Abstract
MicroRNAs (miRNAs) are involved in cancer genesis and progression via acting as tumor suppressors or oncogenes. Previous studies report that miR-1296 shows upregulation in both colorectal cancer (CRC) tissues and plasma samples. However, the accurate clinical significance of miR-1296 and its role in CRC have not been well investigated. The aim of the present study was to disclose the aberrant expression, clinical significance, and the relevant biological function of miR-1296 in CRC. We found a marked upregulation of miR-1296 expression in CRC tissues compared to tumor-adjacent tissues. MiR-1296 overexpression was detected in five CRC cell lines (HCT116, Caco2, HT29, SW620 and SW480). High miR-1296 level was remarkably correlated with tumor size (>5cm), lymph node metastasis and TNM stage (III+IV). Notably. High miR-1296 expression was identified as a predictive factor for poor prognosis of CRC patients by survival analysis. MiR-1296 knockdown inhibited proliferation, migration, invasion capacities of HCT116 and SW480 cells in vitro. Moreover, miR-1296 silencing restrained the growth of CRC cells in vivo. Splicing factor proline and glutamine rich (SFPQ), a novel RNA binding protein, was identified as a direct target gene of miR-1296 in CRC. Downregulation of SFPQ expression was inversely associated with miR-1296 expression in CRC tissues. The Cancer Genome Atlas (TCGA) data revealed the prognostic value of dysregulated SFPQ in CRC patients. Interestingly, our findings established that the oncogenic role of miR-1296 was at least partially mediated by SFPQ in CRC cells. Collectively, these data indicate that miR-1296 accelerates CRC progression possibly by targeting SFPQ and may serve as a potential predictive factor and therapeutic target for CRC.
Keywords: Colorectal cancer, microRNA-1296, Tumor growth, Metastasis, SFPQ
Introduction
Colorectal cancer (CRC) is one of the leading cause for cancer-related death and ranks the third commonest cancer worldwide 1. Surgery remains the most common curative therapy for CRC patients 2. In spite of great advancement in the therapy regimen for CRC, such as systemic chemotherapy, radiotherapy, immunotherapy and targeted therapy, the post-surgical outcome of CRC patients is slightly improved due to the high rate of tumor recurrence and metastasis 3. Accordingly, it is still urgently needed to uncover the molecular pathways underlying CRC progression and identify the novel biomarkers for predicting the post-surgical prognosis of CRC patients.
MicroRNAs (miRNAs) are a family of endogenous small non-coding RNAs (~22 nucleotides in length) that negatively regulate the expression of multiple genes either by inducing translational repression or by causing the degradation of target messenger RNAs (mRNAs), via incompletely base-pairing to a complementary sequence in the 3'-untranslated region (3'-UTR) 4. In some settings, miRNAs also interact with amino acid coding regions of their mRNA targets 5. Increasing evidence show that dysregulated expression of miRNAs is involved in cancer progression and metastasis, and they may be the novel biomarkers and therapeutic targets for cancer treatment 6-8. MiR-1296 is a newly identified cancer-related miRNA. For instance, miR-1296 is down-regulated in prostate cancer and inhibits cell cycle progression of cancer cells via targeting minichromosome maintenance 2 (MCM2) 9. MiR-1296 reduces the expression of PIM2 and promotes apoptosis of HeLa cells 10. MiR-1296 acts as a tumor suppressor by suppressing the expression of Cyclin D1 in triple-negative breast cancer 11. Furthermore, down-regulation of miR-1296 facilitates the invasion and migration capacities of gastric cancer cells via targeting erb-b2 receptor tyrosine kinase 2 (ERBB2) 12. Xu et al. report that miR-1296 restrains the metastatic behaviors of hepatocellular carcinoma cells by inhibiting SRPK1-mediated epithelial-to-mesenchymal transition (EMT) 13. All these studies suggest a tumor suppressive role of miR-1296. However, miR-1296 is found to be a miRNA with prognostic significance in colon cancer and it is up-regulated in the group with disease recurrence 14. Moreover, recent study reports that miR-1296 shows upregulation in both CRC tissues and plasma samples 15. Thus, it is worth to investigate the clinical significance of miR-1296 and its role in CRC.
In this study, we attempted to figure out the potential of miR-1296 to function as a prognostic biomarker and provided a better understanding of its biological function in CRC progression.
Materials and methods
CRC patients and samples
A total of 80 CRC patients, who received surgery in China-Japan Union Hospital of Jilin University, were enrolled in this study. Ethical approval was obtained from the Research Ethics Committee of Jilin University. All the patients signed informed consent forms. None of them had received immunotherapy, radiotherapy or chemotherapy before surgery. All CRC tissues and their corresponding tumor-adjacent tissues were pathologically confirmed and conserved in liquid nitrogen. The clinicopathologic characteristics of patients were shown in Table 1.
Table 1.
