal mucosa specimens. The relative RQ value is used to represent the fold change in quantitative real-time polymerase chain reaction detection. (b) Representative images of NOTCH4 expression in colorectal cancer tissues (i) and their normal control (ii). As visualized in a 200×and 400×magnifications (lower panels).
We further studied the relationship between NOTCH4 protein expression and patients' clinicopathological characteristics in the validation cohort. Representative images of NOTCH4 IHC staining was shown (Fig. 1b). It showed that NOTCH4 expression was significantly higher in cancer tissues than their normal controls (P<0.05). Importantly, the significant correlations were found between high NOTCH4 expression and advanced N stage (P= 0.002), M stage (P= 0.002), lymphovascular invasion (P= 0.026), and CEA status (P= 0.030) (Table 3). The results suggested that NOTCH4 may have a key role in CRC progression.
Table 3.
Association between NOTCH4 expression and clinicpathological factors in the validation cohort.
| Variable | n | NOTCH4 Expression | χ2 Value | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Gender | 1.132 | 0.287 | |||
| Male | 149 | 65(56.5) | 84(63.2) | ||
| Female | 99 | 50(43.5) | 49(36.8) | ||
| Age | 1.394 | 0.238 | |||
| ≦60 | 152 | 75(65.2) | 77(57.9) | ||
| >60 | 96 | 40(34.8) | 56(42.1) | ||
| T category | 4.367 | 0.113 | |||
| T1/2 | 36 | 23(20.2) | 14(10.5) | ||
| T3 | 51 | 22(19.1) | 29(21.8) | ||
| T4 | 160 | 70(60.9) | 90(67.7) | ||
| N stage | 12.795 | 0.002 | |||
| N0 | 107 | 61(53.0) | 46(34.6) | ||
| N1 | 81 | 25(21.7) | 56(42.1) | ||
| N2 | 60 | 29(25.2) | 31(23.3) | ||
| M stage | 9.689 | 0.002 | |||
| M0 | 215 | 108(93.9) | 107(80.5) | ||
| M1 | 33 | 7(6.1) | 26(19.5) | ||
| Pathological grading | 2.225 | 0.329 | |||
| High | 69 | 28(24.3) | 41(30.8) | ||
| Moderate | 125 | 66(57.4) | 59(44.4) | ||
| Poor | 54 | 21(18.3) | 33(24.8) | ||
| Lymphovascular invasion | 4.718 | 0.030 | |||
| Negative | 194 | 97(84.3) | 97(72.7) | ||
| Positive | 54 | 18(15.7) | 36(27.1) | ||
| Perineural invasion | 0.007 | 0.933 | |||
| Negative | 213 | 99(86.1) | 114(85.7) | ||
| Positive | 35 | 16(13.9) | 19(14.3) | ||
| CEA status | 7.272 | 0.026a | |||
| Normal | 157 | 83(72.2) | 74(55.6) | ||
| High | 82 | 29(25.2) | 53(39.8) | ||
| Unknown | 9 | 3(2.6) | 6(4.5) | ||
a exact χ2 test
Upregulation of NOTCH4 is associated with poor prognosis in CRC
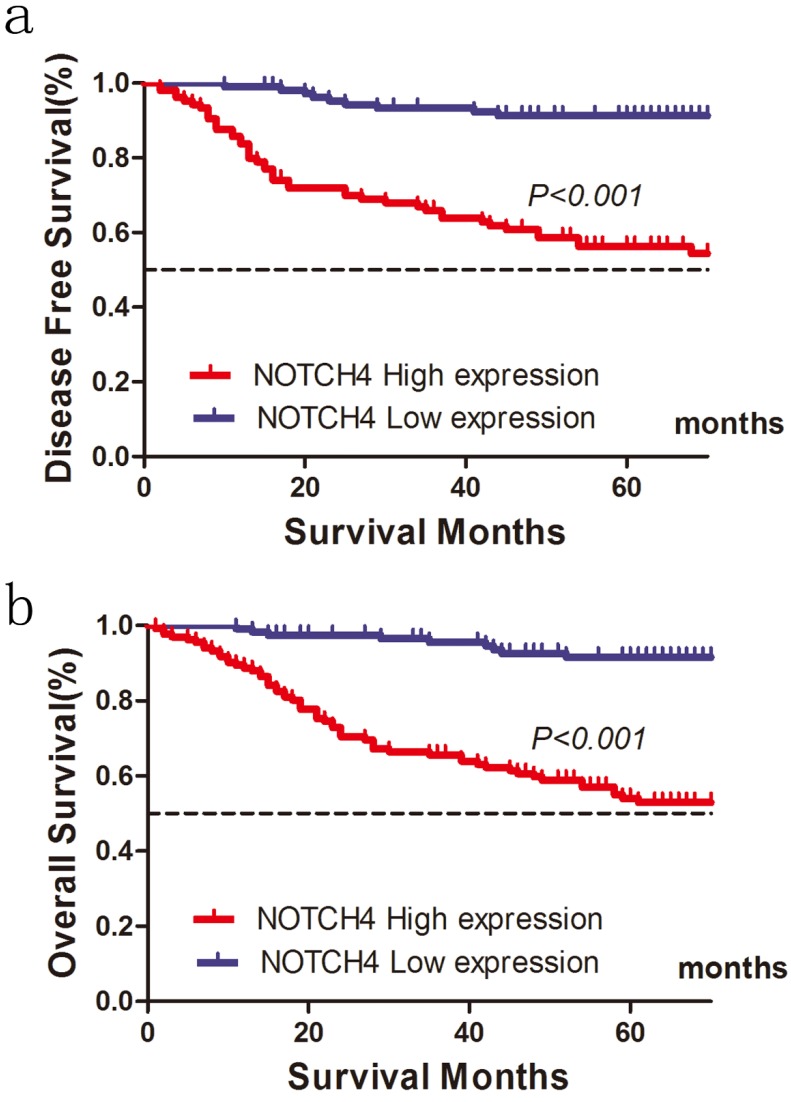
