Abstract
The Rice Yield-related Database (RicyerDB) was created to complement with related research of influence rice (Oryza sativa L.) yield in multiple traits by manually curating the related databases and literature, and genomics and proteomics information that could be useful for comprehensive understanding of the rice biology. RicyerDB provides a more valuable resource in which to efficiently investigate, browse and analyze yield-related genes. The whole data set can be easily queried and downloaded through the webpage. In addition, RicyerDB also constructed a protein-protein interaction network with biological analysis. The combined rice database opens a new path to facilitate researchers achieving information on rice gene in terms of their effects on traits important for rice breeding. The web server is freely available at: http://server.malab.cn/Ricyer/index.html.
Keywords: rice, trait, yield, gene, protein
Introduction
Rice (Oryza sativa L.) is one of the most important food crops worldwide, and more than half of the global population uses it as the main food source 1. In the developing world, rice provides 27% of dietary energy and 20% of dietary protein for people's daily life 2. Moreover, rice has relatively small genome size 3, so it is generally used as a model species in plant biology, especially for studies on monocotyledonous plants. In addition, due to its global importance in food production 4, 5, a number of researches have been published, analyzing the yield associated traits such as grain size 6, 7, grain weight 8, 9, panicle number 10, 11 and so on 12 Besides, until now a huge collection of rice seed carrying useful genes for those traits have been explored by the rice breeders 13.
As increasing rice production is crucial for the farmers rely on it for their livelihood, it has been a longstanding issue for the whole world rice researchers for improving the rice yield through rice breeding 14-16. With the rapid advances in high-throughput technologies, it's possible to use bioinformatics measures emerging multi-omics data to explore the major effect factor of the yield for rice 17, 18. In the molecular level emerge genome and proteome data, increasingly being applied outside of pure research towards support the accelerated breeding of rice. However, due to the size and structure of the biological datasets, working with these data is challenging for many field and bench scientists. The main target of this research is to establish an easy and efficient search and retrieval system that would allow rice researchers and breeders to search the trait-related genes quickly.
Several databases have been published that contain the proteomic and genomic information about rice. The RAP-DB (Rice Annotation Project Database) database was conceptualized in 2004 upon the completion of genome sequencing by the International Rice Genome Sequencing Project with the aim of providing the scientific community with an accurate and timely annotation of the rice genome sequence. One of the major objectives of this project is to facilitate a comprehensive analysis of the genome structure and function of rice on the basis of the annotation. The CRDC (China Rice Data Center) database was constructed by the China Rice Research Institute of Science and Technology Information Center in 2005. This rice gene database mainly collected rice genes (including QTLs) found at domestic and abroad, including gene name, function, location and reference literature. In addition, there're some online rice genomics databases such as the Whole Rice Genome Automated Annotation Database of TIGR and Rice Information System of Beijing Genomics Institute being focus on integrating facility for data-mining and comparative genomics. The above databases primarily focus on single or multiple omics covering relatively complete information about rice. However, to the best of our knowledge, there is currently no consolidated web tool available for collecting all available rice trait-related genes over public databases. Towards this goal, here, we describe the construction and utility of RicyerDB, which will be of use to the rice researchers in general and rice breeders in particular towards successful planning of their breeding objectives.