The correlation between clinicopathological features and miR-1296 expression in colorectal cancer
| Features | n | miR-1296 expression | P | ||
|---|---|---|---|---|---|
| High (n=40) | Low (n=40) | ||||
| Age (years) | <60 | 37 | 16 | 21 | 0.262 |
| ≥60 | 43 | 24 | 19 | ||
| Gender | Male | 48 | 22 | 26 | 0.361 |
| Female | 32 | 18 | 14 | ||
| Differentiation | Well/Moderately-differentiated | 56 | 24 | 32 | 0.051 |
| Poorly-differentiated | 24 | 16 | 8 | ||
| Size (cm) | <5 | 45 | 18 | 27 | 0.043* |
| ≥5 | 35 | 22 | 13 | ||
| Invasion depth | T1+T2 | 27 | 10 | 17 | 0.098 |
| T3+T4 | 53 | 30 | 23 | ||
| Lymph node metastasis | Without | 51 | 21 | 30 | 0.036* |
| With | 29 | 19 | 10 | ||
| TNM stage | I+II | 51 | 20 | 31 | 0.011* |
| III+IV | 29 | 20 | 9 | ||
TNM: tumor-node-metastasis. * Statistically significant.
Cell culture and transfection
Human CRC cell lines including HCT116, HT29, Caco-2, SW620, and SW480 and human intestinal epithelial cell line (HIEC) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in standard growth medium at 37 °C with 5% CO2 in a humidified incubator as previously described 16. MiR-1296 mimic (MC13743) and inhibitor (MH13743) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Small interfering RNA (siRNA) that used to down-regulate splicing factor proline and glutamine rich (SFPQ) was purchased from GenePharma (Shanghai, China). Lipofectamine 3000 (Invitrogen, USA) was employed to perform transfection according to the manufacturer's protocol. The SFPQ siRNA targeting sequence was listed as follow: 5′-GACGACAGGAAGAAUUAAGTT-3′.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA of CRC cells and tissues was extracted using TRIzol reagent according to the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). Total RNA (1 µg) was reversely transcribed into cDNA using a PrimeScript RT reagent kit (Takara, Dalian, China) and a TaqMan miRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). qRT-PCR was carried out in the Applied Biosystems 7500 Sequence Detection system using a miRNA-specific TaqMan miRNA Assay Kit (Applied Biosystems) and a SYBR Premix Ex Taq™ Kit (Takara, Shiga, Japan) with the following primers: miR-1296-5p (forward primer: 5'-TTG TTA GGG CCC TGG CTC-3'; reserve primer: 5'-GTG CAG GGT CCG AGG T-3'), SFPQ (forward primer: 5'-ATG TCT CGG GAT CGG TTC CGG A-3'; reserve primer: 5'-CCA ACA AAC AAC CGA CAT CGC TG-3'), U6 (forward primer: 5'-CGC TTC GGC AGC ACA TAT AC-3'; reverse primer: 5'-CAG GGG CCA TGC TAA TCT T-3'), GAPDH (forward primer: 5'-TGC ACC ACC AAC TGC TTA GC-3'; reverse primer: 5'-GGC ATG GAC TGT GGT CAT GAG-3'). The relative expression of miR-1296 and SFPQ mRNA were normalized to U6 small nuclear RNA and GAPDH, respectively, using 2-△△Ct method.
Immunoblotting analysis
The protocol for Western blotting was described previously 17. Briefly, CRC cells were lysed with RIPA buffer (Beyotime, Haimen, China) on ice. The protein concentration was measured by a Bradford Protein Assay Kit (Beyotime). The protein samples were separated by SDS-PAGE and electro-transferred onto a PVDF membrane. The membranes were blocked with 5% non-fat milk and then were incubated with SFPQ primary antibody (ab177149, Abcam, Cambridge, MA, USA) and GAPDH primary antibody (sc-47724, Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4 ˚C overnight. The second day, the membranes were washed with TBST and incubated with HRP-labeled secondary antibodies (#7074 and 7076, Cell Signaling Technology, Beverly, MA, USA) for 2 hours. The signals were detected with an HRP chemiluminescent kit (Thermo Fisher Scientific, Waltham, MA, USA) and semi-quantified by ImageJ software (1.46; National Institutes of Health, Bethesda, MD, USA).
Proliferation assay
The measurement of cell proliferation was carried out by using Cell Counting Kit-8 (CCK-8; Beyotime, Shanghai, China) assay. CRC cells were plated in a 96-well plate in triplicate with 5×103 cells/well. Then CCK8 solution was added to the well and incubated for 2 hours at 37°C. The absorbance at 490nm was evaluated by VICTOR3™ Multilabel Plate Reader (PerkinElmer Inc., Foster City, CA, USA). For colony formation assay, CRC cells (1×103 cells/well) were plated in a 6-well plate and cultured for 2 weeks. The cell colonies were fixed with methanol and stained with crystal violet. The number of cell colonies were counted by using ImageQuant TL software (GE, USA).
Cell migration and invasion assay
The cell motility capacities were determined using a wound healing assay and a transwell chamber assay respectively, according to standard methods described before 18. SW480 cells with miR-1296 overexpression were treated with mitomycin C (10 μg/ml, Sigma-Aldrich, St. Louis, MO, USA) for 2 h prior to the wound healing and transwell assay.
