Abstract
Resurrection plants possess a unique ability to counteract desiccation stress. Desiccation tolerance (DT) is a very complex multigenic and multifactorial process comprising a combination of physiological, morphological, cellular, genomic, transcriptomic, proteomic, and metabolic processes. Modification in the sugar composition of the hemicellulosic fraction of the cell wall is detected during dehydration. An important change is a decrease of glucose in the hemicellulosic fraction during dehydration that can reflect a modification of the xyloglucan structure. The expansins might also be involved in cell wall flexibility during drying and disrupt hydrogen bonds between polymers during rehydration of the cell wall. Cleavages by xyloglucan-modifying enzymes release the tightly bound xyloglucan-cellulose network, thus increasing cell wall flexibility required for cell wall folding upon desiccation. Changes in hydroxyproline-rich glycoproteins (HRGPs) such as arabinogalactan proteins (AGPs) are also observed during desiccation and rehydration processes. It has also been observed that significant alterations in the process of photosynthesis and photosystem (PS) II activity along with changes in the antioxidant enzyme system also increased the cell wall and membrane fluidity resulting in DT. Similarly, recent data show a major role of ABA, LEA proteins, and small regulatory RNA in regulating DT responses. Current progress in “-omic” technologies has enabled quantitative monitoring of the plethora of biological molecules in a high throughput routine, making it possible to compare their levels between desiccation-sensitive and DT species. In this review, we present a comprehensive overview of structural, physiological, cellular, molecular, and global responses involved in desiccation tolerance.
1. Introduction to Desiccation Tolerance
Plants are sedentary and are most vulnerable to extreme weather conditions especially desiccation (loss of cellular water content equivalent to air dryness) and their associated stress. Among various abiotic stresses encountered by plants, desiccation or dehydration stress due to water deficiency that causes metabolic disruption and mechanical damage to membranes is the most prominent [1, 2]. Desiccation tolerant plants can withstand drying equal to or below that of the absolute water content which is equal to that of complete dryness. Further, these resume normal biological function upon rehydration [3]. Some members of microbial, fungal, and plant kingdoms have evolved DT traits [3, 4]. However, this phenomenon is not common. Some of the land plants possess tolerance to desiccation in vegetative tissues [5] and are called resurrection plants. In the plant kingdom, DT is noted in most of the taxonomic groups ranging from pteridophytes to dicotyledons but absent in gymnosperms [3, 6]. The mechanism of survival under the extreme environmental fluctuations is highly complex, and not all plants have the ability to withstand desiccation. Most important creation that DT plants have to satisfy is to limit the damage to a repairable level. Also, physiological integrity should remain intact under dehydration, and repair mechanisms must exist upon rehydration [7]. Resurrection plants are deemed to be an excellent model to study the mechanisms associated with DT. DT species have been discovered in places with seasonally limited water availability and unreliable rainfalls; they also inhabit soils with minimal water retention as outcrops of rock at low to moderate elevations in tropical and subtropical zones. They have been collected in all the continents [8]. In total, only about 1,300 species of vascular plants are available, of which 135 are flowering plants (termed “resurrection plants”) and display desiccation tolerance in their vegetative tissues [6, 8] The majority of DT plants grow in the tropics and subtropics of East-West Africa and Southern Africa (including Madagascar), Brazil, Australia, North America, and the Western and Eastern Ghats in India [9].
The earliest terrestrial plants were desiccation tolerant at all stages of their life cycle [10]. However, in more complex vascular plants, DT has been retained in only reproductive tissues, but not in vegetative tissues [11]. DT plants today are able to produce vegetative structures like spores, seeds, and pollen, which can keep them viable in the desiccated state for decades, or centuries, for example, the ancient Nelumbo nucifera seed from China [12]. The mechanisms of DT in lower-order resurrection plants like algae, lichens, and bryophytes are not similar to those of angiosperms [13, 14].
The genetic mechanisms required for DT are not only exclusive to resurrection plants but also present in desiccation-sensitive (DS) plants [15]. However, resurrection plants express these genes not only in seed tissues but also in vegetative tissues which help these plants to survive desiccation [16]. For instance, genes encoding LEA (late embryogenesis abundant) proteins that are generally present in the seeds of DS plants at the time of embryo maturation [17] have been isolated from the desiccated vegetative tissues of resurrection plants like C. plantagineum [2] and S. stapfianus [18]. Current phylogenetic data suggest that vascular plants gained the ability to survive desiccation of their vegetative tissues through a mechanism that was first present in spores, and such an evolution has been identified in at least ten independent events within the angiosperms [10]. In addition to angiosperms, DT exists in pteridophytes, mainly Selaginella (Selaginella, Selaginellaceae, class Lycopsida, and order Selaginellales) and its species. Selaginella is an ancient group of lycophytes, a monophyletic equivalent of other vascular plants such as monilophytes (ferns Psilotum and Equisetum) and seed plants (gymnosperms and angiosperms) [19]. The single genus of Selaginella encompasses about 700 species, characterized by strongly flattened, frond-like branching and dimorphic leaves (microphylls) [20]. Some of the DT species of Selaginella are lepidophylla [21], tamariscina [22], and bryopteris [23].
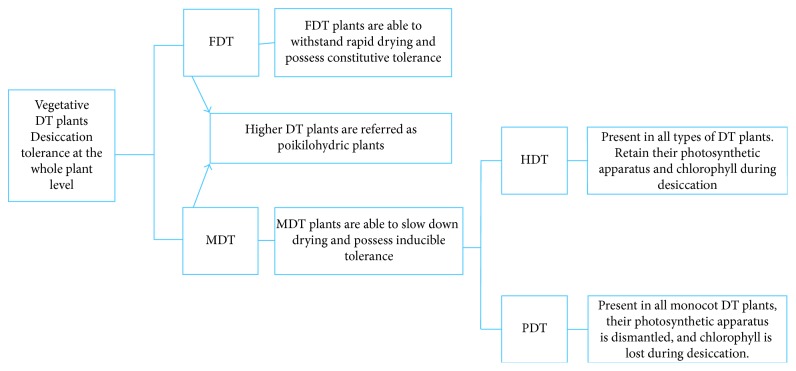
DT plants are classified based on the stress adaptation strategies which are shown in Table 1. They are divided into poikilochlorophyllous desiccation-tolerant (PDT) and homoiochlorophyllous desiccation-tolerant (HDT) plant (Figure 1) [39] types based on the status of the photosynthetic apparatus when dehydrated. During desiccation, HDT species retain their chlorophylls and photosynthetic apparatus in the readily recoverable state; for example, Craterostigma spp. retains the thylakoid and chlorophyll membranes intact through desiccation, although changes in photosynthetic pigment distribution were ascertained [40]. The chloroplasts of these plants have a distinct morphology including the round structure with an elaborate internal membrane organization. During the drying phase in homoiochlorophyllous vascular plants, the photochemical activity is much higher than CO2 absorption [41, 42]. Although the carbon fixation is suppressed through drying, the photoexcitation of chlorophyll in charge of the generation of ROS endures [43]. In C. wilmsii and M. flabellifolius, the photosynthesis is turned off during drying, via chlorophyll shading by leaf folding and anthocyanin accumulation [25, 44]. In HDT plants, rehydration can occur in a leaf disc that is detached from the plant or in a single leaf.
Table 1.
Classification of DT plants according to the types, family, class, and origin.
| Name | Family | Class | Origin | HDT/PDT | References |
|---|---|---|---|---|---|
| Craterostigma plantagineum | Scrophulariaceae | Dicot | Southern Africa | H | [24] |
| Craterostigma wilmsii | Scrophulariaceae | Dicot | Southern Africa | H | [25] |
| Lindernia brevidens | Linderniaceae | Dicot | East Africa | H | [26] |
| Myrothamnus flabellifolius | Myrothamnaceae | Dicot | Southern Africa | H | [27, 28] |
| Boea hygrometrica | Gesneriaceae | Dicot | China | H | [29] |
| Ramonda serbica | Gesneriaceae | Dicot | Serbia | H | [30] |
| Haberlea rhodopensis | Gesneriaceae | Dicot | Balkan mountains | H | [31] |
| Xerophyta viscosa | Velloziaceae | Monocot | Southern Africa | P | [32] |
| Xerophyta humilis | Velloziaceae | Monocot | Southern Africa | P | [33] |
| Sporobolus stapfianus | Poaceae | Monocot | Southern Africa | — | [34] |
| Eragrostis nindensis | Poaceae | Monocot | Southern Africa | P | [35] |
| Selaginella bryopteris | Selaginellaceae | Lycophyte | India | H | [23] |
| Selaginella tamariscina | Selaginellaceae | Lycophyte | China | H | [36] |
| Selaginella lepidophylla | Selaginellaceae | Lycophyte | North and South America | H | [37] |
| Tortula ruralis | Pottiaceae | Bryophyte (moss) | North America | H | [38] |
Figure 1.
Classification of DT plants according to their stress adaptation strategies.
In case of PDT species, chlorophyll loss takes place as a result of desiccation, which is then recovered following rehydration [45]. For example, in Xerophyta, both chlorophyll and photosystem complexes are broken down and the thylakoid membranes are dismantled during the desiccation [44]. Accumulation of toxic ROS is lowered due to the degradation of chlorophyll, and it is an advantage in these plants. Since the chloroplasts lose chlorophylls, the entire thylakoid system, and most carotenoids during dehydration, the entire photosynthetic apparatus has to be reconstructed after rehydration [45, 46]. Loss of pigment and other thylakoid pigments destruction are highly organized in responses to desiccation which occurs through a well-defined metabolic pathway [45]. Thus, homoiochlorophyllous plant species resume photosynthesis faster than poikilochlorophyllous species which need to synthesize all components de novo. When poikilochlorophyllous leaves are detached from the plant, they cannot resurrect in contrast to the leaves of homoiochlorophyllous plants [47].
Taxonomically, PDT are reported from monocots [48]. Poikilochlorophylly is currently well known in eight genera of four families (Liliaceae/Anthericaceae, Cyperaceae, Poaceae, and Velloziaceae). Most of these plants occupy soil less rocky outcrops known as inselbergs in intensely seasonal subtropical and tropical climatic conditions [8]. Well-studied examples include the Australian Borya nitid, an African X. viscosa, X. scabrida, and X. humilis [49]. DT plants can also be subdivided according to the differences in the molecular mechanism of DT. Fully desiccation-tolerant (FDT) plants are capable of withstanding rapid drying and possess constitutive tolerance, while modified desiccation-tolerant (MDT) species have the capacity to survive slow drying and possess inducible tolerance [4, 50].
Some DT monocots developed the strategy of poikilochlorophylly to remain alive and participate in minimal habitats where availability of light is variable [11]. During drying, photosynthetic mechanisms in DT bryophytes are protected, which recovers rapidly resulting in rehydration [10]. The DT grass S. stapfianus is moderately poikilochlorophyllous and retains most of its chlorophyll content during desiccation [117]. Subsequently, a sequence of events takes place: they are dehydration to aridness and rehydration to complete turgor, and S. stapfianus recaptured all of its photosynthetic ability within 24 hours like other plants. The PSII levels in S. stapfianus demonstrated a definite decrease during the dehydration stage. This might be due to the fact that increasing water stress reduces the rate of photosynthesis. In order to retain membrane integrity and protoplast survival by a protective mechanism, toxic oxygen production is carried out by weakened PSII [51]. But only proteins inside the thylakoid membranes of resurrection plants stay constant during rehydration and desiccation; however, DS plants are entirely damaged after a short-term desiccation process [52].
2. Mechanism of Desiccation Tolerance in Resurrection Plants
DT is a very complex multigenic and multifactorial process comprising a combination of structural changes and macromolecular processes [53]. It is not an easy task for organisms to stay alive after losing their 90% of cellular water upon dehydration and to regrow when rehydrated. Cellular damage probability increases during desiccation in resurrection plants, so these plants survive by induction of mechanisms for protection and maintenance of cell integrity [54].
2.1. Leaf Structural Alterations
The scarcity of water leads to changes in morphological traits of resurrection plants. Morphological changes occur in vegetative tissues upon desiccation, and the most important is leaf folding [55]. Folding of leaves during drying occurs in both DT and DS plants. DT dicot leaves of C. wilmsii are completely expanded under well-watered conditions but gradually curl inward during drying and become firmly folded which facilitates the abaxial surfaces of mature leaves exposed to the sun [42]. These changes in the leaf shrink the transpiring surface and minimize oxidative stress damage by ultraviolet radiation [37, 55, 56]. The leaf blades of the DT monocot X. humilis bend into half along the midrib upon dehydration, leaving only the abaxial leaf surface exposed to the sunlight [56]. In the DT grass S. stapfianus, the adaxial side of the leaf rich in the epicuticular wax is exposed to the sun to limit heating of leaf tissues and irradiation [57]. During dehydration, two main events are held simultaneously: cuticular wax covering and closure of stomata, which helps to reduce the rate of water loss from the thylakoid membranes [57]. Closing stomata plants is most likely to minimize the loss of water through transpiration, and hence, the water flux through the plant is limited by reducing leaf growth, which creates a smaller transpiring leaf area as in C. wilmsii, S. stapfianus, and X. humilis [27, 58].
