Abstract
It is a basic task in high-throughput gene expression profiling studies to identify differentially expressed genes (DEGs) between two phenotypes. But the weakly differential expression signals between two phenotypes are hardly detectable with limited sample sizes. To solve this problem, many researchers tried to combine multiple independent datasets using meta-analysis or batch effect adjustment algorithms. However, these algorithms may distort true biological differences between two phenotypes and introduce unacceptable high false rates, as demonstrated in this study. These problems pose critical obstacles for analyzing the transcriptional data in The Cancer Genome Atlas where there are many small-scale batches of data. Previously, we developed RankComp to detect DEGs for individual disease samples through exploiting the incongruous relative expression orderings between two phenotypes and further improved it here to identify DEGs using multiple independent datasets. We demonstrated the improved RankComp can directly analyze integrated cross-site data to detect DEGs between two phenotypes without the need of batch effect adjustments. Its usage was illustrated in detecting weak differential expression signals of breast cancer drug-response data using combined datasets from multiple experiments.
Keywords: Differentially expressed genes, Relative expression orderings, Batch effect, Drug response
Introduction
Detecting differentially expressed genes (DEGs) between two phenotypes is a basic task in high-throughput biological studies. It is often hard to detect DEGs under the circumstances where the genes are differentially expressed only slightly between two phenotypes. Especially when the sample size is not sufficiently large, detecting enough DEGs for subsequent studies is difficult. Nevertheless, it can often be found multiple datasets with regards to the same biological questions from public repositories such as Gene Expression Omnibus (GEO) 1 and ArrayExpress 2. A natural solution to raise statistical power is to combine these datasets produced by different laboratories to effectively detect weak biological signals. However, experimental batch effects hinder the direct combination of different datasets 3. This problem is also a critical obstacle for analyzing transcriptional data in The Cancer Genome Atlas (TCGA) where data scatter over many small-scale batches.
To make the full use of information available from multiple datasets, meta-analysis utilizes statistical techniques to combine results such as p-values 4, effect sizes 5, 6 and ranks 7, 8 obtained from independent studies, but high false-negative rates can arise due to small sample sizes and large scale of heterogeneity 9. More sophisticated hierarchical Bayesian methods 'borrow' information across all genes to sharpen inferences about which genes are differentially expressed 10-13. However, the key assumption of hierarchical models tends to introduce biases when estimating the variances of genes 14. Other researchers considered normalizing data across studies using various batch effect adjustment methods such as DWD 15, PAMR 16, geometric ratio-based method 17, XPN 18, ComBat 19, SVA 20 for microarray data, RUV 21, 22 and svaseq 23 for RNA-sequencing data. However, the normalization of datasets of different batches may distort the true biological signal 24, 25 and even induce spurious group differences, especially when phenotypic groups are not evenly distributed across batches 26, 27.
Recently, based on the finding that the relative expression orderings (REOs) of gene pairs are generally stable in a particular type of normal tissues but widely disrupted in diseased tissues, we have developed an algorithm named RankComp to detect DEGs for individual disease samples by analyzing the disrupted REOs 28. In this article, we extended the application of RankComp to identify genes that exhibit differential expression patterns between distinct groups. The merits that REOs are insensitive to experimental batch effects 29 and highly consistent across different platforms 30 make RankComp feasible for a cross-study comparison of gene expression.
Materials and Methods
Data and pre-processing
All data used in this study were collected from GEO and TCGA as summarized in Table 1. Each of three cancer types included three cancer-normal datasets for evaluating the performance of RankCompV2. In our examples of application below, we gathered five microarray expression datasets of estrogen receptor (ER) negative and positive breast cancer patients treated with neoadjuvant chemotherapy including paclitaxel, 5-fluorouracil, cyclophosphamide and doxorubicin. Responses to preoperative chemotherapy categorized as a pathological complete response (pCR) or residual invasive cancer (RD) were known for every patient.
Table 1.