In vivo nude mice tumorigenesis
1 × 106 SW480 cells with and without miR-1296 knockdown were injected subcutaneously into BALB/c female nude mice (n=5 for each group). Tumor size was measured each 4 days after implantation. Three weeks later, the mice were sacrificed under anesthesia for harvesting xenograft tissues. The tumor tissues were fixed in 10% formalin and then embedded in paraffin. Immunohistochemical staining was employed for detection of Ki-67 (#9449, Cell Signaling Technology) as previously described 16. All animal experiments were approved by the Research Ethics Committee of Jilin University.
Luciferase reporter assay
The 3'UTR of SFPQ containing the putative binding region of miR-1296-5p was amplified from human genomic DNA. Then the sequence was cloned into pGL3 luciferase reporter vector (Promega, Madison, WI, USA). The potential binding sites for miR-1296-5p were mutated by the Quick-change site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). The wild type (wt) SFPQ 3'UTR vector or mutant (mt) SFPQ 3'UTR vector and miR-1296 mimic or inhibitor were co-transfected into HCT116 cells by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) under luminometer (Berthold Detection System, Pforzheim, Germany), and luciferase activity was normalized to Renilla activity.
Statistical analysis
All data were shown as mean ± standard deviation (SD) and analyzed by using GraphPad Prism software version 5.0 (San Diego, CA, USA). Statistical analysis was calculated by Chi-squared test, Student's t-test, ANOVA, Spearman's correlation analysis, Kaplan-Meier method and Log-rank test. P-value < 0.05 was considered as statistical significance. Each experiment was repeated three times.
Results
MiR-1296 expression is up-regulated in CRC and predicts poor prognosis
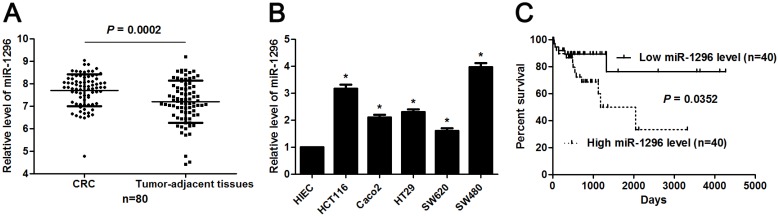
MiR-1296 expression profiles were evaluated in 80 paired CRC and tumor-adjacent tissues by qRT-PCR. Our data revealed that miR-1296 expression was up-regulated in CRC tissues compared to tumor-adjacent tissues (P=0.0002, Figure 1A). The expression difference of miR-1296 between five CRC cell lines (HCT116, Caco2, HT29, SW620 and SW480) and human intestinal epithelial cell line (HIEC) was further disclosed. All CRC cell lines showed a significant high expression of miR-1296 compared to HIEC cells (P<0.05, respectively, Figure 1B). Next, we determined the correlation between miR-1296 expression in tumor tissues and clinicopathological features of CRC patients. Different subgroups (low/high miR-1296 expression) were divided according to the median expression of miR-1296 in the cohort. As shown in Table 1, high miR-1296 expression in CRC tissues was positively associated with tumor size (>5 cm; P=0.043), lymph node metastasis (P=0.036) and TNM stage (III+IV; P=0.011), suggesting a correlation between miR-1296 overexpression and malignant behaviors of CRC. Kaplan-Meier method and the log-rank test were used to determine the role of miR-1296 expression in the post-surgical prognosis of CRC patients. We found that CRC patients with higher miR-1296 expression in tumor tissues suffered from the unfavorable post-surgical survival than those with lower miR-1296 expression (P=0.0352, Figure 1C). Thus, miR-1296 is a potential effective predictive factor for the survival of CRC patients.
Figure 1.
The expression and prognosis of miR-1296 in CRC. (A) MiR-1296 expression was detected in 80 pairs of CRC and tumor-adjacent tissues. qRT-PCR data showed that the expression of miR-1296 was prominently higher in CRC tissues compared to adjacent normal tissues. P<0.05 by Student's t-test. (B) The level of miR-1296 in five CRC cell lines (HCT116, Caco2, HT29, SW620 and SW480) was up-regulated as compared with human intestinal epithelial cell line (HIEC). n=three independent experiments, *P<0.05 by one-way ANOVA. (C) Different subgroups (low/high miR-1296 expression) were plotted according to the median expression of miR-1296 in the cohort. CRC patients with high miR-1296 level had a significant shorter overall survival compared to those with low miR-1296 level. P<0.05 by Log-rank test.