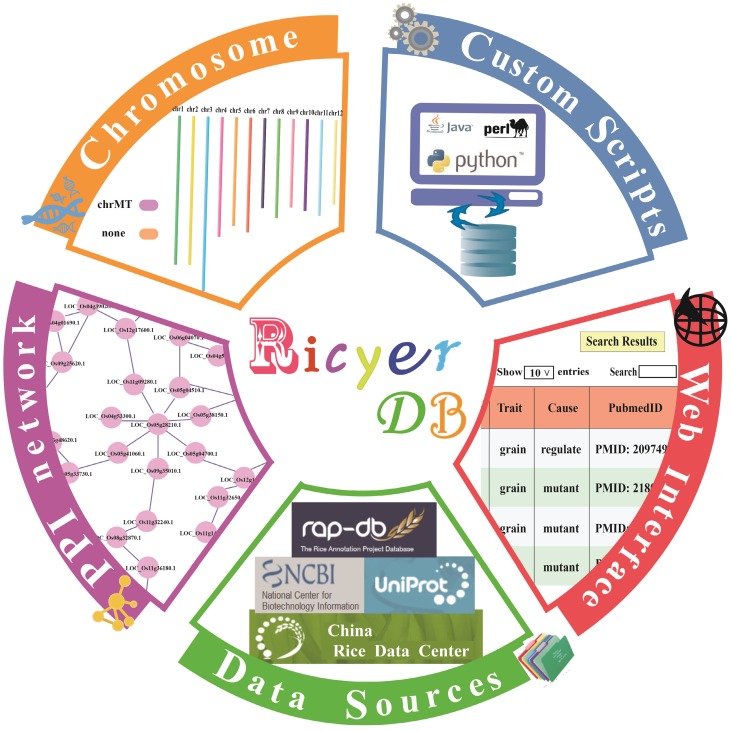
To meet this challenge the RicyerDB and website was developed. RicyerDB integrates diverse data sources to construct a public platform for browsing and interactive visualizations of yield-related genes. Schematic illustration of the overall information of the database is shown in Figure 1. The search tool enables the user to query a particular gene, and even provides insight into the functions/location of overall genes. The whole data set can be easily queried and downloaded through the webpage. Furthermore, the database can perform fast visualization of genome annotation and protein-protein interaction network, as well as providing statistical analysis of PPIN (protein-protein interaction network). In addition, RicyerDB also allows researchers to submit new gene information.
Figure 1.
Schematic illustration of the overall information of RicyerDB.
Materials and Methods
The RicyerDB database integrates diverse publicly available resources to construct a public platform for browsing and interactive visualizations of yield-related genes. We have collected experimental thermodynamic data from PubMed literature and integrated with two public databases RAP-DB and CRDC. The first release of RicyerDB contained more than 400 manually curated gene information entries with literature confirmed, among which 76 come from two databases, the rest come from PubMed. As terminology differs among databases and literature, making cross-comparisons is difficult, therefore data curation from literature requires human extraction and selection. The standard named genes can be retrieval by reference to the NCBI (National Center for Biotechnology Information) Gene.
RicyerDB supplemented yield-related genes with protein sequences and chromosome loci information, which obtained from Uniprot and NCBI Gene, respectively. Uniprot (Universal Protein Resource) is one of the most influentially housing protein information databases. It provides high quality protein sequences and the corresponding ID are freely accessible to the scientific community. Gene is a comprehensive public database, which is maintained and distributed by NCBI (National Center for Biotechnology Information), and contains the gene information about chromosome position as well as gene alias.
To explore the biological function of yield-related genes, information of gene annotation derived from the Gene Ontology Consortium. The Gene Ontology database is a major bioinformatics tool of our evolving knowledge of how genes encode biological functions at the molecular, cellular and tissue levels 19-21. RicyerDB database provides a functional annotation analysis of yield-related genes, and assigns an importance score for each functional annotation. Conversely, for each gene along with annotation information also possesses an importance score.
Meanwhile, a global view of the genes association requires knowledge of interactions between the expressed proteins 22. For each protein-protein interaction stored in STRING (Search Tool for Recurring Instances of Neighboring Genes), a score is provided. The score (i.e., the 'edge weight' in the network) represents confidence score, and is scaled between zero and one. It indicates the estimated likelihood that a given interaction is biologically meaningful, specific and reproducible, given the supporting evidence. These above online information resources are shown in Figure 2.
Figure 2.
The on-line public source databases of RicyerDB.