Description of the datasets used in this study
| Number | Platforms | #Gene | Control | Case | Control vs. Case | Database |
|---|---|---|---|---|---|---|
| GSE23878 | GPL570a | 20486 | 24 | 35 | Normal vs. COAD | GEO |
| GSE37364 | GPL570 | 20486 | 38 | 27 | Normal vs. COAD | GEO |
| GSE20916 | GPL570 | 20486 | 24 | 45 | Normal vs. COAD | GEO |
| GSE29001 | GPL571b | 12432 | 12 | 12 | Normal vs. ESCC | GEO |
| GSE20347 | GPL571 | 12432 | 17 | 17 | Normal vs. ESCC | GEO |
| GSE38129 | GPL571 | 12432 | 30 | 30 | Normal vs. ESCC | GEO |
| Batch93 | RNASeqV2c | 17618 | 16 | 41 | Normal vs. BRCA | TCGA |
| Batch96 | RNASeqV2 | 17675 | 15 | 41 | Normal vs. BRCA | TCGA |
| Batch109 | RNASeqV2 | 17679 | 15 | 70 | Normal vs. BRCA | TCGA |
| The data of ER positive breast cancer | ||||||
| GSE22093 | GPL96d | 12432 | 10 | 32 | pCR vs. RD | GEO |
| GSE23988 | GPL96 | 12432 | 7 | 24 | pCR vs. RD | GEO |
| GSE42822 | GPL96 | 12432 | 7 | 19 | pCR vs. RD | GEO |
| GSE20271 | GPL96 | 12432 | 6 | 83 | pCR vs. RD | GEO |
| GSE20194T | GPL96 | 12432 | 4 | 61 | pCR vs. RD | GEO |
| The data of ER negative breast cancer | ||||||
| GSE22093 | GPL96 | 12432 | 18 | 37 | pCR vs. RD | GEO |
| GSE23988 | GPL96 | 12432 | 13 | 16 | pCR vs. RD | GEO |
| GSE42822 | GPL96 | 12432 | 13 | 15 | pCR vs. RD | GEO |
| GSE20271 | GPL96 | 12432 | 13 | 50 | pCR vs. RD | GEO |
| GSE20194T | GPL96 | 12432 | 16 | 16 | pCR vs. RD | GEO |
Note: a Affymetrix Human Genome U133 Plus 2.0 Array; b Affymetrix Human Genome U133A 2.0 Array; c UNC IlluminaHiSeq_RNASeqV2; d Affymetrix Human Genome U133A Array Abbreviation: COAD: Colon adenocarcinoma; ESCC: Esophageal squamous cell carcinoma; BRCA, Breast invasive carcinoma; pCR: pathologic complete response; RD, residual disease. GEO: Gene Expression Omnibus; TCGA, The Cancer Genome Atlas.
For each microarray expression dataset, quantile normalized expression data were separately acquired through processing the raw intensity files (.CEL) with the RMA algorithm 31. With the custom CDF file, each probe set ID was mapped to Gene ID, and then probe sets that mapped to multiple Gene IDs or did not map to any Gene ID were removed. The expression measurements of all probe sets corresponding to the same Gene ID were averaged to obtain a single measurement (on the log2 scale). For each RNA-seq dataset, the RPKM (reads per kilobase of exon model per million mapped reads) 32 values and raw counts were both download from TCGA. After removing genes with a value of 0 in more than 75% of samples, other zero values were filled with the smallest count in the expression data. The Ensembl gene IDs corresponding to the unique Entrez gene IDs of protein coding genes were used. The RPKM values were used in RankCompV2 and the raw counts were the input of RUV and svaseq method. The measured values, such as RPKM 32, FPKM 33 and TPM 34, normalizing both the gene transcription length and the sequencing depth, can represent the actual gene expression abundance and thus can be applied to the REOs-based methods.
The Rank Comp and RankCompV2 algorithm
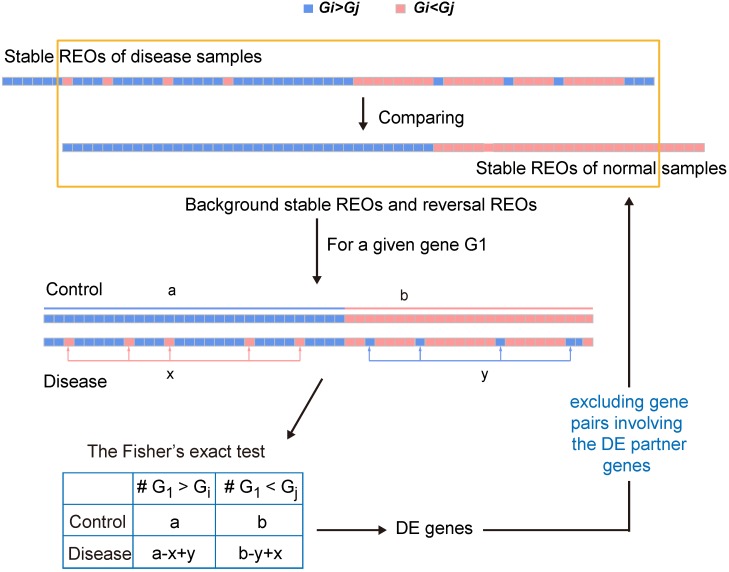
RankComp was developed for identifying DEGs in individual samples by analyzing the disrupted REOs in the disease samples through comparing with the highly stable normal REOs landscape 28, 30. In RankCompV2, we identified the stable gene pairs with significantly consistent REOs 30 in a control group and a disease group, respectively. Here, a parameter φ with default value of 5% was used to screen stable REOs. Focusing on the overlaps of the two lists of stable gene pairs, the concordant and the reversal REOs between two groups were identified. Then Fisher's extract test was used to identify candidate DEGs which may disrupt the gene correlation structure.