MiR-1296 knockdown restrains proliferation, migration and invasion of CRC cells
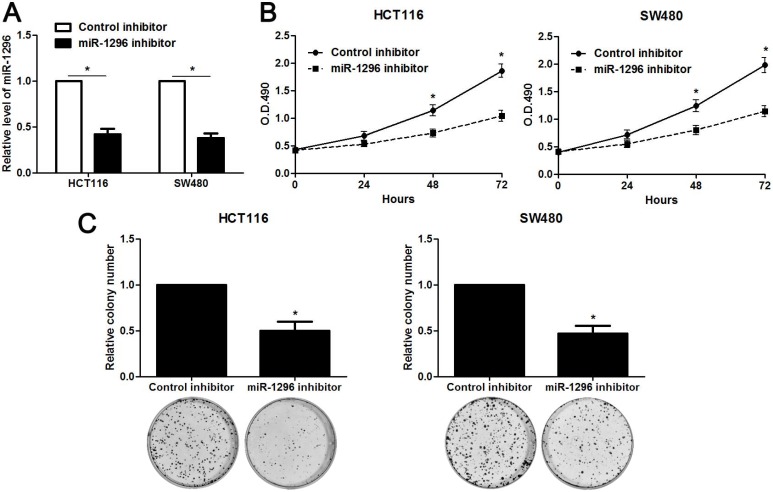
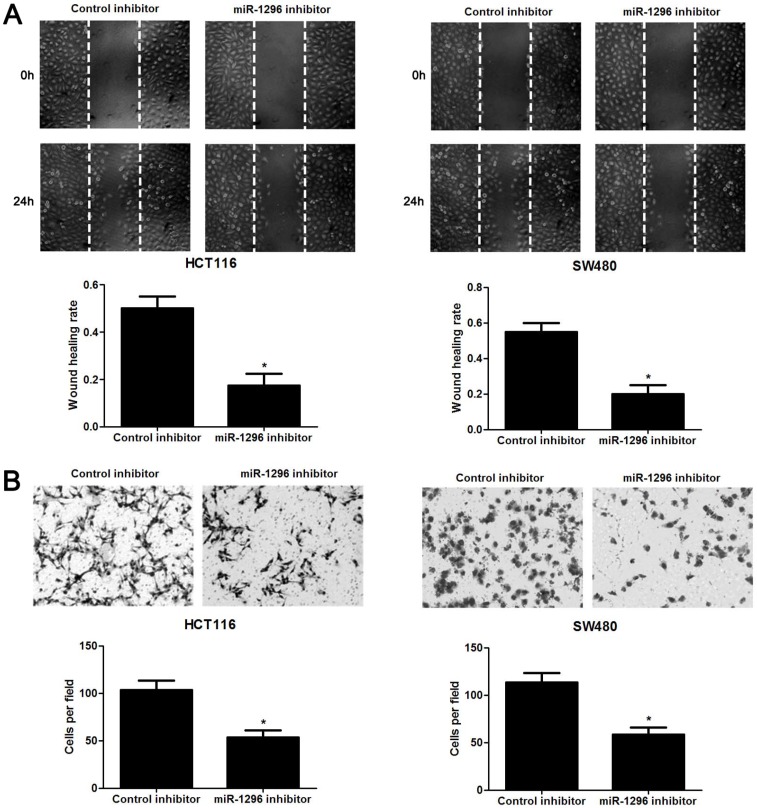
The qRT-PCR analysis confirmed that transfection of miR-1296 inhibitor resulted in miR-1296 knockdown in HCT116 and SW480 cells (P<0.05, respectively, Figure 2A). CCK8 and colony formation assays indicated that miR-1296 knockdown apparently suppressed proliferation of both HCT116 and SW480 cells (P<0.05, respectively, Figure 2B and 2C). Scratching wound healing assay also displayed that miR-1296 silencing markedly prohibited CRC cell migration (P<0.05, respectively, Figure 3A). Additionally, down-regulating miR-1296 expression led to decrease of invasion ability in both HCT116 and SW480 cells (P<0.05, respectively, Figure 3B). To distinguish cell migration and invasion from proliferation, SW480 cells with miR-1296 overexpression were treated with a proliferation blocker mitomycin C (10 μg/ml) for 2 h prior to the wound healing and transwell assay. Our data revealed that miR-1296 overexpression really promoted cell migration and invasion in SW480 cells (P<0.05, respectively, Figure S1).
Figure 2.
MiR-1296 knockdown inhibits proliferation of CRC cells. (A) HCT116 and SW480 cells that were transfected with miR-1296 inhibitor and control inhibitor, respectively, were subjected to qRT-PCR for miR-1296 expression. n=three independent experiments, *P<0.05 by Student's t-test. (B) CCK-8 assays indicated that miR-1296 knockdown suppressed the proliferation of both HCT116 and SW480 cells. n=three independent experiments, *P<0.05 by two-way ANOVA. (C) The number of cell colonies was significantly reduced after miR-1296 knockdown in both HCT116 and SW480 cells. n=three independent experiments, *P<0.05 by Student's t-test.
Figure 3.
MiR-1296 knockdown inhibits migration and invasion of CRC cells. HCT116 and SW480 cells that were transfected with miR-1296 inhibitor and control inhibitor, respectively. (A) Wound healing assays indicated that miR-1296 knockdown suppressed the migration of both HCT116 and SW480 cells. n=three independent experiments, *P<0.05 by Student's t-test. (B) The number of invaded cells was significantly reduced after miR-1296 knockdown in both HCT116 and SW480 cells. n= five fields of three independent experiments, *P<0.05 by Student's t-test.