Results
Implementation
The RicyerDB server consists of two major components: the client web interface and the server backend. The former was implemented using jQuery, Bootstrap, CSS and Html. For the latter, a Java Servlet, for the service connector, responds to the server request. The RicyerDB has been tested in the Google Chrome, Firefox and Internet Explorer web browsers.
Interface
The RicyerDB interface is divided into several sections. Meanwhile, at the bottom of this homepage, existing three friendly links point to the corresponding databases.
Search
A capability to create a valid search query is the key to successful usage of any database. RicyerDB provides an interface for convenient retrieval of all rice trait-related genes and corresponding information in the 'Search' page. With the input of key word in the quick searching box, the search engine will return the brief details of search results as a table. Moreover, users can also search genes through the advanced search. There are two options “smart search” and “regex” in the advanced search part, which can be checked according to the users' need. The queried result table contains gene names, protein sequences, and the supporting literature evidence, and so on. When the user clicks the small triangle on the head of each table column, the results in the table will be resorted in ascending/descending order.
Browse
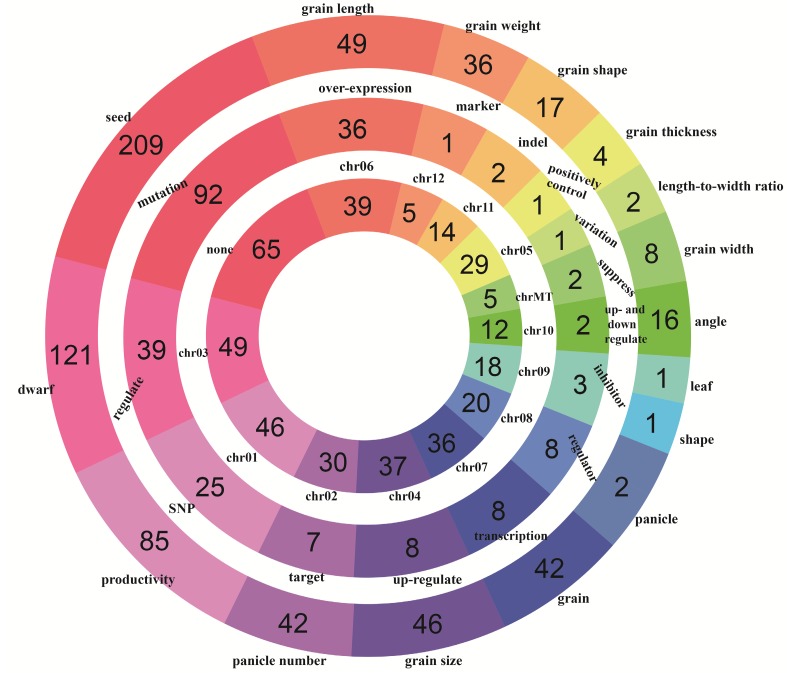
A global overview of all rice yield-related genes from different perspectives can be acquired by browsing the database. In 'Browse' section, users can access RicyerDB in three different paths: 'browse by trait', 'browse by cause' and 'browse by location'. In each path, all genes are classified into several entries. The rings distribution of the three browsing paths is shown in Figure 3.
Figure 3.
The rings distribution of rice yield-related genes. The three rings from the inside to the outside correspond to chromosome, cause and trait, respectively.
JBrowse
JBrowse implements a genome annotation tool that can be used to display an arbitrary set of features on expressed protein, and shows the position of the protein in corresponding chromosome. JBrowse provides multiple configurable levels of zoom, and two scroll speeds. Once an interesting region of the genome is in view, the user can make finer adjustments by scrolling and zooming with the navigation bar, which appears in the upper side of the area.
Interaction
To further explore the relationship between different proteins, 'Interaction' was provided to visualize as a network 23, which nodes present the proteins and edges pre-sent the interactions between proteins. The combined score of each interaction is mapped to the edge thickness. The further analysis results of the network comprised topological and statistical features were also shown in the page.