To minimize the potential effect of other genes' expression changes on the Fisher's exact test for a particular gene, an iterative filter process which is analogous to the strategy used in our previous study 35 was conducted. For each gene after excluding those gene pairs involving the differently expressed partner genes from the concordant and the reversal gene pairs, the Fisher's exact test was performed again until the number of DEGs did not change in two successive iterations. The flow of the algorithm can be found in Figure 1.
Figure 1.
A schematic diagram of the RankCompV2 algorithm. Focusing on the overlaps of the two lists of stable gene pairs, the stable REOs in the control samples are defined as the background stable REOs while the reversely stable REOs in the case samples are defined as the reversal REOs of the disease. For a given gene G1, a indicates the number of gene pairs with the REO pattern (G1 > Gi), and b indicates that with (G1 < Gj) in background stable REOs; x indicates the number of reversal gene pairs with shift (G1 < Gj→G1 > Gj) and y indicates the number of reversal gene pairs with shift (G1 > Gi→G1 < Gi) in reversal REOs of the disease 28, 30, 35.
The filter process is the major difference between the RankCompV2 and the RankComp algorithm. Firstly, RankCompV2 excluded gene pairs composed the gene and the potential DEGs in both consistent gene pairs and reversal gene pairs. While RankComp algorithm only excluded gene pairs composed with the gene and those potential DEGs with the opposite dysregulation direction in reversal gene pairs, which may decrease statistical power. Secondly, RankComp performs only one filtering step, which reduces only partially the influence of other genes' expression changes on the Fisher's exact test for a particular gene 35. RankCompV2 and RankComp are implemented in C language and available at GitHub (https://github.com/pathint/reoa).
Other algorithms
RankProd, a meta-analysis algorithm and available as an R package, was also tested to identify DEGs in merged datasets from different studies 7. The number of permutations was set to 1000. Genes identified as both up- and down-regulated were deleted. Combat uses an empirical Bayesian framework to remove batch effects from microarray expression data 19. The algorithm was performed by the sva package in R 20. The algorithm savseq was also performed with default parameters in the sva package for removing batch effects from sequencing data 23. The RUVSeq package includes three methods to remove batch effects from RNA-Seq Data. RUVg uses negative control genes, assuming constant expression across different samples, RUVs uses replicate control samples and RUVr uses residuals to normalized RNA-Seq read counts among samples. Here only the RUVr method can be performed.
Pathway enrichment analysis
The gene categories for function enrichment analysis were downloaded from Gene Ontology (GO) 36 in March, 2016. The GO-function algorithm 37 was used to determine the significance of biological pathways, molecular functions, and cellular components enriched with a set of interested genes by hypergeometric distribution test.
Reproducibility evaluation of DEGs
We used the POG (Percentage of Overlapping Genes) score 38, 39 and the concordance score to evaluate the reproducibility of DEGs identified from two independent datasets. If two lists of DEGs with length L1 and L2, have n overlaps, among which s have the same dysregulation directions (up- or down-regulation), then the POG score from list 1 (or 2) to list 2 (or 1), denoted as POG12 (or POG21), is calculated as s/L1 (or s/L2), and the concordance score is calculated as s/n. We evaluated whether a concordance score is higher than what expected by chance using a binomial distribution test as follows,
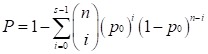 |
where p0(p0=0.05) is the probability of a gene having the concordant dysregulation direction in the two lists by chance.
Results
Performance of RankCompV2 in a single dataset
Three datasets were collected for each of three cancer types (COAD, ESCC and BRCA) to evaluate the performance of RankCompV2 in terms of precision calculated as the concordance score between the identified dysregulation directions and the observed directions based on the average expression values between the cancer and normal samples.
