Abstract
miR-124-3p has been implicated in a variety of cancers. The purpose of the present study was to investigate the expression, prognostic roles and functions of miR-124-3p in gastric cancer. Functional studies indicated that ectopic overexpression of miR-124-3p in gastric cancer cells suppressed cell viability and plate colony formation in vitro and tumor growth in vivo. In situ hybridization analysis demonstrated that decreased expression of miR-124-3p was associated with clinical stage and lymph node metastasis, as well as shorter overall survival and disease-free survival rates. Furthermore, it was observed that miR-124-3p repressed the carcinogenesis of gastric cancer by targeting Ras-related C3 botulinum toxin substrate 1 (Rac1) and specificity protein 1 (SP1). Collectively, these results indicate a potential underlying mechanism for the regulation of gastric cancer by miR-124-3p involving targeting of Rac1 and SP1. Thus, miR-124-3p may be an independent indicator of survival and treatment strategy for patients with gastric cancer.
Keywords: microRNA-124-3p, gastric cancer, invasion, metastasis
Introduction
microRNAs (miRNA/miRs) are endogenously expressed, single-stranded RNAs of approximately 22 nucleotides in length that serve pivotal roles in regulating the expression of target genes post-transcriptionally. miRNAs are involved in critical cellular processes including cell development, proliferation, differentiation and apoptosis (1). Previous results have demonstrated that miRNAs are frequently abnormally expressed in various cancers, and function as oncogenes or tumor suppressors in tumorigenesis and tumor progression (2,3). For instance, the oncogenic miR-21 is commonly overexpressed in gastric cancer and inhibits the tumor suppressors phosphatase and tensin homolog and programmed cell death protein 4 to promote gastric cancer growth and invasion (4).
More recently, studies have demonstrated that miR-124 was downregulated in various cancers, including breast (5), colon (6), prostate (7), stomach (8), liver (9), ovarian (10) and brain (11) cancers. For instance, miR-124 was decreased in breast cancer cell lines and patient specimens; additionally, it was indicated to attenuate cell growth and migration by targeting flotillin-1 (5). miR-124 has also been reported to regulate invasion and metastasis in hepatocellular carcinoma through regulation of rho associated coiled-coil containing protein kinase 2 and enhancer of zeste homolog 2 (EZH2) (9). Furthermore, Kang et al (7), documented that miR-124 may suppress growth and cell invasion in prostate cancer through the proprotein convertase subtilisin/kexin type 6 (also known as PACE4) pathway. These data suggest that miR-124 may act as a tumor suppressor. Our group previously demonstrated that miR-124 suppressed gastric cancer cell proliferation and induced apoptosis by directly targeting EZH2 (8). Xia et al (12) reported that miR-124 inhibited cell proliferation through downregulation of sphingosine kinase 1 in gastric cancer. To date, however, the biological impacts of miR-124 on gastric cancer cell metastasis and the corresponding molecular mechanisms have seldom been investigated.
In the present study, the expression of miR-124 in gastric cancer and its association with clinical parameters were investigated. Furthermore, the association of miR-124 with overall survival (OS) and disease-free survival (DFS) in patients with gastric cancer was evaluated. Functional studies were also performed to investigate the function of miR-124 in gastric cancer cells in vitro and in vivo. Additionally, the potential targeting action of miR-124-3p against Ras-related C3 botulinum toxin substrate 1 (Rac1) and specificity protein 1 (SP1) was assessed. These experiments aimed to indicate the underlying mechanism for the regulation of gastric cancer by miR-124-3p via targeting of Rac1 and SP1.
Materials and methods
Cell culture
The gastric cancer cell lines SGC-7901 and MKN-28 were obtained from the American Type Culture Collection (Manassas, VA, USA). The MKN-28 cell line is a derivative of the MKN-74 gastric tubular adenocarcinoma cell line (13). The cell lines used in the present experiments were reauthenticated by Beijing Microread Genetics Co., Ltd. (Beijing, China) via short tandem repeat profiles analysis every 6 months following resuscitation. The cells were maintained at 37°C in an atmosphere of 5% CO2 in RPMI-1640 medium supplemented with 10% fetal bovine serum, 10,00 U/ml penicillin and 10,000 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Clinical samples
All tissue samples used in the present study were collected from the Hunan Provincial Tumor Hospital (Changsha, China). Written informed consent was obtained from all study participants, and all procedures involving human participants were approved by the Ethics Committee of the University of South China Health Authority (Hengyang, China). The collection and use of tissues followed procedures in accordance with the ethical standards in the Declaration of Helsinki. The tissue microarrays (TMAs) consisted of 121 cases of gastric carcinoma tissues diagnosed by histopathological diagnosis. Specimens were obtained during surgery, fixed in 10% formalin for 24 h at room temperature, embedded in paraffin and cut into sections (4-µm thickness), and stored in the Hunan Provincial Tumor Hospital. The TMAs were used for immunohistochemistry (IHC) and in situ hybridization (ISH) analyses. The sample data, including age, sex, histological grade, tumor size, invasion depth (T stage) and lymph node metastasis, were obtained from clinical and pathological records. Furthermore, following reverse transcription-quantitative polymerase chain reaction (RT-qPCR), the clinical samples were divided into low expression and high expression groups based on miR-124-3p expression scores greater or less than 2 (14).