Submit
It is inevitable that the collections of RicyerDB may not cover all yield-associated genes. So we provide the submission interface to make sure that researchers can submit new genes that are not documented. In the 'Submit' page, RicyerDB invites users to upload novel gene symbol whether through experiment validation associated with rice yield or not. The request to leave the email is convenient for us to further contact you. In most cases, the authors are contacted for missing or ambiguous information and an extensive literature search is performed to complement data.
If a user needs a more complex analysis, the website allows downloading the entire database data. In 'Home' page, the whole data are saved in ZIP formats, users can get them by clicking the 'Download' button.
Except these, RicyerDB also provides a section to facilitate new users quickly access to use, its instruction is in the 'Help' page, including figures and narrative memoranda of it.
Discussion and Future Prospects
Bioinformatics is a rapidly growing field of research that is being driven by the requirement to manage and interrogate the vast quantities of data being generated by 'omics' technologies. Decades of research on rice has generated several known multi-omics resources, such as genome, proteome and transcriptome 23-35, with a sole aim to understand every aspect of rice biology.
Rice is the most important crop consumed all over the world. Several rice genes databases including the annotation as well as mutant information of the rice have been previously constructed, such as JCVI 36, RAD 37, RGKbase 38 and RMD 39. Although, these databases have exhaustive data on rice, they do not precisely catalogue and integrate rice production increase this specific demand. To complement with this absence, we developed the RicyerDB by integrating genome and proteome data. To our knowledge, this is the first database comprehensively focusing on the rice production. We hope this resource will provide effective information and be convenient for the researchers as well as farmers exploit potentials of rice as a major crop to feed the world. A limitation of this database is that it integrates the genome and proteome, which do not cover all omics information of the genes.
Future developments of RicyerDB include regular updates, improving data quantity and quality, and incorporating new types of data, such as epigenome, phenome and other omics data 40, 41. In addition, RicyerDB will collect more rice online database resources and yield-related literature. Our database will be updated periodically in future according to this additional information. In the subsequent modules, an effective prediction algorithm was added to predict the new genes for rice yield based on our database. We also call for worldwide collaborations and look forward to comments and suggestions from researchers and breeders, aiming to build RicyerDB into a more comprehensive knowledgebase of rice production.
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 61771331).
Abbreviations
- RAP-DB
Rice Annotation Project Database
- CRDC
China Rice Data Center
- PPIN
protein-protein interaction network
- Uniprot
Universal Protein Resource
- GO
Gene Ontology
- STRING
Search Tool for Recurring Instances of Neighboring Genes.
References
- 1.Mahender A, Anandan A, Pradhan SK, Pandit E. Rice grain nutritional traits and their enhancement using relevant genes and QTLs through advanced approaches. SpringerPlus. 2016;5:2086. doi: 10.1186/s40064-016-3744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang R, Jiang L, Zheng J, Wang T, Wang H, Huang Y. et al. Genetic bases of rice grain shape: so many genes, so little known. Trends in plant science. 2013;18:218–26. doi: 10.1016/j.tplants.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Seo YS, Chern M, Bartley LE, Han M, Jung KH, Lee I. et al. Towards establishment of a rice stress response interactome. PLoS genetics. 2011;7:e1002020. doi: 10.1371/journal.pgen.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khush GS. What it will take to feed 5.0 billion rice consumers in 2030. Plant molecular biology. 2005;59:1–6. doi: 10.1007/s11103-005-2159-5. [DOI] [PubMed] [Google Scholar]