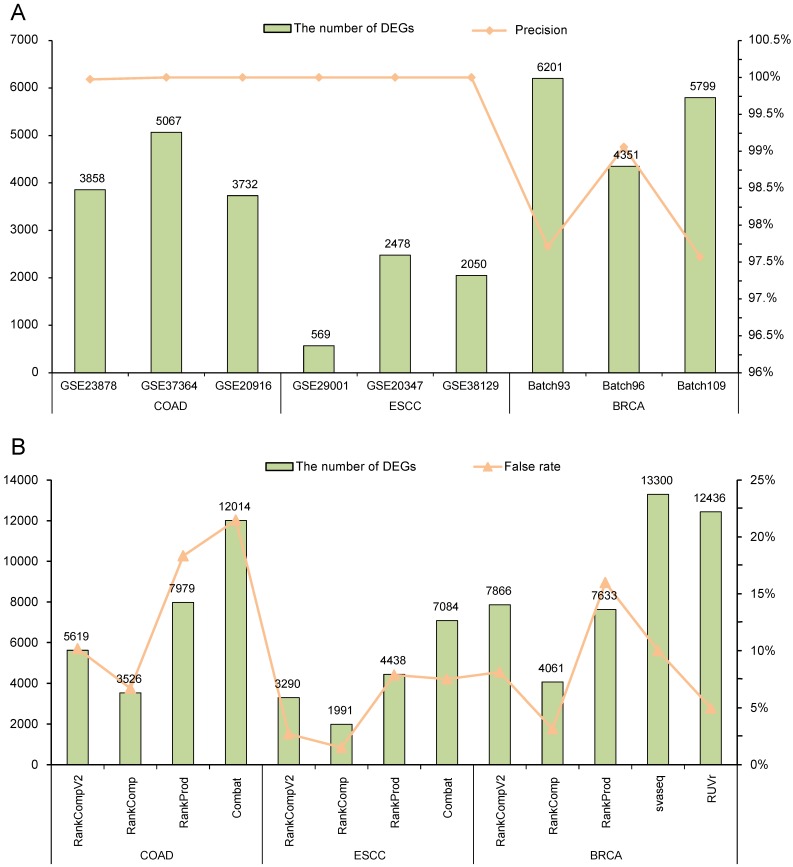
With FDR < 5%, there were 3858, 5067 and 3732 DEGs identified by RankCompV2 in the GSE23878, GSE37364 and GSE20916 datasets of COAD, and the corresponding apparent precisions of these DEGs were 99.97%, 100%, and 100%, respectively (Figure 2A). We also evaluated the reproducibility of these DEGs across datasets. For the three DEG lists, more than 63% DEGs identified in one dataset can be found in another dataset (Supplementary Table S1) and the pair-wise concordance scores are more than 96% (binomial test, p < 1.0E-16). Similar results were observed in datasets of ESCC and BRCA. For ESCC, the DEGs identified from each dataset had a precision of 100%. Not surprisingly, every DEGs lists were highly reproducible (Supplementary Table S1). In three batches of BRCA, RankCompV2 accurately detected DEGs in each dataset with a precision above 97% (Figure 2A), and the concordance scores of every two DEGs lists were 100%. Even 84% DEGs identified in Batch 96 were also identified in Batch 93 (Supplementary Table S1). These results suggest that RankCompV2 can accurately detect DEGs between distinct phenotypic groups in a single dataset.
Figure 2.
Performance evaluation of different algorithms. (A) Performance of RankCompV2 in nine datasets; (B) Comparison of the results of RankCompV2 with other algorithms in merged COAD, ESCC and BRCA datasets, respectively.
Comparing RankCompV2 with other approaches in the merged datasets
We further applied RankCompV2 to identify DEGs by merging the three datasets for each cancer. With FDR < 5%, we obtained 5619, 3290 and 7866 DEGs for COAD, ESCC and BRCA, respectively (Figure 2B). It was obvious that combining multiple datasets led to more discoveries of DEGs than a single dataset with the same FDR control. To judge whether these DEGs are truly dysregulated, we compared them with their original dysregulation directions in each dataset. If original dysregulation directions of the DEG are conflicting with each other in the three datasets of the same cancer type, it is high likely that it is a false discovery. We calculate the false rates of the identified DEGs in the merged datasets. Among DEGs identified by RankCompV2, false rates were 10.16%, 2.71% and 8.12% in the COAD, ESCC and BRCA merged datasets, respectively.
For the original RankComp method, only 3526, 1991 and 4061 DEGs were identified with false rates of 6.69%, 1.51% and 3.15% in the COAD, ESCC and BRCA merged datasets, respectively. RankCompV2 substantially improved the statistical power at the cost of a slight decrease in accuracy. Further comparison found all DEGs identified by RankComp were covered by RankCompV2 and the concordance scores of the two lists were all 100% in the three merged datasets.
RankProd performs a meta-analysis on the merged datasets. Compared with RankCompV2, RankProd identified more DEGs (7979) in the COAD merged dataset, but the false rate was up to 18.34%. For the merged datasets of ESCC and BRCA, 4438 and 7633 DEGs were identified with a false rate of 7.86% and 15.98%, respectively. Fewer DEGs accompanied with lower false rates. A special problem of RankProd is that many genes could be identified as both up- and down- regulated DEGs. In our study we had deleted these conflicting DEGs.