MiR-1296 knockdown prohibits CRC growth in vivo
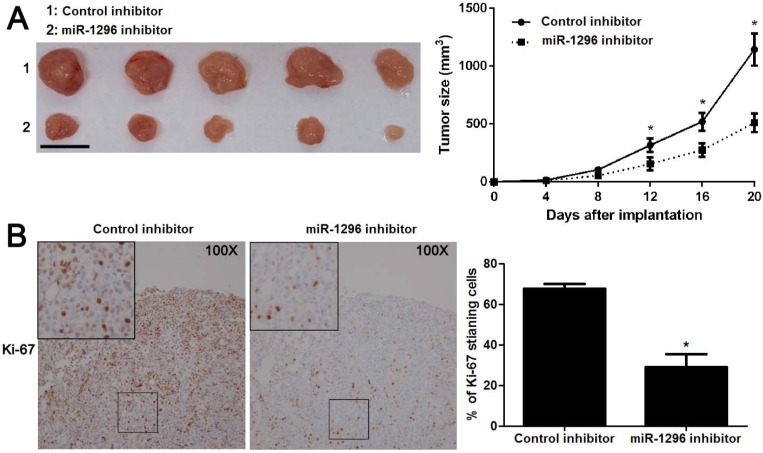
To further substantiate the action of miR-1296 on tumorigenicity in vivo, we carried out the CRC xenograft experiments in a nude mouse model using SW480 cells with or without miR-1296 knockdown. As shown in Figure 4A, the volumes of CRC xenografts in mice inoculated with miR-1296 knockdown SW480 cells were remarkably smaller as compared with controls (P<0.05). Moreover, IHC staining of Ki67 revealed that miR-1296 knockdown suppressed the growth of CRC cells in vivo (P<0.05, Figure 4B). Xenograft tumor tissues arising from miR-1296 knockdown group showed a significant lower miR-1296 expression compared to control group (P<0.05, Figure S2). Collectively, these results indicate that miR-1296 plays a critical role in CRC cell growth in vivo.
Figure 4.
MiR-1296 knockdown restrains SW480 cell growth in vivo. SW480 cells with or without miR-1296 knockdown were implanted into nude mice via subcutaneous injection. (A) Tumor grwoth curves revealed that CRC xenografts arising from miR-1296 knockdown group grew slower than those arising from control group. n=5, *P<0.05 by two-way ANOVA. Scale bar: 1cm. (B) The percentage of Ki-67 staining cancer cells was significant reduced in miR-1296 knockdown group compared to control group. n=5, *P<0.05 by Student's t-test.
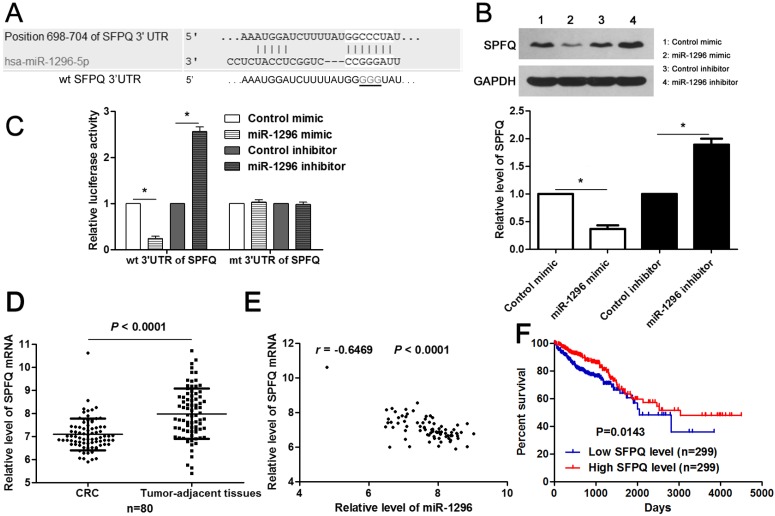
SFPQ is a direct target of miR-1296 in CRC cells
Next, the candidate target genes of miR-1296 were predicted using TargetScan (http://www.targetscan.org) and miRanda (microRNA.org) online databases. We found that the 3'-UTR of SFPQ mRNA contained the complementary sequence of miR-1296 according to public available database analysis (Figure 5A). Thus, SFPQ was selected for further experimental validation. Enforced expression of miR-1296 led to decreased expression of SPFQ in HCT116 cells (P<0.05, Figure 5B). Moreover, knockdown of miR-1296 significantly increased SFPQ expression in HCT116 cells (P<0.05, Figure 5B). Notably, miR-1296 restoration prominently reduced while miR-1296 silencing increased the fluorescence intensity of cells transfected with wt 3'UTR of SFPQ (P<0.05, respectively, Figure 5C). While, the fluorescence intensity of HCT116 cells had no significant variation after being transfected with miR-1296 mimic or inhibitor in the mutant group (Figure 5C). The expression of SFPQ mRNA was determined to be down-regulated in CRC tissues compared to tumor-adjacent tissues (P<0.0001, Figure 5D). An inverse correlation between miR-1296 and SFPQ mRNA expression was detected in CRC tissues (r=-0.6469, P<0.0001, Figure 5E). Different subgroups (low/high SFPQ expression) were divided according to the median expression of SFPQ in the cohort of CRC patients from TCGA database. CRC patients with low SFPQ level had a significant shorter overall survival compared to those with high SFPQ level (P=0.0143, Figure 5F). Thus, miR-1296 is a direct negative regulator of SFPQ in CRC.