2.2. Cell Wall Involvement during Desiccation
The cell wall is a highly dynamic compartment that evolves during cell growth and cell differentiation and in response to biotic and abiotic stresses. The cell wall provides a protective barrier and consists mainly of cellulose microfibrils, hemicelluloses, pectins, and “structural” glycoproteins such as extensin and arabinogalactan proteins [59–61]. Plant cell walls are divided into a primary cell wall and a secondary cell wall. The primary cell wall is present in almost all cells of the plants, whereas the secondary wall is visible only in differentiated tissues. Moreover, two types of cell walls can be differentiated. Type I primary walls found in the eudicotyledons, noncommon in monocotyledons and gymnosperms [62], are composed of a network of cellulose microfibrils, mostly cross-linked with xyloglucans (XyG) and embedded in a matrix of pectic polysaccharides. Type II primary cell walls, characteristic of monocotyledons (grasses and rushes), are composed of glucuronoarabinoxylans (GAXs) and mixed-linkage (1→3),(1→4)-β-D-glucan (β-glucan) polymers that link cellulose microfibrils [63, 64]. Pectic polysaccharides and XyG are generally poorly represented in type II primary cell walls. Furthermore, ferulic and p-coumaric acid arabinosyl esters can cross-link GAX in type II primary walls.
Mechanical stress is one of the more challenging stresses that resurrection plants have to overcome in order to survive desiccation [65]. As water is lost from the cell, plasmolysis occurs resulting in plasma membrane tearing from the more rigid cell wall and cell death. Resurrection plants have developed strategies to minimize the impact of mechanical stress during desiccation and to avoid irreversible damages [66]. Indeed, during the dehydration of the resurrection plant C. wilmsii, a decrease of about 78% of the cellular volume occurs in foliar tissues [44]. This extensive reduction of mesophyll cells is due to a strong folding of cell walls. In S. lepidophylla, an important event of folding of the cell walls and plasmalemma with continuous apposition to the cell wall is visualized during desiccation [67]. In M. flabellifolius, the folding of the cell wall is less distinct, and it is not observed in all cell types [44]. The folding of the cell wall is considered as a strategy developed by cells of DT plants to maintain the contacts between the plasma membrane and the cell wall during dehydration and to avoid the tearing between these structures and hence cell lysis and death. Cell wall modifications do occur in DT plants in response to dehydration. These are listed in Table 2.
Table 2.
Cell wall modifications of DT plants.
| Name | Cell wall modification | References |
|---|---|---|
| Boea hygrometrica | (i) Extensive cell wall folding accompanied by protoplasmic shrinkage | [16, 22, 68] |
| (ii) An increase of pectin and wax/suberin events occurred mainly during the rehydration phase | ||
| (iii) The contents of cell wall-associated lignin were reduced in desiccated leaves | ||
| (iv) Transcripts encoding cell metabolism were induced in rehydrated acclimated plants, indicating cell wall loosening during rehydration | ||
|
| ||
| Craterostigma plantagineum | (i) A marked reduction of the demethylesterification of HG in the dry state | [24, 60, 61, 69] |
| (ii) An upregulation of gene expressions corresponding to expansin and XyG synthesis | ||
| (iii) CpGRP1-CpWAK1 complex could be inducing morphological changes | ||
| (iv) A role for CpCRP1 in the leaf cell wall prior to dehydration stress and in mechanisms which are required for the successful recovery from desiccation | ||
| (v) The transcripts encoding proteins involved in ion transport such as membrane-associated carriers together with proteins involved in cell wall plasticity are abundant in fully hydrated conditions in C. plantagineum | ||
|
| ||
| Craterostigma wilmsii | (i) Decrease about 78% of the cellular volume | [44, 59] |
| (ii) A strong folding of the cell wall | ||
| (iii) A modification in the sugar composition of hemicellulosic fraction | ||
| (iv) An increase of epitopes recognized by the XyG-directed monoclonal antibodies | ||
|
| ||
| Eragrostis nindensis | (i) Arabinoxylans and xylans are involved in the regulation of mechanical properties of cell walls | [61, 70, 71] |
| (ii) Ferulic acid can cross-link neighbouring arabinoxylan molecules or arabinoxylans to enhance cell wall stiffening | ||
|
| ||
| Haberlea rhodopensis | (i) Upregulated transcript HrhDR35 encoding an XyG endotransglucosylase/hydrolase | [31, 72] |
| (ii) Downregulation of many cell wall-related genes including XyG endotransglucosylases and pectate lyases | ||
|
| ||
| Lindernia brevidens | (i) A strong folding of the cell wall | [26] |
|
| ||
| Myrothamnus flabellifolius | (i) Arabinose-enriched primary cell wall | [27, 61, 73, 74] |
| (ii) AGP is a contributor in ensuring flexibility and to facilitate the rehydration | ||
|
| ||
| Ramonda serbica | (i) Activities of nonspecific peroxidases play a role in cell wall remodelling | [75] |
|
| ||
| Selaginella bryopteris | (i) Phospholipase A1 gamma-like protein and glucan endo-1,3-alpha-glucosidase Agn1 have been reported to play a structural role in reinforcing the cell wall during stress | [76] |
|
| ||
| Selaginella lepydophylla | (i) A strong folding of the cell wall | [67] |
| (ii) Plasmalemma with continuous apposition to the cell wall | ||
|
| ||
| Sporobolus stapfianus | (i) A strong folding of the cell wall | [57, 77] |
| (ii) Transcripts encoding enzymes involved in cell wall remodelling are increased in abundance during dehydration | ||
| (iii) A late accumulation of ferulate and caffeate, precursors of cell wall lignin and cross-linking compounds, could enhance cell wall extensibility | ||
|
| ||
| Xerophyta spp. | (i) Highly arabinosylated xylans and arabinogalactan proteins | [61] |
2.2.1. Role of Cell Wall Cellulose in Desiccation Tolerance
One of the major constituents of both types of cell walls is cellulose, which exists as microfibrils composed of β-1,4-linked glucan chains that are linked by hydrogen bonds [78]. Cellulose is synthesized at the plasma membrane by a large multisubunit complex termed the “cellulose synthase” (CESA). The CESA is composed of at least three types of glycosyltransferases arranged into a hexameric rosette [78]. Although the protective function of cellulose during water stress has been well studied, there is no information available so far regarding the function of cellulose during desiccation. Some experiments reported a decrease of cellulose synthesis in response to water stress [79]. For example, in a switchgrass (Panicum virgatum L.), the transcript levels of CesA1, CesA6, and CesA12 encoding a cellulose synthase were suppressed in response to drought stress. However, this effect is reversed upon rehydration [80]. It was proposed that cellulose synthesis is redirected in adapted cells to produce a hemicellulosic compound [81]. In contrast, in other studies carried out on cotton fibers, the abundance of the SuSy, UDP-Glc, and UGPase was enhanced under drought stress. This phenomenon indicates that cotton fibers are able to produce relatively more UDP-Glc for cellulose synthesis under drought stress.
2.2.2. Role of Cell Wall Hemicellulose in Desiccation Tolerance
Hemicelluloses are important polysaccharides in plant cell walls consisting of β-(1→4)-linked backbones. Hemicelluloses include XyG, xylans, mannans, glucomannans, and β-(1→3, 1→4)-glucans. XyG are the major hemicellulosic polymers of dicot plants that strengthen the cell wall by forming a network with cellulose fibrils [87]. Existing models suggest the binding of each XyG polymer to at least two cellulose fibers [82]. This interaction can be modulated by two groups of enzymes: expansins and XyG endotransglucosylase/hydrolases (XTHs). In C. wilmsii, the structure of XyG, the major hemicellulosic compound, is affected by desiccation [59, 83]. Indeed, immunochemical studies revealed an increase of epitopes recognized by the XyG-directed monoclonal antibodies during dehydration. Furthermore, a modification in the sugar composition of the hemicellulosic fraction is detected during dehydration. An important decrease of the glucose in the hemicellulosic fraction during dehydration can reflect a modification of the XyG structure [83]. Expansins are another cell wall protein involved in remodelling of the cell wall. In the related species C. plantagineum, upregulation of gene expressions corresponding to expansin and xyloglucosyl transferase was closely correlated. The expansins might be involved in cell wall flexibility during drying and disrupt hydrogen bonds between polymers in the rehydration of the cell wall [24, 60]. Numerous studies were carried out on cell wall XTH and expansin. In the resurrection plant A. rhodopensis endemic to the Balkan area, upregulation of the transcript HrhDR35 encoding an XTH was observed. This high level of the transcript was induced during the early stage of dehydration and persists in the desiccated state [31]. Another study revealed that overexpression of an XTH gene from hot pepper (CaXTH3) in transgenic Arabidopsis plants confirmed the role of XTH in drought tolerance. However, an abnormal leaf morphology resulting in a severely wrinkled leaf shape along with an irregular pattern of leaf cells was observed in response of the transgenic plants to dehydration. A similar study performed on transgenic tomato plants overexpressing the XTH (CaXTH3) showed that overexpression was able to confer tolerance under severe water-deficit conditions [84]. These results suggest that CaXTH3 may be involved in cell wall remodelling, allowing the protection of the mesophyll cells from full dehydration. Another study suggests that the overexpression of RhEXPA4 in Arabidopsis transgenic plants conferred a dehydration tolerance during the expansion of rose petals [85]. In similar experiments, expansin was overexpressed in tobacco, confirming its role in plant water retention ability and osmotic potential [86]. Together, these data suggest that cleavages by XyG-modifying enzymes release the tightly bound XyG-cellulose network, thus increasing cell wall flexibility required for cell wall folding upon desiccation.
Xylans are one of the major hemicelluloses in the secondary cell walls of dicots and all walls of grasses [87]. Xylans are a diverse group of polysaccharides with a backbone composed of β-(1→4)-linked xylose residues. These xylans usually contain many arabinose residues attached to the backbone and are known as arabinoxylans and GAXs [88]. The DT plants identified as the grass-like Xerophyte sp. and the grass E. nindensis are found to be enriched in arabinose and xylose, suggesting that arabinoxylans are major cell wall components in this species [61, 70, 89]. One can expect that arabinoxylans and xylans are involved in the regulation of mechanical properties of the cell wall during dehydration of E. nindensis. Ferulic acid can cross-link neighbouring arabinoxylan molecules or arabinoxylans to enhance cell wall stiffening [71]. Moreover, xylans are known to cross-link cellulose microfibrils, thus contributing to cell wall mechanical properties. The presence of arabinose substitution on the xylan backbone hinders hydrogen bonding between xylan chains and/or the cross-linking between xylans and cellulose microfibrils [90]. In X. humilis, significant differences are observed for arabinose substitutions between hydrated and desiccated leaf compositions and could indicate that dehydration may cause an increase in the wall arabinoxylan content and/or arabinosylation of wall xylans in this species [61].
2.2.3. Role of Cell Wall Pectic Polysaccharides in Desiccation Tolerance
Pectins are the most complex family of cell wall polysaccharides. These components are enriched in galacturonic acid including homogalacturonan (HG), xylogalacturonan (XGA), rhamnogalacturonan I (RGI), and rhamnogalacturonan II (RGII) [91]. HG is the most abundant polysaccharide and represents 65% of the pectin, while RGI constitutes 20% to 35% [92]. XGA and RGII are minor components, each constituting less than 10% of pectins [92, 93]. HG is composed of free carboxyl groups that are able to cross-link with Ca2+ leading to the formation of a pectic gel [94]. HG can be methyl esterified through the action of PME (pectin methylesterase), which results in contiguous and random patterns of free carboxylic residues [95]. The PME enzyme activity is modulated specifically by inhibitor proteins such as the PME inhibitor (PMEI) [96].
Cell wall composition from the hydrated and dehydrated resurrection plants C. wilmsii and C. plantagineum shows a marked reduction of the demethylesterification of HG in the dry state [61]. A previous study showed that the overexpression of a pepper PMEI protein in Arabidopsis may be involved in drought stress tolerance [96]. Another experiment showed that the overexpression of stress-inducible OsBURP16 in transgenic rice plants, which encodes the beta-subunit of polygalacturonase 1, induces a decrease of pectin content accompanied by sensitivity to drought compared to the wild-type [36]. M. flabellifolius exhibits an unusual arabinose-enriched primary cell wall. However, the monosaccharide composition of the cell wall remained unchanged upon desiccation [27, 61, 73]. It was hypothesized that the high content of arabinan was associated with RGI and/or arabinogalactan proteins. In the roots of cv. Capeiti, ‘‘drought-tolerant,” the number of side chains of RGI and/or RGII significantly increased in response to water stress. One of the possibilities is that these polymers would act as pectin plasticizers and could provide constitutive flexibility of cell walls, protecting the cell walls against water loss [73, 97].