- 5.Huang X, Yang S, Gong J, Zhao Q, Feng Q, Zhan Q. et al. Genomic architecture of heterosis for yield traits in rice. Nature. 2016;537:629–33. doi: 10.1038/nature19760. [DOI] [PubMed] [Google Scholar]
- 6.Xu C, Liu Y, Li Y, Xu X, Xu C, Li X. et al. Differential expression of GS5 regulates grain size in rice. Journal of experimental botany. 2015;66:2611–23. doi: 10.1093/jxb/erv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segami S, Yamamoto T, Oki K, Noda T, Kanamori H, Sasaki H. et al. Detection of Novel QTLs Regulating Grain Size in Extra-Large Grain Rice (Oryza sativa L.) Lines. Rice. 2016;9:34. doi: 10.1186/s12284-016-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han L, Chen J, Mace ES, Liu Y, Zhu M, Yuyama N. et al. Fine mapping of qGW1, a major QTL for grain weight in sorghum. TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2015;128:1813–25. doi: 10.1007/s00122-015-2549-2. [DOI] [PubMed] [Google Scholar]
- 9.Song XJ, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N. et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:76–81. doi: 10.1073/pnas.1421127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das K, Panda BB, Sekhar S, Kariali E, Mohapatra PK, Shaw BP. Comparative proteomics of the superior and inferior spikelets at the early grain filling stage in rice cultivars contrast for panicle compactness and ethylene evolution. Journal of plant physiology. 2016;202:65–74. doi: 10.1016/j.jplph.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Fu Y, Zhao S, Gu P, Zhu Z, Sun C. et al. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant biotechnology journal. 2016;14:377–86. doi: 10.1111/pbi.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar N, Suyal DC, Sharma IP, Verma A, Singh H. Elucidating stress proteins in rice (Oryza sativa L.) genotype under elevated temperature: a proteomic approach to understand heat stress response. 3 Biotech; 2017. p. 7. 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramalingam J, Arul L, Sathishkumar N, Vignesh D, Thiyagarajan K, Samiyappan R. TNAURice: Database on rice varieties released from Tamil Nadu Agricultural University. Bioinformation. 2010;5:264–5. doi: 10.6026/97320630005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang Z, Chen Z, Wang N, Xie G, Lu J, Yan W. et al. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14145–50. doi: 10.1073/pnas.1613792113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka J, Hayashi T, Iwata H. A practical, rapid generation-advancement system for rice breeding using simplified biotron breeding system. Breeding science. 2016;66:542–51. doi: 10.1270/jsbbs.15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swamy BP, Rahman MA, Inabangan-Asilo MA, Amparado A, Manito C, Chadha-Mohanty P. et al. Advances in breeding for high grain Zinc in Rice. Rice. 2016;9:49. doi: 10.1186/s12284-016-0122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helmy M, Tomita M, Ishihama Y. OryzaPG-DB: rice proteome database based on shotgun proteogenomics. BMC plant biology. 2011;11:63. doi: 10.1186/1471-2229-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C, Hu Z, Zheng T, Lu K, Zhao Y, Wang W, RPAN: rice pan-genome browser for approximately 3000 rice genomes. Nucleic acids research; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng L, Chen Z. An integrated framework for functional annotation of protein structural domains. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB) 2015;12:902–13. doi: 10.1109/TCBB.2015.2389213. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Zhang J, Fan C, Tang Y, Deng L. KATZLGO: Large-scale Prediction of LncRNA Functions by Using the KATZ Measure Based on Multiple Networks. IEEE/ACM Transactions on Computational Biology and Bioinformatics %@ 1545-5963; 2017. [DOI] [PubMed] [Google Scholar]
- 21.Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J. et al. The gene ontology annotation (goa) database: sharing knowledge in uniprot with gene ontology. Nucleic acids research. 2004;32(D):262–266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garzón JI, Deng L, Murray D, Shapira S, Petrey D, Honig B. A computational interactome and functional annotation for the human proteome. Elife. 2016;5:e18715. doi: 10.7554/eLife.18715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Zhang Z, Chen Z, Deng L. Integrating Multiple Heterogeneous Networks for Novel LncRNA-disease Association Inference. IEEE/ACM Transactions on Computational Biology and Bioinformatics %@ 1545-5963; 2017. [DOI] [PubMed] [Google Scholar]