Of the 248 patients, 86 patients (34.7 %) were suffered tumor relapse during the median follow-up period of 52 months after surgery (range, 1-89 months), and 68 patients (27.4%) died of the disease. In the NOTCH4 high expression group, recurrence was observed in 70 (52.63 %) of 133 patients, and recurrence was detected in 16(13.91 %) of 115 patients in the NOTCH4 low expression group. High NOTCH4 expression was significantly related with shorter DFS for patients with CRC after colectomy (χ2=42.267, P<0.001; Fig. 2a). In univariate analysis, besides NOTCH4 status, primary tumor stage (T stage) (P<0.001), lymph node metastasis (N stage) (P<0.001), distant metastases (M stage) (P<0.001), and lymphovascular invasion (P<0.001) were significantly correlated with DFS (P<0.05, Table 3). In multivariate analysis, T stage, N stage, M stage, and NOTCH4 expression were independently associated with tumor recurrence (P<0.05, Table 4).
Figure 2.
Influence of NOTCH4 expression patterns on (a) disease free survival and (b) overall survival by Kaplan-Meier analyses in the validation cohort.
Table 4.
Univariate and multivariate Cox proportional hazards analysis of NOTCH4 expression and disease free survival for patients with colorectal cancer in the validation cohort
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Factor | HR (95% CI) | P | HR (95% CI) | P |
| Gender | 0.869(0.506-1.492) | 0.610 | ||
| Age | 1.349(0.7961-2.285) | 0.266 | ||
| T stage | 1.982(1.263-3.110) | 0.003 | 1.694(1.069-2.684) | 0.025 |
| N stage | 2.013(1.455-2.785) | <0.001 | 2.212(1.504-3.254) | <0.001 |
| Grade | 1.364(0.933-1.994) | 0.109 | ||
| Lymphovascular invasion | 2.588(1.474-4.544) | 0.001 | 1.498(0.833-2.693) | 0.177 |
| Perineural invasion | 2.149(1.155-3.998) | 0.016 | 1.206(0.623-2.335) | 0.579 |
| Tumor location | 0.850(0.503-1.436) | 0.542 | ||
| NOTCH4 | 6.809 (3.334-13.904) | <0.001 | 7.848(3.777-16.308) | <0.001 |
Abbreviation: CI, confidence interval; HR, hazard ratio
Bold type indicates statistical significance
The 5-year OS was also worse in patients with high NOTCH4 expression tumors than in those with NOTCH4 low expression tumors (54.9 vs. 91.5%, χ2=39.350, P<0.001) (Fig. 2b; Table 4). In addition, T stage (P=0.003), N stage (P<0.001), lymphovascular invasion (P=0.001), perineural invasion (P=0.016) were associated with OS. The results from the multivariate analysis further confirmed that T stage (P= 0.025), N stage (P<0.001), NOTCH4 expression (P= 0.016) were significantly associated with OS (Table 5).
Table 5.