2.3. Modification of Cell Wall Proteins during Desiccation Tolerance
2.3.1. Hydroxyproline-Rich Glycoproteins (HRGPs)
Plant hydroxyproline-rich glycoproteins (HRGPs) are the major structural proteins in the cell wall [98, 99]. The HRGP family including arabinogalactan proteins (AGPs) and extensins (EXTs) consists of highly O-glycosylated proteoglycans [100]. AGPs are glycosylated with approximately 90% of carbohydrate moieties and only 10% of protein compounds of highly varying length. AGPs consist predominantly of arabinose and galactose residues, although other “minor” sugars including rhamnose, fucose, glucuronic acid, and xylose are also present [101]. AGPs are found inserted into the plasma in the external leaflet of the plasma membrane via a GPI anchor [101]. AGPs are known to be involved in many biological processes including cell development, cell death, cell-to-cell signalling, and cell defence [100]. Moore et al. proposed a model summarizing the importance of “plasticising” components of the cell walls in M. flabellifolius species during desiccation. AGPs are supposed to contribute to the flexibility of the cell walls during dehydration, and consequently, rehydration is facilitated in this plant [74]. The role of AGPs as pectic plasticizers is reviewed in [102], where they are shown to play a role in response to osmotic stress (i.e., salt). A study carried out on rice (Oryza sativa L.) showed that the expression of AGPs encoding genes was modulated in response to drought or salt stress. The expression of two AGPs genes, namely, OsAGP3 andOsAGP24, was upregulated in response to drought stress, whereas the expression of three other genes (OsELA3, OsAGP1, and OsAGP25) was upregulated in response to both drought and salt stresses [103]. The authors hypothesized that deglycosylation of AGPs by glycosidases would result in the release of oligosaccharides, which in turn would increase the intracellular osmotic pressure to reduce the rate of dehydration [103].
EXTs contain mainly hydroxyproline (Hyp), but serine (Ser), lysine (Lys), tyrosine (Tyr), histidine (His), valine (Val), and alanine (Ala) are also constitutive of the protein. The repetitive pattern (Ser-(Hyp)4) and the sequence (Tyr-Lys-Tyr) are characteristic of EXTs [99, 104, 105]. EXTs consist of arabinoside chains limited to 4-5 arabinosyl residues on Hyp residues and single galactose residues attached to Ser residues. EXTs have been extensively studied and shown to fulfill functions related to abiotic stresses in plants [106]. However, there is no information available so far regarding the EXT function during desiccation. Studies carried out on two potato clones (Solanum tuberosum) of the Andean cultivar group with different drought-tolerant phenotypes showed that transcription of one EXT gene was induced in both cultivars [107]. The transcript EXT3 was also shown to be upregulated under mild drought in Arabidopsis. This upregulation was accompanied by loosening of the cell wall allowing for a reduced growth under lower turgor [108].
2.3.2. Role of Glycine-Rich Proteins (GRPs) in Desiccation Tolerance
In plants, glycine-rich proteins (GRPs) are characterized by the presence of more than 60% glycine [109]. There are five classes of GRPs: three classes are based on the pattern of the glycine-rich repeats (class I, GGGX; class II, GGXXXGG; and class III, GXGX) and two other classes are based on the type of functionally conserved motifs (class IV, the oleosin glycine-rich proteins, and class V, the RNA-binding GRPs). Most of the GRPs known to date have been found in the cell walls of many higher plants and form another group of “structural proteins” of the cell wall [110].
GRPs are known to be modulated by abiotic factors. By analogy to HRGPs, those proteins have been proposed to act as a scaffold or agglutinating agents for deposition of cell wall constituents [111]. Analysis of transcripts expressed in desiccated leaves of C. plantagineum identified a gene putatively coding for an apoplastic glycine-rich protein (CpGRP1). CpGRP1 interacts with CpWAK1 which is downregulated in response to dehydration. The CpGRP1-CpWAK1 complex could be inducing morphological changes in the cell wall during dehydration in C. plantagineum. In fact, cell wall pectins and dehydration-induced pectin modifications are predicted to be involved in the activity of the CpGRP1-CpWAK1 complex [69].
2.4. Changes in the Antioxidant Systems during Desiccation Tolerance
During dehydration, resurrection plants produce high amounts of antioxidants [28]. The desiccation process can damage membrane lipids and proteins by producing a number of reactive oxygen species (ROS). By using an effective mechanism, S. stapfianus stimulates free radical scavenging enzymes, for example, ascorbate peroxidase, dehydroascorbate reductase, and glutathione reductase, to eliminate the ROS [112]. It has been shown that more injury occurs during rehydration than in desiccation because of increased oxidative stress at the time of the recovery phase [112]. Desiccation enhances the antioxidant activity in other resurrection plants also [42, 113]. In the DS plant S. stapfianus, when leaves are dried and detached, ascorbate peroxidase activity is decreased which leads to a reduction in antioxidant capacity [112].
The antioxidant enzymes superoxide dismutase (SOD), glutathione reductase (GR), catalase (CAT), and ascorbate peroxidase (APX) show a good response desiccation in DS and DT organisms. However, the response is significantly enhanced during desiccation in DT plants [16]. This phenomenon of higher induced antioxidant enzyme activity is a distinct DT mechanism [16]. Resurrection plants are able to sustain in the desiccated state for many years in relation to DS plants because increases in duration of desiccation will lead to more oxidative damage with gradual loss of the acyl chains of membrane polar lipids. Furthermore, the membranes contain unsaturated double bonds, very sensitive to free radical attack. Resurrection plants contain more number of double bonds in their polar lipids, which is a common characteristic of chloroplasts [113]. The antioxidant system comprising a number of enzymes fails during long duration due to programmed cell death, triggering aging and ultimately plant death as in DS plants [16]. It has been demonstrated that there is a negative association between the longevity of DT tissues and the number of double bonds in the polar lipids of the membranes, which ultimately determines how long resurrection plants are able to survive desiccation.
2.5. Cellular Membranes Lipid Composition Changes during Desiccation Tolerance
In the DT mosses S. lepidophylla and T. ruralis, dehydration results in cell shrinkage leading to highly convoluted membranes and walls [38]. During desiccation, the plasma membrane contains numerous tightly associated lipid droplets similar to a normal lipid bilayer organization. Desiccation normally causes unsaturation, and the level of individual phospholipids within the total lipids decreases in DT vascular plants [114]. However, unusual trends were noticed in the DT plant B. hygroscopica where an increase in unsaturation of all classes of fatty acids during desiccation was observed [115]. It is well known that a high degree of polyunsaturation in phospholipids is responsible for greater membrane fluidity. An increase in membrane fluidity is an important phenomenon for the survival of desiccation. The phospholipid content increases in leaves which are dried and attached to the parent. However, it decreased in leaves that are dried and detached in S. stapfianus during desiccation [116]. The dried leaves attached to the parent improve desiccation tolerance because of an increase in polyunsaturated fatty acids within the plasma membrane. However, in detached and dried leaves, this is not true, and hence, desiccation sensitivity might increase [17]. In S. stapfianus, leaves desiccated on the plant regained almost all of the lipid content, whereas detached dried leaves suffered complete lipid degradation with the loss of polyunsaturated fatty acids when rehydrated [116].
2.6. Signalling Mechanisms Involved in Desiccation Tolerance
During desiccation stress, an increase in the concentration of secondary messengers helps to control the intracellular Ca2+. This results in protein phosphorylation and transcription of stress-controlled genes. Plants under desiccation stress show a marked increase in ABA accumulation as one of the early responses. ABA plays a key role in the initiation of DT resulting in expression of proteins [117, 118]. Several dehydration-regulating genes are linked to ABA in DT plants [72, 119–121]. Regulatory pathways and gene signalling information in resurrection plants are relatively unknown in comparison with Arabidopsis. Phospholipid-based signalling in C. plantagineum is the main primary signalling pathway responsible for downstream mitigation of desiccation [122]. Dehydration induces the activity of two cDNA clones encoding phospholipase D in C. plantagineum but not that of ABA. By generating CpPLD-1 transcripts, secondary messenger molecules are engaged in primary reactions to dehydration. Furthermore, coexpression of CpPLD-2 results in an enhanced metabolism of phospholipids [122]. Dehydration-induced transcription factors regulated by phosphor lipases in C. plantagineum include the myeloblastosis family [123], basic leucine zipper family [124], homeodomain-leucine zipper family [125, 126], and a novel zinc finger factor [127].
2.7. Role of LEA Proteins in Desiccation Tolerance
LEA proteins accumulate to very high levels during the late stage of embryogenesis in seeds when they dry [128]. They also accumulate during dehydration in reproductive and vegetative tissues of plants. Therefore, LEA proteins are adaptive in nature and help to counter cold, drought, dehydration, desiccation, and salt in vegetative tissues [129]. They are also known to respond to ABA, and the plants with a high degree of LEA expression survive desiccation better [130]. Important roles of LEA proteins include DNA repair or unwinding, counteracting the physical stresses imposed from desiccation by stabilizing the filaments of cytoskeletons and chaperone activity to guard protein activity and its conformation [131, 132]. It has also been noticed that these proteins can act collaboratively with sugars, like trehalose, for preventing the aggregation of protein during desiccation [133]. Most of the LEA proteins belong to a more well-known group of proteins called “hydrophilins” characterized by 6% glycine and a high hydrophilicity index making them soluble at high (80°C) temperatures. LEA proteins are ubiquitous in the plant kingdom which are present not only in gymnosperms and angiosperms but also in seedless vascular plants and even in algae, bryophytes, and pteridophytes [54]. Also, these proteins are present in yeasts, bacteria [134, 135], nematodes [136, 137], and fungi [138, 139]. It has also been shown that ABA regulates the LEA protein expression along with dehydration-induced genes [72]. In C. plantagineum, a minimum of two LEA proteins (CDeT 11–24 and CDeT 6–19) are phosphorylated in vivo during desiccation [140]. Similar to LEA proteins, some other molecules like polyphenols (gallolylquinic acids) and small heat shock proteins are also responsive to desiccation [141]. These molecules safeguard membranes against desiccation, which shows that alternate novel elements with LEA like functions are active in resurrection plants. Reviews of Battaglia [142, 143] describe in detail the features and functions of the LEA proteins with respect to DT. LEA proteins perform a function in stabilizing membranes or in the transport of lipids for the reformation of damaged membranes in rehydrating T. ruralis gametophytes [10] or the transport of lipids for reconstitution of broken membranes [10]. Although a number of LEA genes have been isolated, a useful role in desiccation tolerance has been established for very few genes. The expression of the barley LEA sequence, HVA1, increased drought tolerance in transgenic wheat plants. Similarly, in rice, overexpression of HVA1 enhances tolerance to both drought and salt stress [144].
2.8. Role of Small Regulatory RNAs in Desiccation Tolerance
A significant role of small regulatory RNAs in monitoring the plant responses to desiccation stress is widely accepted in the recent scientific literature [119, 145, 146]. It has already been shown that the application of exogenous ABA in C. plantagineum callus was able to induce DT. Induction of DT was mediated by CDT-1 constitutive expression and other ABA-inducible genes. Other reports also suggest that constitutive expression of ABA-responsive transcripts in C. plantagineum is ABA independent [146]. Some other functionally related genes and CDT-1 members have abilities of a short interspersed element retrotransposon. Hence, it has been hypothesized to act as regulatory noncoding RNA molecules which are distinctive in C. plantagineum [147].
3. Functional -omic Studies on Desiccation Tolerance
The term “desiccomics” coined by [148] describes the combined -omic approaches to address and understand the global level changes associated with the dry state of the resurrection plants. Present-day progress in “-omic” technologies has enabled us in quantitative monitoring of the plethora of biological molecules in a high throughput routine, enabling comparison between desiccation-sensitive and desiccation-tolerant species. We summarize transcriptomic, proteomic, metabolomic, and genomic responses in various desiccation-tolerant plants below.
3.1. Transcriptome Analysis in Desiccation-Tolerant Plants
Thousands of ESTs (expressed sequence tags) can be analyzed through transcriptomic approaches. Expression profiling of transcriptomics (mRNA) can capture spatial and temporal gene expression while also quantifiying RNAs under different conditions. Either gene microarray techniques or quantitative reverse transcriptase polymerase chain reaction- (qRT-PCR-) based quantitative analysis of gene expression can be conducted. Current advances in assembly algorithms and sequencing technologies have enabled the reconstruction of the whole transcriptome via deep RNA sequencing (RNA-seq). Using these technologies, resurrection plants without any reference genome can also be analyzed.
In the studies mentioned above in Table 3, cDNA libraries for EST sequencing were generated in either single or two physiological situations (rehydrated and dehydrated fronds/roots/leaves or gametophytes). These studies reflect global transcript changes. Such integrated transcriptome analysis studies have so far been reported for Haberlea rhodopensis and C. plantagineum [24, 161].
Table 3.
Gene expression and EST sequencing studies on various resurrection plants.
The rehydrated moss T. ruralis was subjected to EST sequencing of a cDNA library which resulted in the identification of around 10,368 ESTs with 5,563 genes [152]. Transcriptomic studies of H. rhodopensis and C. plantagineum in different physiological stages (desiccated, control, rehydrated partially, and dehydrated) showed transcripts with the highest match to genes of Vitis vinifera, Populus trichocarpa, and Ricinus communis. In C. plantagineum, transcripts of 182 MB sequences were assembled into 29,000 contigs, which further produced 15,000 more unique individualities. Similar studies showed that 96,353 expressed transcript contigs of H. rhodopensis were characterized [24, 161]. Important knowledge generated from these studies shows that one-third of the contigs from C. plantagineum and around 40% contigs from T. ruralis and H. rhodopensis could not be mapped to UniProt identities. Thus, they are unknown transcripts which are possible sources for future gene detection. Depending on the expression patterns observed for H. rhodopensis and C. plantagineum, transcripts can be divided into two main groups. The first group contains transcripts abundant in control and rehydrated plant tissues, and the second group comprises abundant transcripts in dehydrated and desiccated plant tissues [24, 161].