Batch effect correction by Combat was performed in the COAD and ESCC merged datasets measured by the microarray platforms. Although the false rate for the merged ESCC dataset was 7.51%, it reached up to 21.48% for the merged COAD dataset. For RNA-Seq data of BRCA, edgeR was performed on the adjusted data to identify DEGs after removing the batch effects by svaseq and RUVr, respectively. In the adjusted BRCA data with svaseq and RUVr, 13300 and 12436 DEGs were identified with a false rate of 10.00% and 4.98%, respectively. All results were summarized in Figure 2B.
All these results indicated RankProd and batch effect removal algorithms likely distort true biological differences between two phenotypes and introduce unacceptable high false rates in some datasets. In contrast, RankCompV2 can detect accurate DEGs in these datasets and it is a good choice for identifying DEGs across different studies.
Simulation experiments on null datasets
Basically, RankCompV2 is an empirical algorithm, where the parameter φ is used in the determination of stable REOs for each group. Simulation experiments on null data were performed to evaluate the false discovery rates of DEGs under the default settings φ = 5%.
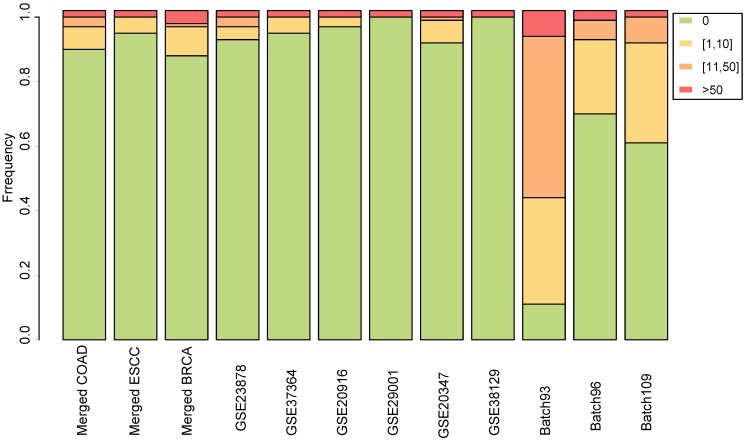
For each dataset, the normal samples were randomly divided into two groups to simulate null datasets, and RankCompV2 was applied to identify DEGs with the default settings. The experiments were repeated 100 times. No DEG was detected in 90, 95 and 88 out of the 100 experiments in the null dataset created from the merged COAD, ESCC and BRCA datasets, respectively. Similar results were observed in the null data simulated from each of the nine single (i.e., non-merged) datasets (Figure 3). The most DEGs identified was in the null data simulated from the Batch 93 dataset where 18 DEGs were found on average but this number was not disastrous compared with 6021 DEGs identified in the real data. The above extensive simulate results indicate that the default parameter setting produce negligible number of false discoveries.
Figure 3.
Stacked bar chart for the distribution of the numbers of DEGs identified from the simulated null datasets among 100 repeated experiments.
Application of RankCompV2 to breast cancer drug-response data
To further illustrate the merits of the RankCompV2, five expression datasets of breast cancer patients receiving the neoadjuvant chemotherapy of paclitaxel, 5-fluorouracil, cyclophosphamide and doxorubicin were collected from the GEO database (Table 1). The expression patterns of ER positive and ER negative subtypes were analyzed separately. Limited or even zero DEGs between pCR and RD groups were identified in each dataset by t-test or Limma 40 with FDR< 10% (data not shown).
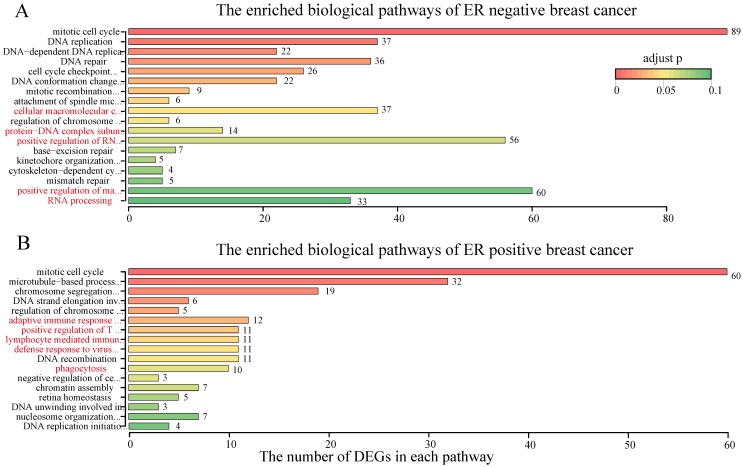
With RankCompV2 applied to the integrated ER negative dataset, we identified 36 up-regulated and 373 down-regulated genes in the RD group. These 409 DEGs were significantly enriched in 18 biological pathways of GO (Figure 4A, Supplementary Table S2). Most of these pathways were known associated with the chemotherapy resistance including cell cycle, DNA replication and DNA repair related pathways 41-46. Other pathways were also enriched, including RNA processing, positive regulation of macromolecule biosynthetic process, cellular macromolecular complex assembly, positive regulation of RNA metabolic process and protein-DNA complex subunit organization, and they serve the future research direction for the chemotherapy resistance mechanism of ER negative breast cancer. These 409 DEGs were also significantly enriched in 6 molecular functions (Supplementary Table S3) and 14 cellular components (Supplementary Table S4) of Gene Ontology.