RNA isolation and RT-qPCR
Total RNA from the gastric cancer cell lines was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and 1 µg of total RNA was reverse transcribed using a Reverse Transcription System kit (Promega Corporation, Madison, WI, USA). RT-qPCR was performed using a Power SYBR-Green PCR Master Mix (Thermo Fisher Scientific, Inc.), with human β-actin amplified as an internal control. For miRNA analysis, reverse transcription and RT-qPCR were performed using an all-in-one miRNA qRT-PCR Detection kit (GeneCopoeia, Inc., Rockville, MD, USA) and U6 small nuclear RNA was used as an endogenous control. For the miRNA and mRNA amplifications, the PCR cycling conditions were as follows: One cycle at 95°C for 3 min, followed by 40 cycles at 95°C for 12 sec and 62°C for 35 sec, and finally 1 cycle at 62-95°C for 15 sec. The relative fold-changes in expression with respect to the endogenous controls were calculated by the 2−ΔΔCq method (15). The primers for SP1, Rac1, U6 and β-actin, synthesized by Invitrogen (Thermo Fisher Scientific, Inc.), were as follows: For SP1, forward, 5′-TGCCTCCACTTCCTCGATTT-3′ and reverse, 5′-TCTGGTGGGCAGTATGTTGT-3′; for U6, forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′; and for β-actin, forward, 5′-AGCGAGCATCCCCCAAAGTT-3′; and reverse, 5′-GGGCACGAAGGCTCATCATT-3′.
Cell viability assay
SGC-7901 and MKN-28 cells were transfected with 50 nM of miR-124 mimics (miR-124: 5′-UAAGGCACGCGGUGAAUGCCAA-3′), miR-124 inhibitors (miR-124-LNA: 5′-UUGGCAUUCACCGCGUGCCUUA-3′) or their scrambled oligonucleotide controls (miR-ctr: 5′-UUCUCCGAACGUGUCACGUTT-3′; and miR-LNA: 5′-CAGUACUUUUGUGUAGUACAA-3′, respectively; Shanghai Jima Industrial Co., Ltd., Shanghai, China) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 6 h, the transfected cells were seeded in 96-well plates at a density of 1,000 cells/well. An MTS assay (Promega Corporation) was performed according to the manufacturer's instructions after 0 and 2 days of incubation at 37°C. Absorbance values were examined at 490 nm using a Spectra Max 250 spectrophotometer (Molecular Devices, LLC, Sunnyvale, CA, USA).
IHC and ISH analysis
For baseline staining, the sections were washed with distilled water, and the cell nuclei were stained with hematoxylin. The sections were then rinsed in running tap water, differentiated with 0.3% acid alcohol, and rinsed again in running tap water. After rinsing in Scott's tap water substitute followed by tap water, the sections were stained with eosin for 2 min. Finally, the sections were dehydrated, cleared and mounted.
IHC and ISH were performed according to standard procedures (14)
For IHC, randomly selected human gastric cancer TMA sections from all patients were incubated with anti-Rac1 (sc-95; 1:500) and anti-SP1 (sc-17824; 1:500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) antibodies at 4°C overnight, then with horseradish peroxidase (HRP)-conjugated rabbit anti-mouse (BA1058; 1:5,000) and HRP-goat anti-rabbit (BA1058; 1:5,000; Boster Biological Technology, Pleasanton, CA, USA) antibodies at room temperature for 1 h. miR-124-3p miRCURY LNA™ custom detection probes (Exiqon A/S, Vedbaek, Denmark) were used for ISH; the 5′-3′ sequences (enhanced with LNA) were UAAGGCACGCGGUGAAUGCC, with a digoxigenin label at both the 5′ and 3′ ends. Hybridization, washing and scanning was performed according to the manufacturer's instructions. The slides were analyzed under a light microscope (Nikon Eclipse 80i; Nikon Corporation, Tokyo, Japan) at ×400 magnification by two independent pathologists.