3.2. Proteomics Studies in Desiccation-Tolerant Plants
Proteins play a major role in plant adjustment to desiccation stress as they are involved directly in the plant metabolism and cell structure. Proteins induced by desiccation consist of regulatory proteins (e.g., signalling proteins, protein kinases, transcription factors, and protein phosphatases). Also, effector proteins that are directly involved in acquisition of desiccation tolerance are LEA proteins, channel proteins, mRNA-binding proteins, components of protein biosynthesis and degradation, water osmolyte synthesis enzymes, detoxification enzymes, and cytoskeletal proteins. Owing to the low abundance of signalling proteins and transcription factors mentioned above, their protein complexes are not easily identified in various states of dehydration and rehydration by classical proteomics. Furthermore, to carry out a specific function, these proteins function as components of larger complexes, and these complexes may be regulated from the stage of its formation. Thus, characterizing these protein complexes will enable vital understanding of these proteins in different stages of desiccation/rehydration.
Reports of proteome analysis in resurrection plants are limited to a very few species. A direct association between protein and transcript richness has been recorded for several gene products with protective functions for dehydration [22, 29, 32, 162]. Quantitative proteome data correlate with transcript data which further confirms that the proteins associated with the carbohydrate metabolism and photosynthesis are abundant in the hydrated tissues of DT plants. Furthermore, termination of and reactivation of photosynthetic activity are major responses observed during desiccation and rehydration, respectively [22, 24, 32, 162].
The decline in the photosynthesis rate is proportional to reduction in abundance of chloroplast photosynthetic proteins, for example, psbO, psbP, the subunit of the F-ATPase, the PSII stability factor HCF136, and the transketolase [32]. In desiccation-tolerant resurrection plants, during desiccation, LEA proteins accumulate abundantly [117, 163–166]. In C. plantagineum, use of 2D-SDS-PAGES and phosphoprotein-specific stain shows that a minimum of two proteins were phosphorylated. Phosphorylation may possibly increase the hydrophilic residues which are necessary for interaction with other macromolecules. Phoshorylation is important for proper subcellular localization as was revealed for LEA proteins in the maize embryo. However, the role of phosphorylation in the LEA proteins CDeT11-24 and CDeT6-19 is yet to be discovered [167]. In resurrection plants, proteome analysis has shown the expression of unknown proteins. In case of S. tamariscinia, functions of 103 unique proteins from 138 protein spots responsive to dehydration could not be ascertained. During dehydration, proteins downregulated in S. tamariscina comprised those involved in the energy and carbohydrate metabolism, photosynthesis, stress signalling, membrane transport, defence proteins, cell division, and cell structure. However, protein abundance increased for antioxidant enzymes [22].
From the leaves of B. hygrometrica, 200 unique proteins were analyzed. Among these proteins, 35% (78) increased in response to desiccation stress, 60% showed decreased levels or remained unchanged, and 50% were induced under rehydration conditions. Many of the proteins associated with the antioxidant and energy metabolism were constitutively expressed which shows the occurrence of constitutive protective mechanisms. Proteins induced due to dehydration in B. hygrometrica are related to GSH, polyphenol metabolism, energy, and metabolism indicating that GSH serves as a key antioxidant. Furthermore, analysis of proteins indicates the photosynthetic degradation-related proteins like a 20 kDa fragment of the RuBisCO large subunit (RbcL) and an oxygen-evolving complex (a 23 kDa polypeptide of the photosystem II). A 20 kDa·RbcL was identified in dehydrated leaf proteins. Protein fragments appearing in B. hygrometrica are assumed to be the consequence of stress-related proteolysis rather than chloroplast-localized protease activity which is ROS-induced [15]. During dehydration, ATP-dependent transport of solutes mediated by ABC transporters was also induced in B. hygrometrica [29]. The putative induction of ATPase subunits identical to a vacuolar H+-ATPase A subunit on dehydration might help in rehydration preparation. Desiccated leaves of X. viscosa and Sporobolus stapfianus showed the same profile of protein as that of B. hygrometrica [168, 169]. Upregulation of enzymes is associated with sugar metabolism like ADP-glucose pyrophosphorylase, sucrose synthase, and GDP-mannose 3,5-epimerase, confirming the importance of sugar metabolism during desiccation stress. Protein expression patterns observed in different resurrection plants show that several proteins are rapidly and massively induced upon dehydration. These proteins continue to exist throughout the period of desiccation and carry out diverse functions like scavenging ROS, protecting proteins, sucrose accumulation, restoration of cell wall proteins and proteins also with unknown functions. Even though the tkt3 transcript levels are expressed constitutively in vegetative tissues of C. plantagineum, in hydrated tissues, the corresponding protein levels are too high, suggesting a slower protein turnover or a high translation rate during hydration. Similarly, in C. plantagineum, tkt7 mRNA abundance during late phases of rehydration does not match the protein abundance [168]. This phenomenon is also true for regulatory genes like transcription factors, which are difficult to investigate because of low abundance.
3.3. Metabolomic Analysis in Desiccation-Tolerant Plants
Metabolomic studies in resurrection plants deal with the quantitative analysis of small molecules in different metabolic states. Different study approaches like mass spectrometry (MS), gas chromatography (GC), liquid chromatography (LC), capillary electrophoresis, and nuclear magnetic resonance (NMR) spectroscopy are used to analyze a large variety of chemical structures. Arabidopsis is the choice model for metabolic studies because of its simple metabolism and the low rate of emission of metabolites during dehydration. Metabolomic studies on DT have been reported for two closely related Sporobolus and Selaginella species which differ in their desiccation tolerance [170, 171]. They showed that metabolite levels vary during desiccation/rehydration which includes lipids, nucleotide derivatives, carbohydrates, amino acids, polyamines, antioxidants, and defence compounds.
A number of significant metabolites have been identified in metabolomic studies on resurrection plants (Table 4). The metabolic state of fully hydrated S. lepidophylla was found to be different from the dehydration state. The mapping of the identified metabolites (66.5%) into the biochemical pathways shows that the dominant metabolites were amino acids (19%) followed by cofactors, carbohydrates, nucleotides, lipids, peptides, and secondary metabolites. However, peptides, amino acids, and nucleotide metabolites were more significant during desiccation. In the hydrated state, carbohydrates such as 4C–6C-containing sugars, lipids, or lipid metabolites (with the exception of choline phosphate), sugar alcohols, and cofactors were also more significant [176]. Among the 251 identified metabolites of S. lepidophylla, 33.4% were unknown metabolites. Seven unknown metabolites were of greater abundance in dehydration conditions than in hydrated states signifying their role in DT. Studies have shown that S. stapfianus metabolically prefers dehydration because of increased concentrations of nitrogen and osmolyte metabolites along with metabolites related to energy in the hydrated state. During dehydration, the metabolism moved towards carbohydrate and nitrogen remobilization, antioxidant production, and ammonia detoxification [162]. This also seems to be the case in C. plantagineum where major metabolite differences were predominantly during dehydration.
Table 4.
| Name of the metabolites | Role of metabolites during desiccation in plants | References |
|---|---|---|
| Carbohydrates: sucrose, raffinose, maltose, verbascose, stachyose, arbutin, glucosylglycerol, trehalose, and glucose | Replacing water on membranes and macromolecules by formation of anhydrous glass vitrification of the cytoplasm filling and stabilization of vacuoles and membrane proteins | [44, 52, 129, 172–175] |
|
| ||
| Amino acids: glutamate, glutamine, arginine, citrulline, aspartate, asparagine, N-6-trimethyllysine, and trans-4-hydroxyproline, and the intermediate metabolites 3-(3-hydroxyphenyl)propionate and the tripeptide ophthalmate (L-Y-glutamyl-L-α-aminobutyrylglycine), quinate, γ-glutamyl, tryptophan, and the derivatives acetyltryptophan or phenylalanine | Biosynthetic precursors for primary and secondary metabolites | [34, 161, 170, 176] |
| These amino acids could function as compatible solutes or as mobile nitrogen reserves for the rehydrating tissues | ||
| Activation of the shikimate pathway which can result in the synthesis of antioxidants | ||
|
| ||
| Nucleotide metabolites: allantoin, 1-methyladenosine, uridine 5′-monophosphate, and inosine | Plant stress protection by influencing ABA production, purine catabolism, and quenching ROS | [170, 176] |
|
| ||
| Lipids: phosphatidylinositol, phosphatidic acid, lysolipids, fatty acids, choline phosphate, and lipoxygenase | Maintenance of membrane integrity and maintenance of membrane fluidity to allow for recovery after dehydration | [122, 129, 176, 177] |
|
| ||
| Polyamines: spermidine and spermine | Membrane stabilization, enzyme activity modulation, plant growth and development, nitrogen assimilation, and respiratory metabolism. Protect ion of macromolecules | [178, 179] |
|
| ||
| Antioxidants: superoxide dismutase, catalase, ascorbate peroxidase, glutathione, etc. | Detoxify the ROS which arises during desiccation stress | [16, 156, 176] |
3.4. Genomic Studies on Desiccation-Tolerant Plants
The availability of genome sequences of DT plants though not on the scale of drought-tolerant plants has enabled a system-level effort to understand the complexities of DT [180]. Genomics is extremely important in order to complement transcriptomic and proteomic studies and also to get better expression profiles in response to desiccation [181]. Extensive studies on the genetic network activated in DT plants will help the scientific community to design mutants in order to evaluate the role of single/multiple gene(s) [182]. So far, genome sequences of only three DT species O. thomaeum, B. hygrometrica, and X. viscosa are available [183–185]. The curated data available do not seem to suggest any typical genomic features that are only specific to DT. Existence of co-linearity between genome structures in DT and non-DT plants shows that no specific genome-level effects occur because of desiccation, and examples of such a co-linearity are known in DT O. thomaeum and other grasses [181]. However, some important genes for protective proteins are present in duplicates indicating a transcribed genome [181]. Chloroplast genomes of T. ruralis are different from those of another moss Physcomitrella patens, both of which have different levels of sensitivity towards desiccation [175]. In addition, the whole-genome sequencing of the desiccation-tolerant grass O. thomaeum can serve as a valuable resource for the plant comparative genomics community [185]. Whole-genome sequence data of X. viscosa revealed that transcripts induced were typically desiccation tolerant in nature. Among the salient features of the genome during dehydration was reduced, transcript abundance of genomic “clusters of desiccation-associated genes” (CoDAGs), which might be due to complete stop of the growth that leads to an increase in expression of desiccation tolerance [183]. The genome of B. hygrometrica is approximately ∼1,548 MB in size. Approximately 85.86% of the assembly is a nongapped sequence. Gene prediction tools show that 49,374 protein-coding genes and 40.68% have been validated by RNA-Seq; among them, 23,250 (47.09%) were found to be similar to database entries resulting in assignment of gene function [184].
4. Conclusion
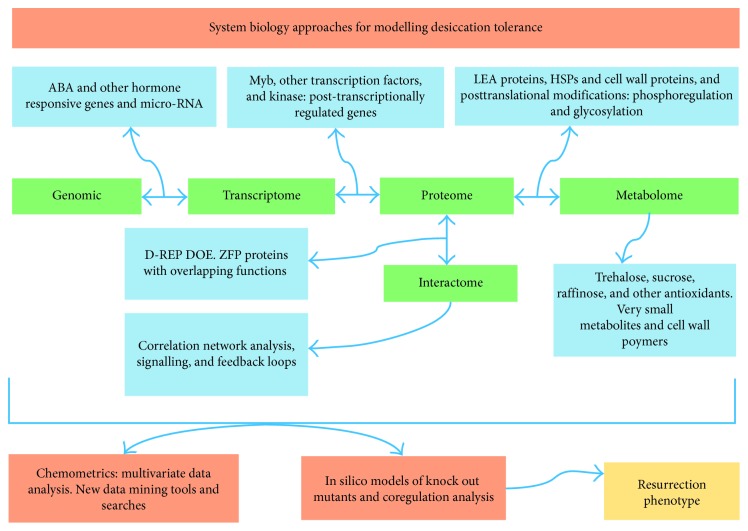
DT plants have extraordinary ability to regrow when rehydrated. This resilience is mainly because of their ability to alter the leaf structure and modify cell wall proteins and polymers during water loss and subsequent recovery. The recovery to the original morphology and physiological state is further aided by changes in photosynthesis, PSII activity, antioxidant systems, and lipid composition of cellular membranes. Also, ABA, LEA proteins, and small regulatory RNAs are responsible for regulating DT responses. Comprehensive and comparative information on changes in proteins/transcripts/metabolites in DT and DS species are now available mainly due to a recent surge in -omic technologies. However, the most challenging in terms of obtaining deep data coverage among these technologies is proteomics due to the low abundance of protein complexes. Recent improvements to various proteomic technologies have increased the sensitivity and robustness of protein identification during DT. In addition to these technological developments in peptide recovery and identification, further developments in bioinformatics and downstream validation technologies are required to make sense of complex data on DT. Therefore, in order to obtain more functional data on DT, a systems biology workflow with a specific focus on DT is the need of the hour. We therefore propose a new systems biology model (Figure 2) by integrating various functional -omic data sets with the aim of identifying new signalling intermediates and feedback loops responsible for overlapping/complex protein interactions resulting in desiccation tolerance. Implementing such a systems biology workflow will enable high confidence comparison of smaller proteomic data with those of the much larger microarray data sets. The strong and repetitive identification of low abundance proteins might just be possible with such a systems biology workflow which is a necessary prerequisite for analyzing global changes in DT plants comprehensively.