- 24.Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y. et al. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant & cell physiology. 2013;54:e6. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu H, Zhu P, Jiao Y, Meng Y, Chen M. PRIN: a predicted rice interactome network. BMC bioinformatics. 2011;12:161. doi: 10.1186/1471-2105-12-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurata N, Yamazaki Y. Oryzabase. An integrated biological and genome information database for rice. Plant physiology. 2006;140:12–7. doi: 10.1104/pp.105.063008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato Y, Namiki N, Takehisa H, Kamatsuki K, Minami H, Ikawa H. et al. RiceFREND: a platform for retrieving coexpressed gene networks in rice. Nucleic acids research. 2013;41:D1214–21. doi: 10.1093/nar/gks1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu B, Zhang D, Xu R, Xu J, Wang X, Chen Q. et al. Combining evolutionary information extracted from frequency profiles with sequence-based kernels for protein remote homology detection. Bioinformatics. 2014;30:472–9. doi: 10.1093/bioinformatics/btt709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Liu F, Wang X, Chen J, Fang L, Chou K-C. Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nucleic Acids Research. 2015;43:W65–W71. doi: 10.1093/nar/gkv458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang T, Tan P, Wang L, Jin N, Li Y, Zhang L. et al. RNALocate: a resource for RNA subcellular localizations. Nucleic acids research. 2017;45:D135–D8. doi: 10.1093/nar/gkw728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang ZY, Lai HY, Yang H, Zhang CJ, Yang H, Wei HH. et al. Pro54DB: a database for experimentally verified sigma-54 promoters. Bioinformatics. 2017;33:467–9. doi: 10.1093/bioinformatics/btw630. [DOI] [PubMed] [Google Scholar]
- 32.Feng P, Ding H, Lin H, Chen W. AOD: the antioxidant protein database. Scientific reports. 2017;7:7449. doi: 10.1038/s41598-017-08115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng P, Ding H, Yang H, Chen W, Lin H, Chou KC. iRNA-PseColl: Identifying the Occurrence Sites of Different RNA Modifications by Incorporating Collective Effects of Nucleotides into PseKNC. Molecular therapy Nucleic acids. 2017;7:155–63. doi: 10.1016/j.omtn.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin H, Ding C, Yuan LF, Chen W, Ding H, Li ZQ, Predicting Subchloroplast Locations Of Proteins Based on the General Form Of Chou's Pseudo Amino Acid Composition: Approached From Optimal Tripeptide Composition. Int J Biomath; 2013. p. 6. [Google Scholar]
- 35.Ding H, Guo SH, Deng EZ, Yuan LF, Guo FB, Huang J. et al. Prediction of Golgi-resident protein types by using feature selection technique. Chemometr Intell Lab. 2013;124:9–13. [Google Scholar]
- 36.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K. et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic acids research. 2007;35:D883–7. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito Y, Arikawa K, Antonio BA, Ohta I, Naito S, Mukai Y. et al. Rice Annotation Database (RAD): a contig-oriented database for map-based rice genomics. Nucleic acids research. 2005;33:D651–5. doi: 10.1093/nar/gki083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, Xia Y, Li X, Hou L, Yu J. The Rice Genome Knowledgebase (RGKbase): an annotation database for rice comparative genomics and evolutionary biology. Nucleic acids research. 2013;41:D1199–205. doi: 10.1093/nar/gks1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q. et al. RMD: a rice mutant database for functional analysis of the rice genome. Nucleic acids research. 2006;34:D745–8. doi: 10.1093/nar/gkj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J, Ru B, Zhu P, Nie F, Yang J, Wang X. et al. MimoDB 2.0: a mimotope database and beyond. Nucleic acids research. 2012;40:D271–7. doi: 10.1093/nar/gkr922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He B, Chai G, Duan Y, Yan Z, Qiu L, Zhang H. et al. BDB: biopanning data bank. Nucleic acids research. 2016;44:D1127–32. doi: 10.1093/nar/gkv1100. [DOI] [PMC free article] [PubMed] [Google Scholar]