Western blot analysis
Western blot analysis was performed using standard procedures. Briefly, total protein from the gastric cancer cell lines was extracted using radioimmunoprecipitation assay lysis buffer with a proteinase inhibitor (P0013B; Beyotime Institute of Biotechnology, Haimen, China). The protein concentrations in the lysates were measured with a Protein BCA Assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Subsequently, 30 µg protein per lane was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to PVDF (polyvinylidene difluoride) membranes. To block nonspecific binding, the membranes were incubated with 5% skimmed milk powder in phosphate-buffered saline at room temperature for 1 h. The membranes were then incubated with primary antibodies overnight at 4°C, followed by horseradish peroxidase-labeled secondary antibodies at room temperature for 1 h, and protein bands were detected by electrochemiluminescence (ECL) with ECL western blotting detection reagents (New England BioLabs, Inc., Ipswich, MA, USA). The antibodies and their dilutions were the same as those used for the IHC analysis. β-actin (4967S; 1:500; Cell Signaling Technology, Inc., Danvers, MA, USA) was used as a protein-loading control.
RNA interference assay
SP1 short hairpin (sh)RNA (HSH054522-CH1), Rac1 shRNA (HSH016063-CH1; GeneCopoeia, Inc.) and control shRNA (sc-108080; Santa Cruz Biotechnology, Inc.) lentiviral particles were used for knockdown experiments. SGC-7901 cells were incubated with the lentiviral vectors (40 µg/l) and 5 mg/ml polybrene (Shanghai Yi Sheng Biotechnology Co., Ltd., Shanghai, China) with Lipofectamine 2000 overnight. After 48 h, the cells were harvested for use in cell viability and expression assays.
SP1 and Rac1 vector construction
SP1 and Rac1-expressing vectors were constructed. Full-length SP1 and Rac1 cDNAs were purchased from GeneCopeia, Inc. and were individually subcloned into the eukaryotic expression vector pcDNA3.1(+) (V79020; Invitrogen; Thermo Fisher Scientific, Inc.). pcDNA3.1 (+) was used as a negative control. These vectors (40 nM) were transfected into cells using Lipofectamine 2000 for overexpression experiments. At 24 h after transfection, the cells were harvested and analyzed for cell viability.
Luciferase assays
The sequence of the 3′ untranslated region (UTRs) of the SP1 gene was obtained from the TargetScan database version 7.1 (http://www.targetscan.org/vert_71/). The 3′UTR was amplified by PCR (14) from the genomic DNA of the SGC-7901 cell line and inserted into a pGL3 control vector (Promega Corporation) using the XBA1 site immediately downstream from the stop codon of luciferase. The SP1 primer sets used for PCR were as follows: Forward 5′-CCTTCAGGGATTTCCAACTG-3′ and reverse, 5′-GTCCAAAAGGCATCAGGGTA-3′. A mutant insert was also generated in which the first four nucleotides of the miR-124 binding site the SP1 gene (AUGT-GCAC; predicted by TargetScan) were mutated using a QIAGEN XL-site directed Mutagenesis kit (Qiagen, Inc. Valencia, CA, USA). SGC-7901 and MKN-28 cells were cotransfected by nucleoporation (Amaxa Nucleofector™; Lonza Group, Ltd., Basel, Switzerland) (16) with 5 µg firefly luciferase reporter vector (Promega Corporation) and 0.5 μg control vector containing Renilla luciferase (pRL-TK; Promega Corporation). For each nucleoporation, 50 nM of the miR-124-3p mimic, miR-124-LNA (inhibitor), miR-ctr or miR-LNA was used. Firefly and Renilla luciferase activities were measured consecutively using a dual luciferase assay (Promega Corporation) at 48 h after transfection.
Mouse xenograft model
A gastric cancer model in male BALC/c mice (Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) was established. A total of 12 mice (6 weeks old; 20-22 g) were used, which were housed under controlled conditions at 25°C, 60% humidity and a 12-h light/dark cycle, with food and water available ad libitum. A total of 5×105 SGC-7901 cells were inoculated subcutaneously into the dorsal flanks of the mice. After 10 days of tumor growth, the tumors were infected with the miR-124 mimic, miR-124-LNA or their controls (n=3 mice per group). Tumor size was measured every 4 days. The maximum tumor size permitted was 1.2 cm3. The mice were sacrificed and necropsies were performed after 28 days, and the tumors were isolated and weighed. Additionally, the tumor volumes were calculated according to the formula: AxB2/2 (A: the largest diameter, B: the diameter perpendicular to A). All of the animal procedures were performed in accordance with the Animal Research: Reporting of In Vivo Experiments guidelines and National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978). Ethical approval was obtained for the animal experiments from the Ethics Committee of the University of South China Health Authority. All possible steps were taken to avoid animal suffering at each stage of the experiment.