Figure 5.
MiR-1296 directly regulates SFPQ expression in CRC cells. (A) The complementary sequences of miR-1296 were discovered in 3'UTR of SFPQ mRNA using TargetScan (http://www.targetscan.org) and miRanda (microRNA.org). The mutagenesis was performed in the complementary sites for the seed region of miR-1296 (wt, wild type; mt, mutant type). (B) HCT116 cells were transfected with miR-1296 mimic and miR-1296 inhibitor, respectively. qRT-PCR results indicated that miR-1296 overexpression reduced while miR-1296 silencing increased the expression of SFPQ. n=three independent experiments, *P<0.05 by Student's t-test. (C) MiR-1296 inversely modulated the luciferase activity of plasmids that carried wt 3'UTR of SFPQ rather than mt 3'UTR of SFPQ. n=three independent experiments, *P<0.05 by Student's t-test. (D) Down-regulation of SFPQ mRNA was detected in CRC tissues compared to tumor-adjacent tissues. P<0.05 by Student's t-test. (E) MiR-1296 was negatively correlated with SFPQ mRNA expression in CRC tissues. n=80, P<0.05 by Spearman's correlation test. (F) Different subgroups (low/high SFPQ expression) were divided according to the median expression of SFPQ in the cohort of CRC patients from TCGA database. CRC patients with low SFPQ level had a significant shorter overall survival compared to those with high SFPQ level. P<0.05 by Log-rank test.
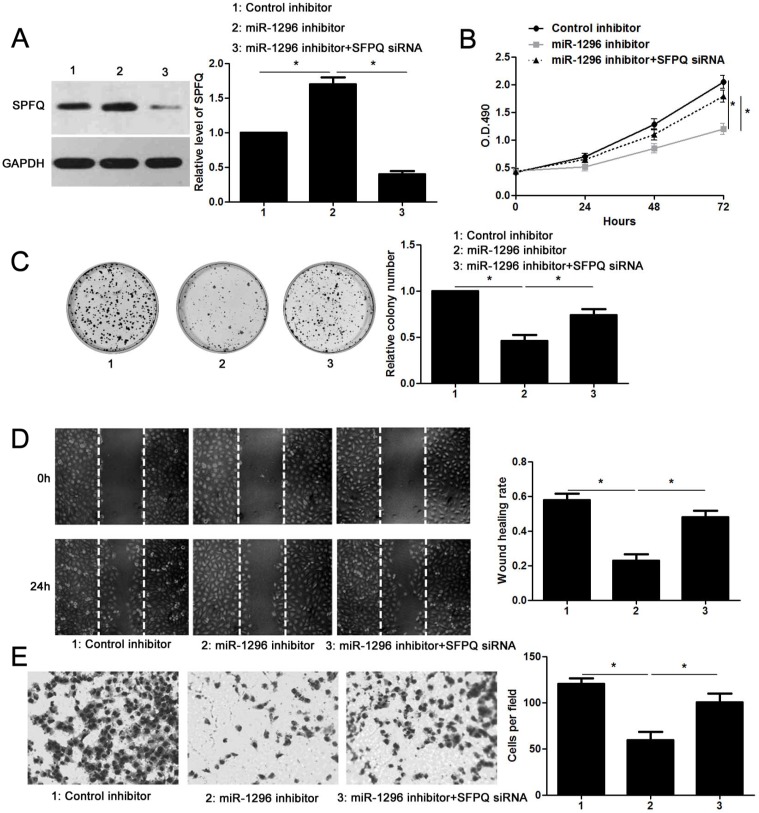
SFPQ is essential for miR-1296 associated phenotypes in CRC cells
Since SFPQ was a direct target of miR-1296, we then carried out rescue experiments to confirm that SFPQ was required for miR-1296-associated phenotypes in CRC cells. SW480 cells with miR-1296 knockdown were transfected with a specific siRNA targeting SFPQ (P<0.05, Figure 6A), and the cell proliferation and metastasis abilities were measured. We found that silencing SFPQ expression led to an obvious increased proliferation rate in SW480 cells with miR-1296 knockdown (P<0.05, respectively, Figure 6B and 6C). Similarly, SFPQ knockdown dramatically enhanced the migration and invasion capacities of SW480 cells with miR-1296 knockdown, as determined by wound healing and transwell invasion assays, respectively (P<0.05, respectively, Figure 6D and 6E). These data confirm that SFPQ is a downstream effector of miR-1296 in CRC cells.
Figure 6.
SFPQ silencing reverses the effect of miR-1296 knockdown in SW480 cells. (A) SW480 cells that were transfected with indicated plasmids were subjected to western blotting for SFPQ expression. n=three independent experiments, *P<0.05 by one-way ANOVA. (B) MiR-1296 knockdown decreased SW480 cell proliferation. While, SFPQ silencing enhanced the proliferation of SW480 cells with miR-1296 knockdown. n=three independent experiments, *P<0.05 by two-way ANOVA. (C) SFPQ silencing abolished the inhibitory effect of miR-1296 knockdown on colony formation of SW480 cells. n=three independent experiments, *P<0.05 by one-way ANOVA. (D) and (E) The number of migrated and invaded SW480 cells was reduced by miR-1296 knockdown and subsequently reversed by SFPQ silencing. n=three independent experiments, *P<0.05 by one-way ANOVA.