Univariate and multivariate Cox proportional hazards analysis of NOTCH4 expression and overall survival for patients with colorectal cancer in the validation cohort
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Factor | HR (95% CI) | P | HR (95% CI) | P |
| Gender | 0.558 (0.527-1.413) | 0.558 | ||
| Age | 1.381(0.857-2.226) | 0.185 | ||
| Grade | 1.211(0.862-1.701) | 0.270 | ||
| T stage | 2.419(1.500-3.902) | <0.001 | 1.785(1.102-2.890) | 0.019 |
| N stage | 2.091 (1.547-2.825) | <0.001 | 1.686(1.172-2.425) | 0.005 |
| M stage | 8.486(5.080-14.175) | <0.001 | 3.571(1.995-6.394) | <0.001 |
| Lymphovascular invasion | 2.761(1.690-4.511) | <0.001 | 1.068(0.614-1.858) | 0.816 |
| Perineural invasion | 1.480(0.809-2.710) | 0.204 | ||
| Tumor location | 0.669 (0.415-1.078) | 0.099 | ||
| NOTCH4 | 6.476(3.307-12.689) | <0.001 | 5.323(2.668-10.623) | <0.001 |
Abbreviation: CI, confidence interval; HR, hazard ratio
Bold type indicates statistical significance
Discussion
There are numerous evidences indicated that members of the Notch families played important role in regulating a series of cellular processes, including tumorigenesis and progression, indicating that they might serve as biomarkers for diagnosis and prognosis of cancer. However, their roles in CRC have not been fully studied. In order to identify new biomarker of NOTCH family for CRC, we first investigated NOTCH families in TCGA and GSE39582 database. The results showed that the NOTCH3 and NOTCH4 were upregulated and implied poor prognosis in both TCGA and GSE39582 database. NOTCH3 has been studied in CRC in previous studies, high NOTCH3 expression is associated with tumor recurrence after surgical resection of CRC and promoted growth both in vitro and in vivo 13, 19. So, we choose NOTCH4 for further study.
For TCGA and GSE39582 database lacks some important information, such as the quality of surgery and the adjunctive therapeutic strategies. Additionally, the OS information was not available in GSE39592 and some RFS information also missing in TCGA database, we then validated the significance of NOTCH4 using our own cases. The NOTCH4 was significantly upregulated in cancer tissues than adjacent normal colonic tissues. The expression of NOTCH4 was gradually increased from normal colonic tissues to primary cancer tissues to liver metastases. Higher NOTCH4 expression implied advanced N stage, distant metastases and present of lymphovascular invasion. Survival analysis indicated that NOTCH4 status is an independently prognostic factor for both OS and RFS in external validation cohort. NOTCH4 can induces epithelial-mesenchymal transition (EMT) by upregulated HEY1 in head and neck squamous cell carcinoma20. Knockdown NOTCH4 inhibits prostate cancer growth and EMT via the NF-κB pathway21. So, it is not surprised that NOTCH4 play critical role in CRC carcinogenesis and progression.
In summary, our study for the first time indicated that NOTCH4 is ectopic expression in CRC and is a potential new biomarker for survival for CRC, which provides valuable information for guiding target therapeutic strategy. Our results warrant further studies of the mechanisms by which NOTCH4 promotes tumor progression and metastases in CRC.
Acknowledgments
This study was partially supported by Medical and Health Program of Zhejiang Province, China (Grant No. 2018KY905).
Authors' contributions
GW, ZC, XZ and SY participated in the design of the study and performed the statistical analysis and manuscript. JL, HD, QF, FY and GC carried out the data extraction. JL, SY and XZ conceived of the study, and participated in its design and coordination and helped to draft the manscripts. JL, GW and ZC performed the experiment and sample collection. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Institutional Review Boards of Taizhou Hospital approved the study protocol and all subjects provided written informed consent.