Statistical analysis
Comparisons between groups were performed with Student's t-test and χ2 test. OS and DFS curves were plotted for the patients according to the Kaplan-Meier method, with the log-rank test used for comparison. Survival was recorded from the day of the surgery in which specimens were extracted. Differences were considered statistically significant at P<0.05. The statistical analyses were performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA).
Results
miR-124-3p affects cell viability and plate colony formation in vitro and tumor growth in vivo in gastric cancer cells
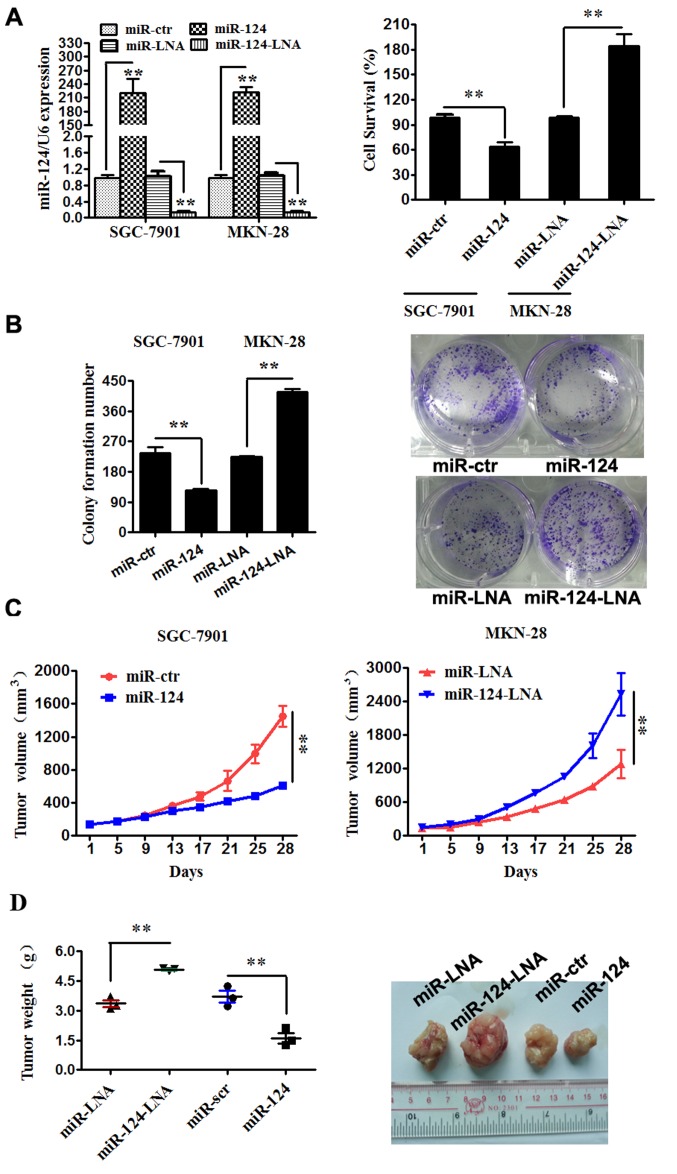
Previous studies demonstrated that miR-124-3p was downregulated in gastric cancer tissues and cell lines, particularly in SGC-7901 cells, while it was relatively highly expressed in MKN-28 cells. Therefore, the present study transfected SGC-7901 cells with miR-124-3p mimics (miR-124) and MKN-28 cells with miR-124-3p inhibitor (miR-124-LNA) to investigate the changes in cell viability and colony formation abilities in vitro and tumor growth abilities in nude mice.
Firstly, SGC-7901 and MKN-28 cells were respectively transfected with miR-124-3p mimics, miR-124-3p inhibitor and the corresponding controls, and the transfection was determined to be successful (Fig. 1A). The results of subsequent assays indicated that miR-124-3p significantly suppressed SGC-7901 cell viability (P<0.01; Fig. 1A), plate colony formation (P<0.01; Fig. 1B) and tumor growth in nude mice (P<0.01; Fig. 1C and D). By contrast, miR-124-3p inhibitor promoted MKN-28 cell proliferation (P<0.01; Fig. 1A), plate colony formation (P<0.01; Fig. 1B) and tumor growth in nude mice (P<0.01; Fig. 1C and D).