Discussion
MiR-1296 has been reported to have a close correlation with human cancers, such as breast cancer, hepatocellular carcinoma, gastric cancer and prostate cancer 9, 11-13. MiR-1296 usually functions as a tumor suppressor by directly targeting oncogenic genes 9, 11-13. However, recent studies report that miR-1296 is over-expressed in both CRC tissues and plasma samples, and its up-regulation is correlated with colon cancer recurrence 14, 15. Thus, the expression and role of miR-1296 in human cancer is a controversial topic. In the present study, we found that miR-1296 expression level was higher in CRC tissues than that in tumor adjacent tissues, indicating that miR-1296 may serve as an oncogenic miRNA in CRC progression. The observation was also confirmed in CRC cell lines compared to HIEC cell line. Notably, CRC patients with high miR-1296 level had larger tumor size, more lymph node metastasis, advanced TNM tumor stage and shorter survival. Based on these observations, we further carried out the gain- and loss-of-function analyses to clarify the biological functions of miR-1296 in terms of CRC cell proliferation, migration and invasion. The results indicated that miR-1296 knockdown suppresses CRC cell growth and metastasis in vitro and in vivo.
To understand the molecular mechanisms of miR-1296 as an oncogene in CRC, we searched for putative target genes of miR-1296 by using public available database and SFPQ was selected out for subsequent research in this study. SFPQ was identified as a direct target of miR-1296 by using luciferase reporter assay. Furthermore, miR-1296 inversely modulated SFPQ abundance in CRC cells and negatively correlated with SFPQ mRNA expression in CRC tissues. Survival analysis of CRC patients from TCGA database revealed that CRC patients with low SFPQ level had a significant shorter overall survival compared to those with high SFPQ level. Torres et al. investigate the prognostic value of SFPQ in CRC patients using the GSE17538 and GSE39582 data set and find a consistent result with us 19. SFPQ, containing an RNA-binding domain and a DNA-binding domain, is implicated in multiple cellular functions including DNA repair, pre-mRNA splicing and transcription regulation 20, 21. Previous studies have recognized SFPQ as a tumor-suppressor protein 22, 23. SFPQ regulates the oncogenic role of polypyrimidine tract binding protein 2 (PTBP2) by forming a complex with PTBP2 24. Long non-coding RNA (lncRNA) metastasis associated with lung adenocarcinoma transcript-1 (MALAT1) contributes to the growth and metastasis of CRC by binding to SFPQ and subsequently releasing proto-oncogene PTBP2 25. SFPQ knockdown prominently facilitates the proliferation and migration ability of CRC cells, and it is essential for the oncogenic role of MALAT1 25. Furthermore, lncRNA gastric adenocarcinoma predictive long intergenic noncoding (GAPLINC) binds to SFPQ and abrogates the pro-metastatic role of GAPLINC in CRC cells 26. Next, we proceeded to address the biological significance of the miR-1296-SFPQ axis in the regulation of CRC cell phenotypes. SW480 cells with miR-1296 knockdown were transfected with a specific siRNA targeting SFPQ and then examined for cell proliferation, migration and invasion. Silencing of SFPQ significantly enhanced both proliferation and metastasis abilities of SW480 cells with miR-1296 knockdown, suggesting that miR-1296 exerts its function, at least in part, through regulating SFPQ expression.
In summary, we demonstrate that upregulation of miR-1296 acts as a promising indicator for poor prognosis of CRC patients. Moreover, miR-1296 facilitates CRC cell proliferation and metastasis, at least partially, by regulating SFPQ expression. These findings reveal a new mechanism by which miR-1296 may act as a candidate oncogene in CRC. Our data suggest that miR-1296 may have great potential in controlling tumorigenesis and metastasis, and further confirm that miR-1296 is a promising target for CRC prevention and therapy.
Supplementary Material
Supplementary figures.