Figure 2.
Systems biology framework for DT studies.
Acknowledgments
The authors Sharathchandra RG and Driouich A are grateful to CEFIPRA/IFCPAR for funding support to the research project (Project no. IFC/5300-BA/2015/36) entitled “A Comparative Systems Biology Approach for Understanding Desiccation Tolerance in Forage Grasses and Selaginella sps.”
Abbreviations
- ABA:
Abscisic acid
- AGPs:
Arabinogalactan proteins
- Ala:
Alanine
- APX:
Ascorbate peroxidase
- CESA:
Cellulose synthase
- DS:
Desiccation sensitive
- DT:
Desiccation tolerance
- EXTs:
Extensins
- FDT:
Fully desiccation tolerant
- GAX:
Glucuronoarabinoxylans
- GR:
Glutathione reductase
- GRPs:
Glycine-rich proteins
- HG:
Homogalacturonan
- His:
Histidine
- HRGPs:
Hydroxyproline-rich glycoproteins
- Hyp:
Hydroxyproline
- LEA:
Late embryogenesis abundant
- Lys:
Lysine
- MDT:
Modified desiccation tolerant
- PME:
Pectin methylesterase
- PMEI:
PME inhibitor
- PS:
Photosystem
- RGI:
Rhamnogalacturonan
- IRGII:
Rhamnogalacturonan II
- ROS:
Reactive oxygen species
- Ser:
Serine
- SOD:
Superoxide dismutase
- Tyr:
Tyrosine
- Val:
Valine
- XGA:
Xylogalacturonan
- XTH:
XyG endotransglucosylase/hydrolase
- XyG:
Xyloglucans.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Levitt J. Responses of Plants to Environmental Stresses: Water, Radiation, Salt and Other Stresses. Vol. 2. New York, NY, USA: Academic Press; 1980. [Google Scholar]
- 2.Bohnert H. J., Nelson D. J., Jensen R. G. Adaptations to environmental stresses. Plant Cell. 1995;7(7):1099–1111. doi: 10.2307/3870060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alpert P. The discovery, scope and puzzle of desiccation tolerance in plants. Plant Ecology. 2000;151(1):5–17. doi: 10.1023/a:1026513800380. [DOI] [Google Scholar]
- 4.Farrant J. M. Mechanisms of desiccation tolerance in angiosperm resurrection plants. In: Jenks A., Wood A. J., editors. Plant Desiccation Tolerance. Wallingford, UK: CAB International Press; 2007. pp. 51–90. [Google Scholar]
- 5.Bartels D. Desiccation tolerance studied in the resurrection plant Craterostigma plantagineum. Integrative and Comparative Biology. 2005;45(5):696–701. doi: 10.1093/icb/45.5.696. [DOI] [PubMed] [Google Scholar]
- 6.Gaff D. F. Desiccation tolerant plants in Southern Africa. Science. 1971;174(4013):1033–1034. doi: 10.1126/science.174.4013.1033. [DOI] [PubMed] [Google Scholar]
- 7.Bewley J. D. Physiological aspects of desiccation tolerance. Annual Review of Plant Physiology. 1979;30(1):195–238. doi: 10.1146/annurev.pp.30.060179.001211. [DOI] [Google Scholar]
- 8.Porembski S., Barthlott W. Granitic and gneissic outcrops (inselbergs) as centres of diversity for desiccation tolerant vascular plants. Plant Ecology. 2001;151(1):19–28. [Google Scholar]
- 9.Kappen L., Valladares F. Opportunistic growth and desiccation tolerance: the ecological success of poikilohydrous autotrophs. In: Pugnaire F. I., Valladares F., editors. Handbook of Functional Plant Ecology. New York, NY, USA: Marcel Dekker; 1999. pp. 9–80. [Google Scholar]
- 10.Oliver M. J., Velten J., Mishler B. D. Desiccation tolerance in bryophytes: a reflection of the primitive strategy for plant survival in dehydrating habitats? Integrative and Comparative Biology. 2005;45(5):788–799. doi: 10.1093/icb/45.5.788. [DOI] [PubMed] [Google Scholar]
- 11.Oliver M. J., Tuba Z., Mishler B. D. The evolution of vegetative desiccation tolerance in land plants. Plant Ecology. 2000;151(1):85–100. doi: 10.1023/a:1026550808557. [DOI] [Google Scholar]
- 12.Miller S. J., Mudgett M. B., Schopf J. W., Clarke S., Berger R. Exceptional seed longevity and robust growth ancient sacred lotus from China. American Journal of Botany. 1995;82:1367–1380. [Google Scholar]
- 13.Oliver M. J., Bewley J. D. Desiccation tolerance of plant tissues: a mechanistic overview. Horticultural Reviews. 1997;18:171–214. [Google Scholar]
- 14.Oliver M. J., Wood A. J., O’Mahony P. To dryness and beyond preparation for the dried state and rehydration in vegetative desiccation tolerant plants. Plant Growth Regulation. 1998;24(3):193–201. doi: 10.1023/a:1005863015130. [DOI] [Google Scholar]
- 15.Bartels D., Salamini F. Desiccation tolerance in the resurrection plant Craterostigma plantagineum. A contribution to the study of drought tolerance at the molecular level. Plant Physiology. 2001;127(4):1346–1353. doi: 10.1104/pp.127.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Illing N., Denby K. J., Collett H., Shen A., Farrant J. M. The signature of seeds in resurrection plants: a molecular and physiological comparison of desiccation tolerance in seeds and vegetative tissues. Integral and Comparative Biology. 2005;45(5):771–787. doi: 10.1093/icb/45.5.771. [DOI] [PubMed] [Google Scholar]
- 17.Close T. J. Dehydrins emergence of a biochemical role of a family of plant dehydration proteins. Physiologia Plantarum. 1996;97(4):795–803. doi: 10.1111/j.1399-3054.1996.tb00546.x. [DOI] [Google Scholar]
- 18.Neale A. D., Blomstedt C. K., Bronso P., et al. The isolation of genes from the resurrection grass Sporobolus stapfianus which are induced during severe drought stress. Plant Cell Environment. 2000;23(3):265–277. doi: 10.1046/j.1365-3040.2000.00548.x. [DOI] [Google Scholar]
- 19.Banks J. A. Selaginella and 400 million years of separation. Annual Review of Plant Biology. 2009;60(1):223–238. doi: 10.1146/annurev.arplant.59.032607.092851. [DOI] [PubMed] [Google Scholar]
- 20.Korall P., Kenrick P. Phylogenetic relationships in Selaginellaceae based on RBCL sequences. American journal of Botany. 2002;89(3):506–517. doi: 10.3732/ajb.89.3.506. [DOI] [PubMed] [Google Scholar]
- 21.Pampurova S., Dijck P. V. The desiccation tolerant secrets of Selaginella lepidophylla: what we have learned so far. Plant Physiology and Biochemistry. 2014;80:285–290. doi: 10.1016/j.plaphy.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Chen S., Zhang H., et al. Desiccation tolerance mechanism in resurrection fern ally Selaginella tamariscina revealed by physiological and proteomic analysis. Journal of Proteome Research. 2010;9(12):6561–6577. doi: 10.1021/pr100767k. [DOI] [PubMed] [Google Scholar]
- 23.Pandey V., Ranjan S., Deeba F., et al. Desiccation induced physiological and biochemical changes in resurrection plant, Selaginella bryopteris. Journal of Plant Physiology. 2010;167(16):1351–1359. doi: 10.1016/j.jplph.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez M. C. S., Edsgard D., Hussain S. S., et al. Transcriptomes of the desiccation tolerant resurrection plant Craterostigma plantagineum. Plant Journal. 2010;63(2):212–228. doi: 10.1111/j.1365-313X.2010.04243.xx. [DOI] [PubMed] [Google Scholar]
- 25.Cooper K., Farrant J. M. Recovery of the resurrection plant Craterostigma wilmsii from desiccation: protection versus repair. Journal of Experimental Botany. 2002;53(375):1805–1813. doi: 10.1093/jxb/erf028. [DOI] [PubMed] [Google Scholar]
- 26.Phillips J. R., Fischer E., Baron M., et al. Lindernia brevidens: a novel desiccation tolerant vascular plant, endemic to ancient tropical rainforests. Plant Journal. 2008;54(5):938–948. doi: 10.1111/j.1365-313x.2008.03478.x. [DOI] [PubMed] [Google Scholar]
- 27.Moore J. P., Nguema-Ona E., Chevalier L., et al. Response of the leaf cell wall to desiccation in the resurrection plant Myrothamnus flabellifolius. Plant Physiology. 2006;141(2):651–662. doi: 10.1104/pp.106.077701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kranner I., Beckett R. P., Wornik S., Zorn M., Pfeihofer H. W. Revival of a resurrection plant correlates with its antioxidant status. Plant Journal. 2002;31(1):13–24. doi: 10.1046/j.1365-313x.2002.01329.x. [DOI] [PubMed] [Google Scholar]
- 29.Jiang G., Wang Z., Shang H., Yang W., Hu Z., Phillips J., et al. Proteome analysis of leaves from the resurrection plant Boea hygrometrica in response to dehydration and rehydration. Planta. 2007;225(6):1405–1420. doi: 10.1007/s00425-006-0449-z. [DOI] [PubMed] [Google Scholar]
- 30.Jovanovic V. S., Kukavica B., Stevanovic B., Navari-Izzo F. Senescence and drought related changes in peroxidase and superoxide dismutase isoforms in leaves of Ramonda serbica. Journal of Experimental Botany. 2006;57(8):1759–1768. doi: 10.1093/jxb/erl007. [DOI] [PubMed] [Google Scholar]
- 31.Georgieva K., Sarvari E., Keresztes A. V. Protection of thylakoids against combined light and drought by a luminal substance in the resurrection plant Haberlea rhodopensis. Annals of Botany. 2010;105(1):117–126. doi: 10.1093/aob/mcp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingle R. A., Schmidt U. G., Farrant J. M., Thomson J. A., Mundree S. G. Proteomic analysis of leaf proteins during dehydration of the resurrection plant Xerophyta viscose. Plant Cell Environment. 2007;30(4):435–446. doi: 10.1111/j.1365-3040.2006.01631.x. [DOI] [PubMed] [Google Scholar]
- 33.Collett H., Butowt R., Smith J., et al. Photosynthetic genes are differentially transcribed during the dehydration-rehydration cycle in the resurrection plant, Xerophyta humilis. Journal of Experimental Botany. 2003;54(392):2593–2595. doi: 10.1093/jxb/erg285. [DOI] [PubMed] [Google Scholar]
- 34.Martinelli T. In situ localization of glucose and sucrose in dehydrating leaves of Sporobolus stapfianus. Journal of Plant Physiology. 2008;165(6):580–587. doi: 10.1016/j.jplph.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 35.Willigen C. V., Pammenter N. W., Jaffer M. A., Mundree S. G., Farrant J. M. An ultrastructural study using anhydrous fixation of Eragrostis nindensis, a resurrection grass with both desiccation tolerant and sensitive tissues. Functional Plant Biology. 2003;30(3):1–10. doi: 10.1071/fp02221. [DOI] [PubMed] [Google Scholar]
- 36.Liu H., Ma Y., Chen N., et al. Overexpression of stress-inducible OsBURP16, the β subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environment. 2014;37(5):1144–1158. doi: 10.1111/pce.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brighigna L., Bennici A., Tani C., Tani G. Structural and ultrastructural characterization of Selaginella lepidophylla, a desiccation tolerant plant, during the rehydration process. Flora. 2002;197(2):81–91. doi: 10.1078/0367-2530-00018. [DOI] [Google Scholar]
- 38.Platt K. A., Oliver M. J., Thomson W. W. Membranes and organelles of dehydrated Selaginella and Tortula retain their normal configuration and structural integrity freeze fracture evidence. Protoplasma. 1994;178(1-2):57–65. doi: 10.1007/bf01404121. [DOI] [Google Scholar]
- 39.Toldi O., Tuba Z., Scott P. Vegetative desiccation tolerance: is it a goldmine for bioengineering crops. Plant Science. 2009;176(2):187–199. doi: 10.1016/j.plantsci.2008.10.002. [DOI] [Google Scholar]
- 40.Alamillo J. M., Bartels D. Effects of desiccation on photosynthesis pigments and the ELIP like dsp22 protein complex in the resurrection plant Craterostigma plantagineum. Plant Science. 2001;160(6):1161–1170. doi: 10.1016/s0168-9452(01)00356-9. [DOI] [PubMed] [Google Scholar]
- 41.Schwab K. B., Schreiber U., Heber U. Response of photosynthesis and respiration of resurrection plants to desiccation and rehydration. Planta. 1989;177(2):217–227. doi: 10.1007/bf00392810. [DOI] [PubMed] [Google Scholar]
- 42.Sherwin H. W., Farrant J. M. Protection mechanisms against excess light in the resurrection plants Craterostigma wilmsii and Xerophyta viscosa. Plant Growth Regulation. 1998;24(3):203–210. doi: 10.1023/a:1005801610891. [DOI] [Google Scholar]
- 43.Augusti A., Scartazza F., Izzo N. F., Sgherri C. L., Stevanovic B., Brugnoli E. Photosystem II photochemical efficiency, zeaxanthin and antioxidant contents in the poikilohydric Ramonda serbica during dehydration and rehydration. Photosynthsie Research. 2001;67(1-2):79–88. doi: 10.1023/a:1010692632408. [DOI] [PubMed] [Google Scholar]
- 44.Farrant J. M. A comparison of mechanisms of desiccation tolerance among three angiosperm resurrection plant species. Plant Ecology. 2000;151(1):29–39. doi: 10.1023/a:1026534305831. [DOI] [Google Scholar]
- 45.Tuba M., Proctor C. F., Csintalan Z. Ecophysiological responses of homoiochlorophyllous and poikilochlorophyllous desiccation tolerant plants: a comparison and ecological perspective. Plant Growth Regulation. 1998;24(3):211–217. doi: 10.1023/a:1005951908229. [DOI] [Google Scholar]
- 46.Tuba Z., Lichtenthaler H. K., Csintalan Z., Nagy Z., Szente K. Reconstitution of chlorophylls and photosynthetic CO2 assimilation upon rehydration of the desiccated poikilochlorophyllous plant Xerophyta scabrida (Pax) Th. Dur. Et. Schinz. Planta. 1994;192(3):414–420. doi: 10.1007/bf00198578. [DOI] [Google Scholar]