Figure 4.
The enriched biological pathways by the DEGs in RD groups.
For the ER positive breast cancer, RankCompV2 identified 63 up-regulated and 141 down-regulated genes in the RD group. Only 40 of the 204 DEGs were included in the 409 DEGs of the ER negative dataset. These 204 DEGs were significantly enriched in 17 pathways (Figure 4B, Supplementary Table S2). Besides cell cycle and DNA replication related pathways, immune related pathways including lymphocyte mediated immunity, adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains, positive regulation of T cell activation and defense response to virus were also enriched. Furthermore, the phagocytosis pathway was also significantly enriched. These novel pathways suggest a different resistance mechanism in ER positive breast cancer. Simultaneously, 6 molecular functions (Supplementary Table S3) and 12 cellular components (Supplementary Table S4) of Gene Ontology were enriched with the 204 DEGs.
The two applications demonstrate the power of RankCompV2 in using public data for the discovery of signals that are too weak to be identified in a single experiment.
Discussion
Differing from traditional meta-analysis and batch effect adjustment algorithms, we provided a new perspective for differential expression analysis of merged datasets. The RankCompV2 method, based on the REOs of gene pairs within samples which were insensitive to batch effects and measurement platforms, can enrich DEGs, significantly and accurately, from the integrated cross-site datasets. RankCompV2 is an improved version of the original RankComp. But the validation results suggest that each has its own merits in analyzing the merged datasets. RankComp has a low statistical power but a high accuracy, while RankCompV2 has a higher statistical power but a slightly lower accuracy. For other algorithms, the normalization procedure and the batch effect adjustment algorithms may distort true biological differences between two phenotypes which induce many false discoveries 24-27.
The RankCompV2 method facilitates the analysis of mRNA expression data in TCGA, where each batch has a small sample size and even only one-phenotype data, which cannot be dealt with by other methods. It is valuable to reuse the one-phenotype data, particularly the control datasets, since there lack the control samples in many disease studies, for example normal heart and brain tissues 47, 48.Our laboratory also evaluated the effect of biological factors such as gender, ethnicity, smoking status and age and find that these disease-related confounding factors might have an influence on REOs 49. When applying the RnankCompV2 to two different groups, it is better to balance the confounding factors between the two groups.
One flaw of RankCompV2 is its low statistical power. On one hand, none of approaches proposed to adjust the P-values for discrete statistics is widely accepted for application 50-55, we used the Benjamini-Hochberg FDR control approach for selecting DEGs tested by the Fisher's exact test, which tends to have insufficient power for discrete data because the null distribution of the p-values of discrete tests is not the uniform distribution required by the BH procedure 50, 56, 57. Further studies on improving RankCompV2 is a work in progress. On the other hand, the RankCompV2 method tends to identify genes with large expression changes, which could form many reversal gene pairs. Such DEGs can disrupt the gene correlation structure between two phenotypes and might be of special biological significance because functionally related genes tend to express coordinately in a stable physiology or pathology state 24.
Supplementary Material
Supplementary tables.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [21534008, 81372213, 81572935, 81602738 and 61602119]; and the Joint Scientific and Technology Innovation Fund of Fujian Province [2016Y9044].
Abbreviations
- DEGs
differentially expressed genes
- GEO
Gene Expression Omnibus
- TCGA
The Cancer Genome Atlas
- REOs
relative expression orderings
- ER
estrogen receptor
- pCR
pathological complete response
- RD
residual invasive cancer
- RPKM
reads per kilobase of exon model per million mapped reads
- COAD
Colon adenocarcinoma
- ESCC
Esophageal squamous cell carcinoma
- BRCA
Breast invasive carcinoma
- GO
Gene Ontology.