Abbreviations
- CRC
colorectal cancer
- TCGA
the Cancer Genome Atlas
- HR
hazard ratio
- 95% CI
95% confidential interval
- OS
overall survival
- DFS
disease free survival
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IHC
immunohistochemistry.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin; 2017. [DOI] [PubMed] [Google Scholar]
- 2.Li Q, Qin Y, Wei P, Lian P, Li Y, Xu Y, Gas1 Inhibits Metastatic and Metabolic Phenotypes in Colorectal Carcinoma. Mol Cancer Res; 2016. [DOI] [PubMed] [Google Scholar]
- 3.Han J, Hendzel MJ, Allalunis-Turner J. Notch signaling as a therapeutic target for breast cancer treatment? Breast Cancer Res. 2011;13:210. doi: 10.1186/bcr2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji Y, Chen S, Xiang B, Li Y, Li L, Wang Q. Jagged1/Notch3 Signaling Modulates Hemangioma-Derived Pericyte Proliferation and Maturation. Cell Physiol Biochem. 2016;40:895–907. doi: 10.1159/000453148. [DOI] [PubMed] [Google Scholar]
- 5.Lim JS, Ibaseta A, Fischer MM, Cancilla B, O'Young G, Cristea S. et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. 2017;545:360–4. doi: 10.1038/nature22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowell CS, Radtke F. Notch as a tumour suppressor. Nat Rev Cancer. 2017;17:145–59. doi: 10.1038/nrc.2016.145. [DOI] [PubMed] [Google Scholar]
- 7.Sun DW, Zhang HD, Mao L, Mao CF, Chen W, Cui M. et al. Luteolin Inhibits Breast Cancer Development and Progression In Vitro and In Vivo by Suppressing Notch Signaling and Regulating MiRNAs. Cell Physiol Biochem. 2015;37:1693–711. doi: 10.1159/000438535. [DOI] [PubMed] [Google Scholar]
- 8.Bellavia D, Checquolo S, Campese AF, Felli MP, Gulino A, Screpanti I. Notch3: from subtle structural differences to functional diversity. Oncogene. 2008;27:5092–8. doi: 10.1038/onc.2008.230. [DOI] [PubMed] [Google Scholar]
- 9.Chu D, Zhang Z, Zhou Y, Wang W, Li Y, Zhang H. et al. Notch1 and Notch2 have opposite prognostic effects on patients with colorectal cancer. Ann Oncol. 2011;22:2440–7. doi: 10.1093/annonc/mdq776. [DOI] [PubMed] [Google Scholar]
- 10.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004;14:779–86. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- 11.Bonyadi Rad E, Hammerlindl H, Wels C, Popper U, Ravindran Menon D, Breiteneder H. et al. Notch4 Signaling Induces a Mesenchymal-Epithelial-like Transition in Melanoma Cells to Suppress Malignant Behaviors. Cancer Res. 2016;76:1690–7. doi: 10.1158/0008-5472.CAN-15-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paiva TF Jr, de Jesus VH, Marques RA, da Costa AA, de Macedo MP, Peresi PM. et al. Angiogenesis-related protein expression in bevacizumab-treated metastatic colorectal cancer: NOTCH1 detrimental to overall survival. BMC Cancer. 2015;15:643. doi: 10.1186/s12885-015-1648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozawa T, Kazama S, Akiyoshi T, Murono K, Yoneyama S, Tanaka T. et al. Nuclear Notch3 expression is associated with tumor recurrence in patients with stage II and III colorectal cancer. Ann Surg Oncol. 2014;21:2650–8. doi: 10.1245/s10434-014-3659-9. [DOI] [PubMed] [Google Scholar]
- 14.Li Q, Qin Y, Wei P, Lian P, Li Y, Xu Y. et al. Gas1 Inhibits Metastatic and Metabolic Phenotypes in Colorectal Carcinoma. Mol Cancer Res. 2016;14:830–40. doi: 10.1158/1541-7786.MCR-16-0032. [DOI] [PubMed] [Google Scholar]
- 15.Li Q, Li Y, Xu J, Wang S, Xu Y, Li X. et al. Aldolase B Overexpression is Associated with Poor Prognosis and Promotes Tumor Progression by Epithelial-Mesenchymal Transition in Colorectal Adenocarcinoma. Cell Physiol Biochem. 2017;42:397–406. doi: 10.1159/000477484. [DOI] [PubMed] [Google Scholar]
- 16.Li Q, Wu J, Wei P, Xu Y, Zhuo C, Wang Y. et al. Overexpression of forkhead Box C2 promotes tumor metastasis and indicates poor prognosis in colon cancer via regulating epithelial-mesenchymal transition. Am J Cancer Res. 2015;5:2022–34. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Jiang Z, Zhang Y, Wang X, Liu L, Fan Z. RNA sequencing analysis reveals protective role of kruppel-like factor 3 in colorectal cancer. Oncotarget. 2017;8:21984–93. doi: 10.18632/oncotarget.15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S, Ma C, Shan S, Zhou L, Li W. High expression of matrix metalloproteinases 16 is associated with the aggressive malignant behavior and poor survival outcome in colorectal carcinoma. Sci Rep. 2017;7:46531. doi: 10.1038/srep46531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serafin V, Persano L, Moserle L, Esposito G, Ghisi M, Curtarello M. et al. Notch3 signalling promotes tumour growth in colorectal cancer. J Pathol. 2011;224:448–60. doi: 10.1002/path.2895. [DOI] [PubMed] [Google Scholar]
- 20.Fukusumi T, Guo TW, Sakai A, Ando M, Ren S, Haft S. et al. The NOTCH4-HEY1 Pathway Induces Epithelial-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma. Clin Cancer Res. 2018;24:619–33. doi: 10.1158/1078-0432.CCR-17-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Kuang Y, Wang Y, Xu Q, Ren Q. Notch-4 silencing inhibits prostate cancer growth and EMT via the NF-kappaB pathway. Apoptosis. 2017;22:877–84. doi: 10.1007/s10495-017-1368-0. [DOI] [PubMed] [Google Scholar]