- 47.Dinakar C., Bartels D. Desiccation tolerance in resurrection plants: new insights from transcriptome, proteome and metabolome analysis. Frontiers in Plant Science. 2013;4:p. 482. doi: 10.3389/fpls.2013.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaff D. F. Responses of desiccation tolerant “resurrection” plants to water stress. In: Kreeb K. H., Richter H., Hinckley T. M., editors. Structural and Functional Responses to Environmental Stresses: Water Shortages. The Hague, Netherlands: SPB Academic Publishing; 1989. pp. 264–311. [Google Scholar]
- 49.Proctor M. C. F., Tuba Z. Poikilohydry and homoihydry, antithesis or spectrum of possibilities? New Phytologist. 2002;156(3):327–349. doi: 10.1046/j.1469-8137.2002.00526.x. [DOI] [PubMed] [Google Scholar]
- 50.Alpert P. Constrains of tolerance: why are desiccation tolerant organisms so small or rare? Journal of Experimental Biology. 2006;209(9):1575–1584. doi: 10.1242/jeb.02179. [DOI] [PubMed] [Google Scholar]
- 51.Blasi D. S., Puliga S., Losi L., Vazzana C. S. stapfianus and E. curvula cv. Consol in vivo photosynthesis, PSII activity and ABA content during dehydration. Plant Growth Regulation. 1998;25(2):97–104. [Google Scholar]
- 52.Deng X., Hu Z. A., Wang H. X., Wen X. G., Kuang T. Y. A comparison of photosynthetic apparatus of the detached leaves of the resurrection plant Boea hygrometrica with its non-tolerant relative Chirita heterotrichia in response to dehydration and rehydration. Plant Science. 2003;165(4):851–861. doi: 10.1016/s0168-9452(03)00284-x. [DOI] [Google Scholar]
- 53.Moore J. P., Le N. T., Brandt W. F., et al. Towards a systems-based understanding of plant desiccation tolerance. Trends in Plant Science. 2009;14(2):110–117. doi: 10.1016/j.tplants.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 54.Dinakar C., Djilianov D., Bartels D. Photosynthesis in desiccation tolerant plants: energy metabolism and antioxidative stress defense. Plant Science. 2012;182:29–41. doi: 10.1016/j.plantsci.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 55.Nar H., Saglam A., Terzi R., Varkonyi Z., Kadioglu A. Leaf rolling and photosystem II efficiency in Ctenanthe setose exposed to drought stress. Photosynthetica. 2009;47(3):429–436. doi: 10.1007/s11099-009-0066-8. [DOI] [Google Scholar]
- 56.Jones A. M., Bennett M. H., Mansfield J. W., Grant M. Analysis of the defence phosphoproteome of Arabidopsis thaliana using differential mass tagging. Proteomics. 2006;6(14):4155–4165. doi: 10.1002/pmic.200500172. [DOI] [PubMed] [Google Scholar]
- 57.Vecchia D. F., Asmar E. T., Calamassi R., Rascio N., Vazzana C. Morphological and ultrastructural aspects of dehydration and rehydration in leaves of Sporobolus stapfianus. Plant Growth Regulation. 1998;24(3):219–228. doi: 10.1023/a:1005853527769. [DOI] [Google Scholar]
- 58.Tardieu F. Plant tolerance to water deficit: physical limits and possibilities for progress. Plant tolerance to water deficit: physical limits and possibilities for progress. Comptes Rendus Geoscience. 2005;337(1-2):57–67. doi: 10.1016/j.crte.2004.09.015. [DOI] [Google Scholar]
- 59.Vicré M., Farrant J. M., Driouich A. Insights into the cellular mechanisms of desiccation tolerance among angiosperm resurrection plant species. Plant Cell & Environment. 2004;27(11):1329–1340. doi: 10.1111/j.1365-3040.2004.01212.x. [DOI] [Google Scholar]
- 60.Jones L., McQueen-Mason S. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Letters. 2004;559(1–3):61–65. doi: 10.1016/s0014-5793(04)00023-7. [DOI] [PubMed] [Google Scholar]
- 61.Moore J. P., Nguema-Ona E. E., Vicré-Gibouin M., et al. Arabinose-rich polymers as an evolutionary strategy to plasticize resurrection plant cell walls against desiccation. Planta. 2013;237(3):739–754. doi: 10.1007/s00425-012-1785-9. [DOI] [PubMed] [Google Scholar]
- 62.Minic Z., Jouanin L. Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiology and Biochemistry. 2006;44(7–9):435–449. doi: 10.1016/j.plaphy.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 63.Vogel J. Unique aspects of the grass cell wall. Current Opinion in Plant Biology. 2008;11(3):301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 64.Le Gall H., Philippe F., Domon J.-M., et al. Cell wall metabolism in response to abiotic stress. Plants. 2015;4(1):112–166. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iljin W. S. Drought resistance in plants and physiological processes. Annual Review of Plant Physiology. 1957;8(1):257–274. doi: 10.1146/annurev.pp.08.060157.001353. [DOI] [Google Scholar]
- 66.Farrant J., Willigen V. C., Loffell D., et al. An investigation into the role of light during desiccation of three angiosperm resurrection plants. Plant Cell and Environment. 2003;26(8):1275–1286. doi: 10.1046/j.0016-8025.2003.01052.x. [DOI] [Google Scholar]
- 67.Thomson W. W., Platt K. A. Conservation of cell order in desiccated mesophyll of Selaginella lepidophylla [Hook and Grev.] Spring. Annals of Botany. 1997;79(4):439–447. doi: 10.1006/anbo.1996.0375. [DOI] [Google Scholar]
- 68.Mitra J., Xu G., Wang B., Li M., Deng X. Understanding desiccation tolerance using the resurrection plant Boea hygrometrica as a model system. Frontiers in Plant Science. 2013;4 doi: 10.3389/fpls.2013.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giarola V., Krey S., Driesch B., Bartels D. The Craterostigma plantagineum glycine-rich protein CpGRP1 interacts with a cell wall-associated protein kinase 1 (CpWAK1) and accumulates in leaf cell walls during dehydration. New Phytologist. 2016;210(2):535–550. doi: 10.1111/nph.13766. [DOI] [PubMed] [Google Scholar]
- 70.Plancot B., Vanier G., Maire F., et al. Structural characterization of arabinoxylans from two African plant species Eragrostis nindensis and Eragrostis tef using various mass spectrometric methods. Rapid Communications in Mass Spectrometry. 2014;28(8):908–916. doi: 10.1002/rcm.6859. [DOI] [PubMed] [Google Scholar]
- 71.Grabber J. H., Ralph J., Lapierre C., Barrière Y. Genetic and molecular basis of grass cell-wall degradability. I. Lignin–cell wall matrix interactions. Comptes Rendus Biologies. 2004;327(5):455–465. doi: 10.1016/j.crvi.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 72.Phillips J. R., Oliver M. J., Bartels D. Molecular genetics of desiccation tolerant systems. In: Black M., Pritchard H. W., editors. Desiccation and Survival in Plants: Drying without Dying. Wallingford, UK: CABI Publishing; 2002. pp. 19–341. [Google Scholar]
- 73.Moore J. P., Vicré-Gibouin M., Farrant J. M., Driouich A. Adaptations of higher plant cell walls to water loss: drought vs desiccation. Physiologia Plantarum. 2008;134(2):237–245. doi: 10.1111/j.1399-3054.2008.01134.x. [DOI] [PubMed] [Google Scholar]
- 74.Moore J. P., Lindsey G. G., Farrant J. M., Brandt W. F. An overview of the biology of the desiccation-tolerant resurrection plant Myrothamnus flabellifolia. Annals of Botany. 2007;99(2):211–217. doi: 10.1093/aob/mcl269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rakić T., Lazarević M., Jovanović Ž. S., et al. Resurrection plants of the genus Ramonda: prospective survival strategies–unlock further capacity of adaptation, or embark on the path of evolution? Frontiers in Plant Science. 2013;4:p. 550. doi: 10.3389/fpls.2013.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deeba F., Pandey A. K., Pandey V. Organ specific proteomic dissection of Selaginella bryopteris undergoing dehydration and rehydration. Frontiers in Plant Science. 2016;7 doi: 10.3389/fpls.2016.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yobi A., Schlauch K. A., Tillett R. L., et al. Sporobolus stapfianus: insights into desiccation tolerance in the resurrection grasses from linking transcriptomics to metabolomics. BMC Plant Biology. 2017;17(1) doi: 10.1186/s12870-017-1013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Somerville C., Bauer S., Brininstool G., et al. Toward a systems approach to understanding plant cell walls. Science. 2004;306(5705):2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- 79.Iraki N. M., Bressan R. A., Hasegawa P. M., Carpita N. C. Alteration of the physical and chemical structure of the primary cell wall of growth-limited plant cells adapted to osmotic stress. Plant Physiology. 1989;91(1):39–47. doi: 10.1104/pp.91.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang Y., Yao Y., Wang Y. Physiological response, cell wall components, and gene expression of switchgrass under short-term drought stress and recovery. Crop Science. 2012;52(6):p. 2718. doi: 10.2135/cropsci2012.03.0198. [DOI] [Google Scholar]
- 81.Iraki N. M., Singh N., Bressan R. A., Carpita N. C. Cell walls of tobacco cells and changes in composition associated with reduced growth upon adaptation to water and saline stress. Plant Physiology. 1989;91(1):48–53. doi: 10.1104/pp.91.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cosgrove D. J., Jarvis M. C. Comparative structure and biomechanics of plant primary and secondary cell walls. Frontiers in Plant Science. 2012;3:1–6. doi: 10.3389/fpls.2012.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vicré M., Sherwin H. W., Driouich A., et al. Cell wall characteristics and structure of hydrated and dry leaves of the resurrection plant Craterostigma wilmsii, a microscopical study. Journal of Plant Physiology. 1999;155(6):719–726. doi: 10.1016/s0176-1617(99)80088-1. [DOI] [Google Scholar]
- 84.Choi J. Y., Seo Y. S., Kim S. J., et al. Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants (Solanum lycopersicum cv. Dotaerang) Plant Cell Reports. 2011;30(5):867–877. doi: 10.1007/s00299-010-0989-3. [DOI] [PubMed] [Google Scholar]
- 85.Dai F., Zhang C., Jiang X., et al. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant physiology. 2012;160(4):2064–2082. doi: 10.1104/pp.112.207720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han Y. Y., Li A. X., Zhao M. R., Wang W. Characterization of a wheat (Triticum. aestivum L.) expansin gene, TaEXPB23, involved in the abiotic stress response and phytohormone regulation. Plant Physiology and Biochemistry. 2012;54:49–58. doi: 10.1016/j.plaphy.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 87.Faik A. Xylan biosynthesis: news from the grass. Plant Physiology. 2010;153(2):396–402. doi: 10.1104/pp.110.154237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scheller H. V., Ulvskov P. Hemicelluloses. Annual Review of Plant Biology. 2010;61(1):263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 89.Gaff D. F., Ellis R. P. Southern African grasses with foliage that revives after dehydration. Bothalia. 1974;11(3):305–308. doi: 10.4102/abc.v11i3.1476. [DOI] [Google Scholar]
- 90.Christensen U., Alonso-Simon A., Scheller H. V., et al. Characterization of the primary cell walls of seedlings of Brachypodium distachyon–a potential model plant for temperate grasses. Phytochemistry. 2010;71(1):62–69. doi: 10.1016/j.phytochem.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 91.Ridley B. L., O’Neill M. A., Mohnen D. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry. 2001;57(6):929–967. doi: 10.1016/s0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- 92.Mohnen D. Pectin structure and biosynthesis. Current Opinions in Plant Biology. 2008;11(3):266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 93.Zandleven J., Sørensen S. O., Harholt J., et al. Xylogalacturonan exists in cell walls from various tissues of Arabidopsis thaliana. Phytochemistry. 2007;68(8):1219–1226. doi: 10.1016/j.phytochem.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 94.Carpita N. C., Gibeaut D. M. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant Journal. 1993;3(1):1–30. doi: 10.1046/j.1365-313x.1993.00999.x. [DOI] [PubMed] [Google Scholar]