References
- 1.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M. et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic acids research. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A. et al. ArrayExpress-a public database of microarray experiments and gene expression profiles. Nucleic acids research. 2007;35:D747–50. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE. et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nature reviews Genetics. 2010;11:733–9. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer research. 2002;62:4427–33. [PubMed] [Google Scholar]
- 5.Choi JK, Yu U, Kim S, Yoo OJ. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics. 2003;19(Suppl 1):i84–90. doi: 10.1093/bioinformatics/btg1010. [DOI] [PubMed] [Google Scholar]
- 6.Hu P, Greenwood CM, Beyene J. Integrative analysis of multiple gene expression profiles with quality-adjusted effect size models. BMC bioinformatics. 2005;6:128. doi: 10.1186/1471-2105-6-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–7. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 8.Zintzaras E, Ioannidis JP. Meta-analysis for ranked discovery datasets: theoretical framework and empirical demonstration for microarrays. Computational biology and chemistry. 2008;32:38–46. doi: 10.1016/j.compbiolchem.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Cahan P, Rovegno F, Mooney D, Newman JC, St Laurent G 3rd, McCaffrey TA. Meta-analysis of microarray results: challenges, opportunities, and recommendations for standardization. Gene. 2007;401:12–8. doi: 10.1016/j.gene.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conlon EM, Song JJ, Liu JS. Bayesian models for pooling microarray studies with multiple sources of replications. BMC bioinformatics. 2006;7:247. doi: 10.1186/1471-2105-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scharpf RB, Tjelmeland H, Parmigiani G, Nobel AB. A Bayesian model for cross-study differential gene expression. Journal of the American Statistical Association. 2009;104:1295–310. doi: 10.1198/jasa.2009.ap07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan L, Yuan M. An empirical Bayes' approach to joint analysis of multiple microarray gene expression studies. Biometrics. 2011;67:1617–26. doi: 10.1111/j.1541-0420.2011.01602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conlon EM, Postier BL, Methe BA, Nevin KP, Lovley DR. A Bayesian model for pooling gene expression studies that incorporates co-regulation information. PloS one. 2012;7:e52137. doi: 10.1371/journal.pone.0052137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Sun Z, He Q, Zhu Y, Qin ZS. Bayesian inference with historical data-based informative priors improves detection of differentially expressed genes. Bioinformatics. 2016;32:682–9. doi: 10.1093/bioinformatics/btv631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benito M, Parker J, Du Q, Wu J, Xiang D, Perou CM. et al. Adjustment of systematic microarray data biases. Bioinformatics. 2004;20:105–14. doi: 10.1093/bioinformatics/btg385. [DOI] [PubMed] [Google Scholar]
- 16.Sims AH, Smethurst GJ, Hey Y, Okoniewski MJ, Pepper SD, Howell A. et al. The removal of multiplicative, systematic bias allows integration of breast cancer gene expression datasets - improving meta-analysis and prediction of prognosis. BMC medical genomics. 2008;1:42. doi: 10.1186/1755-8794-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo J, Schumacher M, Scherer A, Sanoudou D, Megherbi D, Davison T. et al. A comparison of batch effect removal methods for enhancement of prediction performance using MAQC-II microarray gene expression data. The pharmacogenomics journal. 2010;10:278–91. doi: 10.1038/tpj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shabalin AA, Tjelmeland H, Fan C, Perou CM, Nobel AB. Merging two gene-expression studies via cross-platform normalization. Bioinformatics. 2008;24:1154–60. doi: 10.1093/bioinformatics/btn083. [DOI] [PubMed] [Google Scholar]
- 19.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 20.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nature biotechnology. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peixoto L, Risso D, Poplawski SG, Wimmer ME, Speed TP, Wood MA. et al. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic acids research. 2015;43:7664–74. doi: 10.1093/nar/gkv736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic acids research; 2014. p. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Cheng L, Zhang Y, Wu R, Wang M, Gu Y. et al. Extensive up-regulation of gene expression in cancer: the normalised use of microarray data. Molecular bioSystems. 2012;8:818–27. doi: 10.1039/c2mb05466c. [DOI] [PubMed] [Google Scholar]
- 25.Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB. et al. Revisiting global gene expression analysis. Cell. 2012;151:476–82. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazar C, Meganck S, Taminau J, Steenhoff D, Coletta A, Molter C. et al. Batch effect removal methods for microarray gene expression data integration: a survey. Briefings in bioinformatics. 2013;14:469–90. doi: 10.1093/bib/bbs037. [DOI] [PubMed] [Google Scholar]
- 27.Nygaard V, Rodland EA, Hovig E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics. 2016;17:29–39. doi: 10.1093/biostatistics/kxv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Sun Q, Zhao W, Qi L, Gu Y, Li P. et al. Individual-level analysis of differential expression of genes and pathways for personalized medicine. Bioinformatics. 2015;31:62–8. doi: 10.1093/bioinformatics/btu522. [DOI] [PubMed] [Google Scholar]