Abbreviations
- CRC
colorectal cancer
- miRNAs
microRNAs
- 3'-UTR
3'-untranslated region
- MCM2
minichromosome maintenance 2
- ERBB2
erb-b2 receptor tyrosine kinase 2
- EMT
epithelial-to-mesenchymal transition
- SFPQ
Splicing factor proline and glutamine rich
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- CCK-8
Cell Counting Kit-8
- PTBP2
polypyrimidine tract binding protein 2
- LncRNA
long non-coding RNA
- MALAT1
metastasis associated with lung adenocarcinoma transcript-1
- GAPLINC
gastric adenocarcinoma predictive long intergenic noncoding
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression Omnibus.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Li L, Ma BB. Colorectal cancer in Chinese patients: current and emerging treatment options. Onco Targets Ther. 2014;7:1817–28. doi: 10.2147/OTT.S48409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet. 2016;7:105–14. [PMC free article] [PubMed] [Google Scholar]
- 4.Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q. et al. miR-187-3p inhibits the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting S100A4. Cancer Lett. 2016;381:380–90. doi: 10.1016/j.canlet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuo H, Wang Y, Wang L, Yao B, Li Q, Wang C. et al. MiR-324-3p promotes tumor growth through targeting DACT1 and activation of Wnt/beta-catenin pathway in hepatocellular carcinoma. Oncotarget. 2017;8:65687–98. doi: 10.18632/oncotarget.20058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao J, Liu Z, Wang Y, Wang L, Yao B, Li Q. et al. MiR-542-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting UBE3C. Biomed Pharmacother. 2017;93:420–8. doi: 10.1016/j.biopha.2017.06.070. [DOI] [PubMed] [Google Scholar]
- 8.Tao Y, Song Y, Han T, Wang C, Zhao T, Gu Y. miR-205 regulation of ICT1 has an oncogenic potential via promoting the migration and invasion of gastric cancer cells. Biomed Pharmacother. 2017;96:191–7. doi: 10.1016/j.biopha.2017.09.147. [DOI] [PubMed] [Google Scholar]
- 9.Majid S, Dar AA, Saini S, Chen Y, Shahryari V, Liu J. et al. Regulation of minichromosome maintenance gene family by microRNA-1296 and genistein in prostate cancer. Cancer Res. 2010;70:2809–18. doi: 10.1158/0008-5472.CAN-09-4176. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, He W, Gao J, Luo J, Huang X, Gao C. Computational prediction and experimental validation of a novel synthesized pan-PIM inhibitor PI003 and its apoptosis-inducing mechanisms in cervical cancer. Oncotarget. 2015;6:8019–35. doi: 10.18632/oncotarget.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phan B, Majid S, Ursu S, de Semir D, Nosrati M, Bezrookove V. et al. Tumor suppressor role of microRNA-1296 in triple-negative breast cancer. Oncotarget. 2016;7:19519–30. doi: 10.18632/oncotarget.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shan X, Wen W, Zhu D, Yan T, Cheng W, Huang Z. et al. miR 1296-5p Inhibits the Migration and Invasion of Gastric Cancer Cells by Repressing ERBB2 Expression. PLoS One. 2017;12:e0170298. doi: 10.1371/journal.pone.0170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Q, Liu X, Liu Z, Zhou Z, Wang Y, Tu J. et al. MicroRNA-1296 inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting SRPK1-mediated PI3K/AKT pathway. Mol Cancer. 2017;16:103. doi: 10.1186/s12943-017-0675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bobowicz M, Skrzypski M, Czapiewski P, Marczyk M, Maciejewska A, Jankowski M. et al. Prognostic value of 5-microRNA based signature in T2-T3N0 colon cancer. Clin Exp Metastasis. 2016;33:765–73. doi: 10.1007/s10585-016-9810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy ZB, Bartak BK, Kalmar A, Galamb O, Wichmann B, Dank M, Comparison of Circulating miRNAs Expression Alterations in Matched Tissue and Plasma Samples During Colorectal Cancer Progression. Pathol Oncol Res; 2017. [DOI] [PubMed] [Google Scholar]
- 16.Tao Y, Han T, Zhang T, Sun C. Sulfatase-2 promotes the growth and metastasis of colorectal cancer by activating Akt and Erk1/2 pathways. Biomed Pharmacother. 2017;89:1370–7. doi: 10.1016/j.biopha.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Tao Y, Sun C, Zhang T, Song Y. SMURF1 promotes the proliferation, migration and invasion of gastric cancer cells. Oncol Rep. 2017;38:1806–14. doi: 10.3892/or.2017.5825. [DOI] [PubMed] [Google Scholar]
- 18.Tao Y, Yang S, Wu Y, Fang X, Wang Y, Song Y. et al. MicroRNA-216a inhibits the metastasis of gastric cancer cells by targeting JAK2/STAT3-mediated EMT process. Oncotarget. 2017;8:88870–81. doi: 10.18632/oncotarget.21488. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Torres S, Garcia-Palmero I, Marin-Vicente C, Bartolome RA, Calvino E, Fernandez-Acenero MJ. et al. Proteomic Characterization of Transcription and Splicing Factors Associated with a Metastatic Phenotype in Colorectal Cancer. J Proteome Res. 2018;17:252–64. doi: 10.1021/acs.jproteome.7b00548. [DOI] [PubMed] [Google Scholar]
- 20.Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S. et al. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol. 2005;25:6734–46. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urban RJ, Bodenburg Y, Kurosky A, Wood TG, Gasic S. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol Endocrinol. 2000;14:774–82. doi: 10.1210/mend.14.6.0485. [DOI] [PubMed] [Google Scholar]
- 22.Wang G, Cui Y, Zhang G, Garen A, Song X. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc Natl Acad Sci U S A. 2009;106:16794–8. doi: 10.1073/pnas.0909022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc Natl Acad Sci U S A. 2005;102:12189–93. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meissner M, Dechat T, Gerner C, Grimm R, Foisner R, Sauermann G. Differential nuclear localization and nuclear matrix association of the splicing factors PSF and PTB. J Cell Biochem. 2000;76:559–66. [PubMed] [Google Scholar]
- 25.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z. et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–48. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7:42183–94. doi: 10.18632/oncotarget.9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.