- 95.Harholt J., Suttangkakul A., Scheller H. V. Biosynthesis of Pectin. Plant Physiology. 2010;153(2):384–395. doi: 10.1104/pp.110.156588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.An S. H., Sohn K. H., Choi H. W., et al. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta. 2008;228(1):61–78. doi: 10.1007/s00425-008-0719-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leucci M. R., Lenucci M. S., Piro G., Dalessandro G. Water stress and cell wall polysaccharides in the apical root zone of wheat cultivars varying in drought tolerance. Journal of Plant Physiology. 2008;165(11):1168–1180. doi: 10.1016/j.jplph.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 98.Showalter A. M. Structure and function of plant cell wall proteins. Plant Cell. 1993;5(1):9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cassab G. I. Plant cell wall proteins. Annual Review of Plant Physiology. 1998;49(1):281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- 100.Zagorchev L., Kamenova P., Odjakova M. The role of plant cell wall proteins in response to salt stress, the role of plant cell wall proteins in response to salt stress. Scientific World Journal. 2014;2014:9. doi: 10.1155/2014/764089.e764089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nguema-Ona E., Coimbra S., Vicré-Gibouin M., et al. Arabinogalactan proteins in root and pollen-tube cells: distribution and functional aspects. Annals of Botany. 2012;110(2):383–404. doi: 10.1093/aob/mcs143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lamport D. T. A., Kieliszewski M. J., Showalter A. M. Salt stress upregulates periplasmic arabinogalactan proteins: using salt stress to analyse AGP function∗. New Phytologist. 2006;169(3):479–492. doi: 10.1111/j.1469-8137.2005.01591.x. [DOI] [PubMed] [Google Scholar]
- 103.Ma H., Zhao J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.) Journal of Experimental Botany. 2010;61(10):2647–2668. doi: 10.1093/jxb/erq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kieliszewski M. J., Lamport D. T. A. Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant Journal. 1994;5(2):157–172. doi: 10.1046/j.1365-313x.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- 105.Kieliszewski M. J., Shpak E. Synthetic genes for the elucidation of glycosylation codes for arabinogalactan-proteins and other hydroxyproline-rich glycoproteins. Cellular and Molecular Life Sciences. 2001;58(10):1386–1398. doi: 10.1007/pl00000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oka T., Saito F., Shimma Y.-I., et al. Characterization of endoplasmic reticulum-localized UDP-D-galactose: hydroxyproline O-galactosyltransferase using synthetic peptide substrates in arabidopsis. Plant Physiology. 2010;152(1):332–340. doi: 10.1104/pp.109.146266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Evers D., Lefèvre I., Legay S., et al. Identification of drought-responsive compounds in potato through a combined transcriptomic and targeted metabolite approach. Journal of Experimental Botany. 2010;61(9):2327–2343. doi: 10.1093/jxb/erq060. [DOI] [PubMed] [Google Scholar]
- 108.Clauw P., Coppens F., De Beuf K., et al. Leaf responses to mild drought stress in natural variants of arabidopsis. Plant Physiology. 2015;167(3):800–816. doi: 10.1104/pp.114.254284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mangeon A., Junqueira R. M., Sachetto-Martins G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signaling & Behavior. 2010;5(2):99–104. doi: 10.4161/psb.5.2.10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mousavi A., Hotta Y. Glycine-rich proteins: a class of novel proteins. Applied Biochemistry and Biotechnology. 2005;120(3):169–174. doi: 10.1385/abab:120:3:169. [DOI] [PubMed] [Google Scholar]
- 111.Fusaro A., Mangeon A., Junqueira R. M., et al. Classification, expression pattern and comparative analysis of sugarcane expressed sequences tags (ESTs) encoding glycine-rich proteins (GRPs) Genetics and Molecular Biology. 2001;24(1–4):263–273. doi: 10.1590/s1415-47572001000100035. [DOI] [Google Scholar]
- 112.Sgherri C. L. M., Loggini B., Bochicchio A., Navari-Izzo F. Antioxidant system in Boea hygroscopica: changes in response to desiccation and rehydration. Phytochemistry. 1994;37(2):377–381. doi: 10.1016/0031-9422(94)85063-1. [DOI] [Google Scholar]
- 113.Hoekstra F. A. Differential longevities in desiccated anhydrobiotic plant systems. Integrative and Comparative Biology. 2005;45(5):725–733. doi: 10.1093/icb/45.5.725. [DOI] [PubMed] [Google Scholar]
- 114.Quartacci M. F., Pinzino C., Sgherri C. L. M., Vecchia D. F., NavariIzzo F. Growth in excess copper induces changes in the lipid composition and fluidity of PSII-enriched membranes in wheat. Physiologia Plantarum. 2002;108(1):87–93. doi: 10.1034/j.1399-3054.2000.108001087.x. [DOI] [Google Scholar]
- 115.Navari-Izzo F., Ricci F., Vazzana C., Quartacci M. F. Unusual composition of thylakoid membranes of the resurrection plant Boea hygroscopica changes in lipids upon dehydration and rehydration. Physiologia Plantarum. 1995;94(1):135–142. doi: 10.1111/j.1399-3054.1995.tb00794.x. [DOI] [Google Scholar]
- 116.Quartacci M. F., Forli M., Rascio N., Dallavecchia F., Bochicchio A., Navari-Izzo F. Desiccation tolerant Sporobolus stapfianus: lipid composition and cellular ultrastructure during dehydration and rehydration. Journal of Experimental Botany. 1997;48(6):1269–1279. doi: 10.1093/jxb/48.6.1269. [DOI] [Google Scholar]
- 117.Ingram J., Bartels D. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47(1):377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 118.Hilbricht T., Varotto S., Sgaramella V., Bartels D., Salamini F., Furini A. Retro transposon and siRNA have a role in the evolution of desiccation tolerance leading to resurrection of the plant Craterostigma plantagineum. New Phytologist. 2008;179(3):877–887. doi: 10.1111/j.1469-8137.2008.02480.x. [DOI] [PubMed] [Google Scholar]
- 119.Bartels D., Schneider K., Terstappen G., Piatkowski D., Salamini F. Molecular cloning of abscisic acid modulated genes which are induced during desiccation of the resurrection plant Craterostigma plantagineum. Planta. 1990;181(1):27–34. doi: 10.1007/bf00202321. [DOI] [PubMed] [Google Scholar]
- 120.Piatkowski D., Schneider K., Salamini F., Bartels D. Characterization of five abscisic acid responsive cDNA clones isolated from the desiccation tolerant plant Craterostigma plantagineum and their relationship to other water stress genes. Plant Physiology. 1990;94(4):1682–1688. doi: 10.1104/pp.94.4.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Michel D., Salamini F., Bartels D., Dale P., Baga M., Szalay A. Analysis of a desiccation and ABA-responsive promoter isolated from the resurrection plant Craterostigma plantagineum. Plant Journal. 1993;4(1):29–40. doi: 10.1046/j.1365-313x.1993.04010029.x. [DOI] [PubMed] [Google Scholar]
- 122.Frank W., Munnik T., Kerkmann K., Salamini F., Bartels D. Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell. 2000;12(1):111–123. doi: 10.1105/tpc.12.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Iturriaga G., Leyns L., Villegas A., Gharaibeh R., Salamini F., Bartels D. A family of novel myb-related genes from the resurrection plant Craterostigma plantagineum are specifically expressed in callus and roots in response to ABA or desiccation. Plant Molecular Biology. 1996;32(4):707–716. doi: 10.1007/bf00020211. [DOI] [PubMed] [Google Scholar]
- 124.Ditzer A., Bartels D. Identification of a dehydration and ABA-responsive promoter regulon and isolation of corresponding DNA binding proteins for the group 4 LEA gene CpC2 from Craterostigma plantagineum. Plant Molecular Biology. 2006;61(4-5):643–663. doi: 10.1007/s11103-006-0038-3. [DOI] [PubMed] [Google Scholar]
- 125.Deng X., Phillips J., Brautigam A., et al. A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses. Plant Molecular Biology. 2006;61(3):469–489. doi: 10.1007/s11103-006-0023-x. [DOI] [PubMed] [Google Scholar]
- 126.Deng X., Phillips J., Meijer A. H., Salamini F., Bartels D. Characterization of five novel dehydration responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum. Plant Molecular Biology. 2002;49(6):601–610. doi: 10.1023/a:1015501205303. [DOI] [PubMed] [Google Scholar]
- 127.Hilbricht T., Salamini F., Bartels D. CpR18, a novel SAP-domain plant transcription factor, binds to a promoter region necessary for ABA mediated expression of the CDeT27-45 gene from the resurrection plant Craterostigma plantagineum Hochst. Plant Journal. 2002;31(3):293–303. doi: 10.1046/j.1365-313x.2002.01357.x. [DOI] [PubMed] [Google Scholar]
- 128.Dure L. Structural motifs of LEA proteins in higher plants. In: Close T. J., Bray E. A., editors. Responses of Plants to Cellular Dehydration during Environmental Stress. Rockville, MD, USA: American Society for Plant Physiology; 1993. pp. 104–118. [Google Scholar]
- 129.Hoekstra F. A., Golovina E. A., Tetteroo F. A., Wolkers W. F. Induction of desiccation tolerance in plant somatic embryos: how exclusive is the protective role of sugars? Cryobiology. 2001;43(2):140–150. doi: 10.1006/cryo.2001.2358. [DOI] [PubMed] [Google Scholar]
- 130.Crowe J. H., Hoekstra F. A., Crowe L. M. Anhydrobiosis. Annual Review of Physiology. 1992;54(1):579–599. doi: 10.1146/annurev.physiol.54.1.579. [DOI] [PubMed] [Google Scholar]
- 131.Wise M. J., Tunnacliffe A. POPP the question: what do LEA proteins do? Trends Plant Science. 2004;9(1):13–17. doi: 10.1016/j.tplants.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 132.Reyes J. L., Rodrigo M. J., Cokmenero-Flores J. M., et al. Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environment. 2005;28(6):709–718. doi: 10.1111/j.1365-3040.2005.01317.x. [DOI] [Google Scholar]
- 133.Goyal K., Walton L. J., Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochemistry Journal. 2005;388(1):151–157. doi: 10.1042/bj20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stacy R. A. P., Aalen R. B. Identification of sequence homology between the internal hydrophilic repeated motifs of group 1 late-embryogenesis abundant proteins in plants and hydrophilic repeats of the general stress protein GsiB of Bacillus subtilis. Planta. 1998;206(3):476–478. doi: 10.1007/s004250050424. [DOI] [PubMed] [Google Scholar]
- 135.Garay A. A., Flores C. J. M., Garciarrubio A., Covarrubias A. A. Highly hdrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. Journal of Biological Chemistry. 2000;275(8):5668–5674. doi: 10.1074/jbc.275.8.5668. [DOI] [PubMed] [Google Scholar]
- 136.Solomon A., Salomon R., Paperna I., Glazer I. Desiccation stress of ento-mopathogenic nematodes induces the accumulation of a novel heat stable protein. Parasitology. 2000;121(4):409–416. doi: 10.1017/s0031182099006563. [DOI] [PubMed] [Google Scholar]
- 137.Browne J. A., Dolan K. M., Tyson T., Goyal K., Tunnacliffe A., Burnell A. M. Dehydration specific induction of hydrophilic protein genes in the anhydro biotic nematode Aphelenchus avenae. Eukaryotic Cell. 2004;3(4):966–975. doi: 10.1128/ec.3.4.966-975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mtwisha L., Brandt W., McCready S., Lindsey G. G. HSP 12 is a LEA like protein in Saccharomyces cerevisiae. Plant Molecular Biology. 1998;37(3):513–521. doi: 10.1023/a:1005904219201. [DOI] [PubMed] [Google Scholar]
- 139.Abba S., Ghignone S., Bonfante P. A dehydration-inducible gene in the truffle Tuber borchii identifies a novel group of dehydrins. BMC Genomics. 2006;7:p. 39. doi: 10.1186/1471-2164-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rohrig H., Schmidt J., Colby T., Brautigam A., Hufnagel P., Bartels D. Desiccation of the resurrection plant Craterostigma plantagineum induces dynamic changes in protein phosphorylation. Plant Cell Environment. 2006;29(8):1606–167. doi: 10.1111/j.1365-3040.2006.01537.x. [DOI] [PubMed] [Google Scholar]
- 141.Moore J. P., Westall K. L., Ravenscroft N., et al. The predominant polyphenol in the leaves of the resurrection plant Myrothamnus flabellifolius, 3, 4, 5 tri-O-galloylquinic acid, protects membranes against desiccation and free radical-induced oxidation. Biochemistry Journal. 2005;385(1):301–308. doi: 10.1042/bj20040499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Battaglia M., Carrillo Y. O., Garciaarrubio A., Campos F., Covarrubias A. A. The enigmatic LEA proteins and other hydrophilins. Plant Physiology. 2008;148(1):6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tunnacliffe A., Wise M. J. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94(10):791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- 144.Sivamani E., Bahieldin A., Wraith J. M., et al. Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Science. 2000;155(1):1–9. doi: 10.1016/s0168-9452(99)00247-2. [DOI] [PubMed] [Google Scholar]