- 29.Geman D, d'Avignon C, Naiman DQ, Winslow RL. Classifying gene expression profiles from pairwise mRNA comparisons. Statistical applications in genetics and molecular biology. 2004;3:Article19. doi: 10.2202/1544-6115.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan Q, Chen R, Yan H, Cai H, Guo Y, Li M. et al. Differential expression analysis for individual cancer samples based on robust within-sample relative gene expression orderings across multiple profiling platforms. Oncotarget. 2016;7:68909–20. doi: 10.18632/oncotarget.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 32.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 33.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory in biosciences = Theorie in den Biowissenschaften. 2012;131:281–5. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Cai H, Wang X, Ao L, Guo Y, He J, A rank-based algorithm of differential expression analysis for small cell line data with statistical control. Briefings in bioinformatics; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Zhou X, Zhu J, Gu Y, Zhao W, Zou J. et al. GO-function: deriving biologically relevant functions from statistically significant functions. Briefings in bioinformatics. 2012;13:216–27. doi: 10.1093/bib/bbr041. [DOI] [PubMed] [Google Scholar]
- 38.Zhang M, Yao C, Guo Z, Zou J, Zhang L, Xiao H. et al. Apparently low reproducibility of true differential expression discoveries in microarray studies. Bioinformatics. 2008;24:2057–63. doi: 10.1093/bioinformatics/btn365. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M, Zhang L, Zou J, Yao C, Xiao H, Liu Q. et al. Evaluating reproducibility of differential expression discoveries in microarray studies by considering correlated molecular changes. Bioinformatics. 2009;25:1662–8. doi: 10.1093/bioinformatics/btp295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 41.Tordai A, Wang J, Andre F, Liedtke C, Yan K, Sotiriou C. et al. Evaluation of biological pathways involved in chemotherapy response in breast cancer. Breast cancer research: BCR. 2008;10:R37. doi: 10.1186/bcr2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen K, Rice SD, Gingrich DA, Wang D, Mi Z, Tian C. et al. Distinct genes related to drug response identified in ER positive and ER negative breast cancer cell lines. PloS one. 2012;7:e40900. doi: 10.1371/journal.pone.0040900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray S, Briasoulis E, Linardou H, Bafaloukos D, Papadimitriou C. Taxane resistance in breast cancer: mechanisms, predictive biomarkers and circumvention strategies. Cancer treatment reviews. 2012;38:890–903. doi: 10.1016/j.ctrv.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Edwardson DW, Narendrula R, Chewchuk S, Mispel-Beyer K, Mapletoft JP, Parissenti AM. Role of Drug Metabolism in the Cytotoxicity and Clinical Efficacy of Anthracyclines. Current drug metabolism. 2015;16:412–26. doi: 10.2174/1389200216888150915112039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nature reviews Cancer. 2003;3:330–8. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 46.Chien AJ, Moasser MM. Cellular mechanisms of resistance to anthracyclines and taxanes in cancer: intrinsic and acquired. Seminars in oncology. 2008;35:S1–S14. doi: 10.1053/j.seminoncol.2008.02.010. quiz S39. [DOI] [PubMed] [Google Scholar]
- 47.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5923–8. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reifenberger G, Weber RG, Riehmer V, Kaulich K, Willscher E, Wirth H. et al. Molecular characterization of long-term survivors of glioblastoma using genome- and transcriptome-wide profiling. International journal of cancer. 2014;135:1822–31. doi: 10.1002/ijc.28836. [DOI] [PubMed] [Google Scholar]
- 49.Hong G, Li H, Zhang J, Guan Q, Chen R, Guo Z. Identifying disease-associated pathways in one-phenotype data based on reversal gene expression orderings. Scientific reports. 2017;7:1348. doi: 10.1038/s41598-017-01536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilbert PB. A modified false discovery rate multiple-comparisons procedure for discrete data, applied to human immunodeficiency virus genetics. Journal of the Royal Statistical Society: Series C. 2005;54(1):143–58. [Google Scholar]
- 51.Tarone RE. A modified Bonferroni method for discrete data. Biometrics. 1990;46:515–22. [PubMed] [Google Scholar]
- 52.Fellows I. The minimaxity of the mid p-value under linear and squared loss functions. Communications in Statistics-Theory and Methods. 2010;40(2):244–54. [Google Scholar]
- 53.Austin SR, Dialsingh I, Altman N. Multiple Hypothesis Testing: A review. Indian Soc Of Agricultural Stat. 2014;68:303–14. [Google Scholar]
- 54.Muralidharan O. An empirical Bayes mixture method for effect size and false discovery rate estimation. The Annals of Applied Statistics; 2010. pp. 422–38. [Google Scholar]
- 55.Martin R, Tokdar ST. A nonparametric empirical Bayes framework for large-scale multiple testing. Biostatistics. 2012;13:427–39. doi: 10.1093/biostatistics/kxr039. [DOI] [PubMed] [Google Scholar]
- 56.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of statistics; 2001. pp. 1165–88. [Google Scholar]
- 57.Heller R, Gur H. False discovery rate controlling procedures for discrete tests. arXiv preprint arXiv; 2011. p. 1112. 4627. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.