- 145.Sunkar R., Chinnusamy V., Zhu J., Zhu J. K. Small RNAs as big players in plant abiotic stress responses. Trends Plant Science. 2007;12(7):301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 146.Phillips J. R., Dalmay T., Bartels D. The role of small RNAs in abiotic stress. FEBS Letters. 2007;581(19):3592–3597. doi: 10.1016/j.febslet.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 147.Furini F., Koncz C., Salamini F., Bartels D. High level transcription of a member of a repeated gene family confers dehydration tolerance to callus tissue of Craterostigma plantagineum. EMBO Journal. 1997;16(12):3599–3608. doi: 10.1093/emboj/16.12.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Leprince O., Buitink J. Desiccation tolerance: from genomics to the field. Plant Science. 2010;179(6):554–564. doi: 10.1016/j.plantsci.2010.02.011. [DOI] [Google Scholar]
- 149.Scott H. B., Oliver M. J. Accumulation and polysomal recruitment of transcripts in response to desiccation and dehydration of the moss Tortula ruralis. Journal of Experimental Botany. 1994;45(5):577–583. doi: 10.1093/jxb/45.5.577. [DOI] [Google Scholar]
- 150.Wood A. J., Oliver M. J. Translational control in plant stress: formation of messenger ribonucleoprotein complexes (mRNPs) in Tortula ruralis in response to desiccation. Plant Journal. 1999;18(4):359–370. doi: 10.1046/j.1365-313x.1999.00458.x. [DOI] [Google Scholar]
- 151.Zeng O., Chen X. B., Wood A. J. Two early light inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. Journal of Experimental Botany. 2002;53(371):1197–1205. doi: 10.1093/jexbot/53.371.1197. [DOI] [PubMed] [Google Scholar]
- 152.Oliver M. J., Dowd S., Zaragoza J., Mauget S., Payton P. The rehydration transcriptome of the desiccation tolerant bryophyte Tortula ruralis: transcript classification and analysis. BMC Genomics. 2004;5(1):p. 89. doi: 10.1186/1471-2164-5-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zentella R., Gallardo J. O., van Dijck P., et al. A Selaginella lepidophylla trehalose-6-phosphate synthase complements growth and stress-tolerance defects in a yeast tsp1 mutant. Plant Physiology. 1999;119(4):1473–1482. doi: 10.1104/pp.119.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Le T. N., Blomstedt C. K., Kuang J., et al. Genes differentially expressed in desiccation tolerant leaf tissue of the resurrection grass Sporobolus stapfianus. Functional Plant Biology. 2007;34(7):589–600. doi: 10.1071/fp06231. [DOI] [PubMed] [Google Scholar]
- 155.Mundree S. G., Whittaker A., Thomson J. A., Farrant J. M. An aldose reductase homolog from the resurrection plant Xerophyta viscosa Baker. Planta. 2000;211(5):693–700. doi: 10.1007/s004250000331. [DOI] [PubMed] [Google Scholar]
- 156.Mowla S. B., Thomson J. A., Farrant J. M., Mundree S. G. A novel stress inducible antioxidant enzyme identified from the resurrection plant Xerophyta viscosa Baker. Planta. 2002;215(5):716–726. doi: 10.1007/s00425-002-0819-0. [DOI] [PubMed] [Google Scholar]
- 157.Lehner A., Chopera D., Peter S., et al. Protection mechanisms in the resurrection plant Xerophyta viscosa: cloning, expression, characterisation and role of XvINO1, a gene coding for a myo-inositol 1-phosphate synthase. Functional Plant Biology. 2008;35(1):26–39. doi: 10.1071/fp07142. [DOI] [PubMed] [Google Scholar]
- 158.Mulako I., Farrant J. M., Collett H., Illing N. Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is up-regulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) Dur and Schinz. Journal of Experimental Botany. 2008;59(14):3885–3901. doi: 10.1093/jxb/ern226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Collett H. M., Shen A., Gardner M., Farrant J. M., Denby K. J., Illing N. Towards transcript profiling of desiccation tolerance in Xerophyta humilis: construction of a normalized 11 k X. humilis cDNA set and microarray expression analysis of 424 cDNAs in response to dehydration. Physiologia Plantarum. 2004;122(1):39–53. doi: 10.1111/j.1399-3054.2004.00381.x. [DOI] [Google Scholar]
- 160.Bockel C., Salamini F., Bartels D. Isolation and characterization of genes expressed during early events of the dehydration process in the resurrection plant Craterostigma plantagineum. Journal of Plant Physiology. 1998;152(2-3):158–166. doi: 10.1016/s0176-1617(98)80127-2. [DOI] [Google Scholar]
- 161.Gechev T. S., Benina M., Obata T., et al. Molecular mechanisms of desiccation tolerance in the resurrection glacial relic Haberlea rhodopensis. Cellular and Molecular Life Sciences. 2013;70(4):689–709. doi: 10.1007/s00018-012-1155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Oliver M. J., Jain R., Balbuena T. S., Agrawal G., Gasulla F., Thelen J. J. Proteome analysis of leaves of the desiccation tolerant grass, Sporobolus stapfianus, in response to dehydration. Phytochemistry. 2011;72(10):1273–1284. doi: 10.1016/j.phytochem.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 163.Michel D., Furini A., Salamini F., Bartels D. Structure and regulation of an ABA-and desiccation responsive gene from the resurrection plant Craterostigma plantagineum. Plant Molecular Biology. 1994;24(4):549–560. doi: 10.1007/bf00023553. [DOI] [PubMed] [Google Scholar]
- 164.Velasco R., Salamini F., Bartels D. Dehydration and ABA increase mRNA levels and enzyme activity of cytosoli GAPDH in the resurrection plant Craterostigma plantagineum. Plant Molecular Biology. 1994;26(1):541–546. doi: 10.1007/bf00039567. [DOI] [PubMed] [Google Scholar]
- 165.Alamillo J. M., Bartels D. Light and stage of development influence the expression of desiccation induced genes in the resurrection plant Craterostigma plantagineum. Plant Cell Environment. 1996;19(3):300–310. doi: 10.1111/j.1365-3040.1996.tb00252.x. [DOI] [Google Scholar]
- 166.Ndimba T., Farrant J. M., Thomson J., Mundree S. Molecular characterization of XVT8, a stress-responsive gene from the resurrection plant Xerophyta viscosa Baker. Plant Growth Regulation. 2001;35(2):137–145. [Google Scholar]
- 167.Goday A., Martinez S. D., Gómez J., Puigdomènech P., Pagès M. Gene expression in developing Zea mays embryos. Regulation by abscisic acid of a highly phosphorylated 23- to 25-kD group of proteins. Plant Physiology. 1988;88(3):564–569. doi: 10.1104/pp.88.3.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Blomstedt C. K., Gianello R. D., Gaff D. F., Hamill J. D., Neale A. D. Differential gene expression in desiccation tolerant and desiccation sensitive tissue of the resurrection grass, Sporobolus stapfianus. Australian Journal of Plant Physiology. 1998;25(8):937–946. doi: 10.1071/pp98113. [DOI] [Google Scholar]
- 169.Marais S., Thomson J. A., Farrant J. M., Mundree S. G. VATP1XV–a novel stress responsive V-ATPase subunit c homologue isolated from the resurrection plant Xerophyta viscosa Baker. Physiologia Plantarum. 2004;122(1):54–61. doi: 10.1111/j.1399-3054.2004.00389.x. [DOI] [Google Scholar]
- 170.Oliver M. J., Guo L., Alexander D., Ryals J., Wone B., Cushman J. A sister group metabolomic contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfinus. Plant Cell. 2011;23(4):1231–1248. doi: 10.1105/tpc.110.082800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yobi A., Wone B. W. M., Xu W., et al. Comparative metabolic profiling between desiccation sensitive and desiccation tolerant species of Selaginella reveals insights into the resurrection trait. Plant Journal. 2012;72(6):983–999. doi: 10.1111/tpj.12008. [DOI] [PubMed] [Google Scholar]
- 172.Leopold A. C., Vertucci C. W. Physical attributes of desiccated seeds. In: Leopold A. C., editor. Membranes, metabolism and Dry Organisms. New York, NY, USA: Cornell University Press; 1986. pp. 22–34. [Google Scholar]
- 173.Vertucci C. W., Farrant J. M. Acquisition and loss of desiccation- tolerance. In: Kigel J., Galili G., editors. Seed Development and Germination. New York, NY, USA: Marcel Dekker; 1995. pp. 237–0271. [Google Scholar]
- 174.Hartung W., Schiller P., Karl-Josef D. Physiology of poikilohydric plants. Progress in Botany. 1998;59:299–327. doi: 10.1007/978-3-642-80446-5_11. [DOI] [Google Scholar]
- 175.Oliver M. J., Murdock A. G., Mishler B. D., et al. Chloroplast genome sequence of the moss Tortula ruralis: gene content, polymorphism, and structural arrangement relative to other green plant chloroplast genomes. BMC Genomics. 2010;11(1):p. 143. doi: 10.1186/1471-2164-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yobi A., Wone B. W., Xu W., et al. Metabolomic profiling in Selaginella lepidophylla at various hydration states provides new insights into the mechanistic basis of desiccation tolerance. Molecular Plant. 2013;6(2):369–385. doi: 10.1093/mp/sss155. [DOI] [PubMed] [Google Scholar]
- 177.Gasulla F., Vom Dorp K., Dombrink I., et al. The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: a comparative approach. Plant Journal. 2013;75(5):726–741. doi: 10.1111/tpj.12241. [DOI] [PubMed] [Google Scholar]
- 178.Alcázar R., Altabella T., Marco F., et al. Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta. 2010;231(6):1237–1249. doi: 10.1007/s00425-010-1130-0. [DOI] [PubMed] [Google Scholar]
- 179.Alcázar R., Bitrián M., Bartels D., Koncz C., Altabella T., Tiburcio A. F. Polyamine metabolic canalization in response to drought stress in Arabidopsis and the resurrection plant Craterostigma plantagineum. Plant Signaling and Behaviour. 2011;6(2):243–250. doi: 10.4161/psb.6.2.14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Appels R., Nystrom-Persson J., Keeble-Gagnere G. Advances in genome studies in plants and animals. Functional & Integrative Genomics. 2014;14(1):1–9. doi: 10.1007/s10142-014-0364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Giarola V., Hou Q., Bartels D. Angiosperm plant desiccation tolerance: hints from transcriptomics and genome sequencing. Trends Plant Science. 2017;22(8):705–717. doi: 10.1016/j.tplants.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 182.Bennett M. D., Leitch I. J. Nuclear DNA amounts in angiosperms: targets trends and tomorrow. Annuals of Botany. 2011;107(3):467–590. doi: 10.1093/aob/mcq258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Costa M. D., Artur M. A., Maia J., et al. A footprint of desiccation tolerance in the genome of Xerophyta viscosa. Nature Plants. 2017;3(4):p. 17038. doi: 10.1038/nplants.2017.38. [DOI] [PubMed] [Google Scholar]
- 184.Xiao L., Yang G., Zhang L., et al. The resurrection genome of Boea hygrometrica: a blueprint for survival of dehydration. Proceedings of the National Academy of Sciences. 2015;112(18):5833–5837. doi: 10.1073/pnas.1505811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Buren V. R., Bryant D., Edger P. P., et al. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature. 2015;527:508–511. doi: 10.1038/nature15714. [DOI] [PubMed] [Google Scholar]
- 186.Schneider K., Wells B., Schmelzer E., Salamini F., Bartels D. Desiccation leads to the rapid accumulation of both cytosolic and chloroplastic proteins in the resurrection plant Craterostigma plantagineum Hochst. Planta. 1993;189(1):120–131. doi: 10.1007/bf00201352. [DOI] [Google Scholar]
- 187.Ekmekci Y., Bohms A., Thomson J. A., Mundree S. G. Photochemical and antioxidant responses in the leaves of Xerophyta viscosa Baker and Digitaria sanguinalis L. under water deficit. Zeitschrift für Naturforschung C. 2005;60(5-6):435–443. doi: 10.1515/znc-2005-5-612. [DOI] [PubMed] [Google Scholar]
- 188.Cho S. K., Kim J. E., Park J.-A., et al. Constitutive expression of a biotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Letters. 2006;580(13):3136–3144. doi: 10.1016/j.febslet.2006.04.062. [DOI] [PubMed] [Google Scholar]
- 189.Majeran W., Zybailov B., Ytterberg A. J., Dunsmore J., Sun Q., Wijk V. K. J. Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Molecular Cell Proteomics. 2008;7(9):1609–1638. doi: 10.1074/mcp.m800016-mcp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Turck C. W., Falick A. M., Kowalak J. A., et al. The association of biomolecular resource facilities proteomics research group 2006 study: relative protein quantitation. Molecular Cell Proteomics. 2007;6(8):1291–1298. doi: 10.1074/mcp.m700165-mcp200. [DOI] [PubMed] [Google Scholar]
- 191.Bernacchia G., Schwall G., Lottspeich F., Salamini F., Bartels D. The transketolase gene fily of the resurrection plant Craterostigma plantagineum: differential expression during the rehydration phase. EMBO Journal. 1995;14(3):610–618. doi: 10.1002/j.1460-2075.1995.tb07037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhu Y., Wang B., Phillips J., et al. Global transcriptome analysis reveals acclimation-primed processes involved in the acquisition of desiccation tolerance in Boea hygrometrica. Plant Cell Physiology. 2015;56(7):1429–1441. doi: 10.1093/pcp/pcv059. [DOI] [PubMed] [Google Scholar]