Figure 1.
miR-124-3p expression in gastric cancer cells affects proliferation and plate colony formation in vitro and tumor growth in vivo. (A) SGC-7901 and MKN-28 cells were transfected with miR-124 mimics, miR-124 inhibitors or their scrambled oligonucleotides controls. An MTS assay was used to analyze the viability of the SGC-7901 and MKN-28 cells. **P<0.01. (B) SGC-7901 and MKN-28 cells were transfected with miR-124 mimics, miR-124 inhibitors or their controls, and plate colony formation abilities of the cell lines were analyzed. (C) Tumor growth in mouse xenograft models. SGC-7901 and MKN-28 cells transfected with miR-124, miR-124 inhibitor or their controls were injected subcutaneously into BALC/c mice (n=3 per group). Tumor size was measured every 4 days. All data (A-C) are presented as means ± standard error of the mean. (D) After 28 days, all mice were sacrificed and dissected, and the tumors were weighed; data are presented as the median and range. **P<0.01 as indicated. miR, microRNA; -LNA, inhibitor; -ctr, control.
miR-124-3p is downregulated and correlates with advanced clinical stage, lymph node metastases and poor clinical outcomes in gastric cancer
ISH was performed to evaluate miR-124-3p levels in 121 gastric tumors with a TMA. The clinical samples were divided into low expression and high expression groups based on miR-124-3p expression scores greater or less than 2 (14). The results indicated that miR-124-3p was markedly downregulated in stomach tumors, with low miR-124-3p expression determined in 57.9% (70/121) of the specimens. Subsequently, the potential clinicopathological implications of altered miR-124-3p expression were determined. It was observed that miR-124-3p expression was inversely associated with histological grade, TNM stage and lymph node metastasis (P=0.037, P=0.005 and P=0.002, respectively). However, no significant associations between miR-124-3p expression and age, gender or T stage were identified (Table I).
Table I.
Analysis of the association between miR-124 expression and clinicopathological parameters in primary gastric cancer.
| miR-124 expression | ||||
|---|---|---|---|---|
| Variable | Cases, n | Low | High | P-value |
| Age, years | ||||
| <60 | 69 | 40 | 29 | 1.000 |
| ≥60 | 52 | 30 | 22 | |
| Gender | ||||
| Male | 65 | 41 | 24 | 0.268 |
| Female | 56 | 29 | 27 | |
| Histological grade (differentiation) | ||||
| Well and moderate | 31 | 23 | 8 | 0.037 |
| Poor and other | 90 | 47 | 43 | |
| T stage | ||||
| T1-T2 | 66 | 34 | 32 | 0.142 |
| T3-T4 | 55 | 36 | 19 | |
| TNM stage | ||||
| I–II | 46 | 19 | 27 | 0.005 |
| III–IV | 75 | 51 | 24 | |
| Lymph node metastasis | ||||
| Present | 88 | 59 | 29 | 0.002 |
| Absent | 33 | 11 | 22 | |
Clinicopathological features were analyzed by χ2 test.
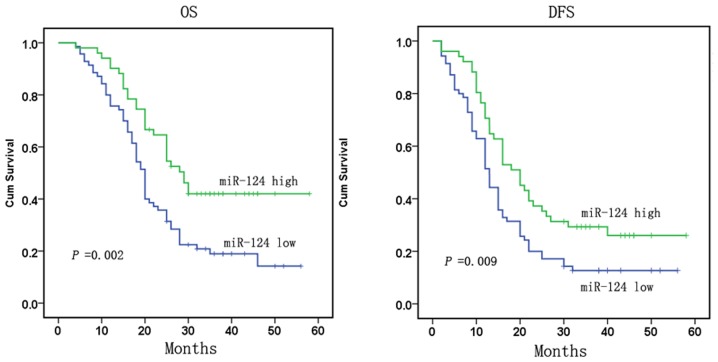
To analyze the significance of miR-124-3p in terms of clinical prognosis, Kaplan-Meier survival analysis was performed based on patient OS and DFS rates
The results demonstrated that patients with low miR-124-3p expression had lower mean OS and DFS rates compared with patients with high miR-124-3p expression (P=0.002 for OS and P=0.009 for DFS; Fig. 2). Collectively these results suggest that miR-124-3p may serve critical roles in the carcinogenesis and progression of gastric cancer.
Figure 2.
miR-124-3p is downregulated and associated with advanced clinical stage, lymph node metastases and poor clinical outcomes in gastric cancer. Low expression levels of miR-124-3p correlated with shorter survival; OS and DFS curves for 121 studied patients with high or low miR-124-3p expression are shown. miR, microRNA; OS, overall survival; DFS, disease-free survival.
SP1 and Rac1 are direct targets of miR-124-3p
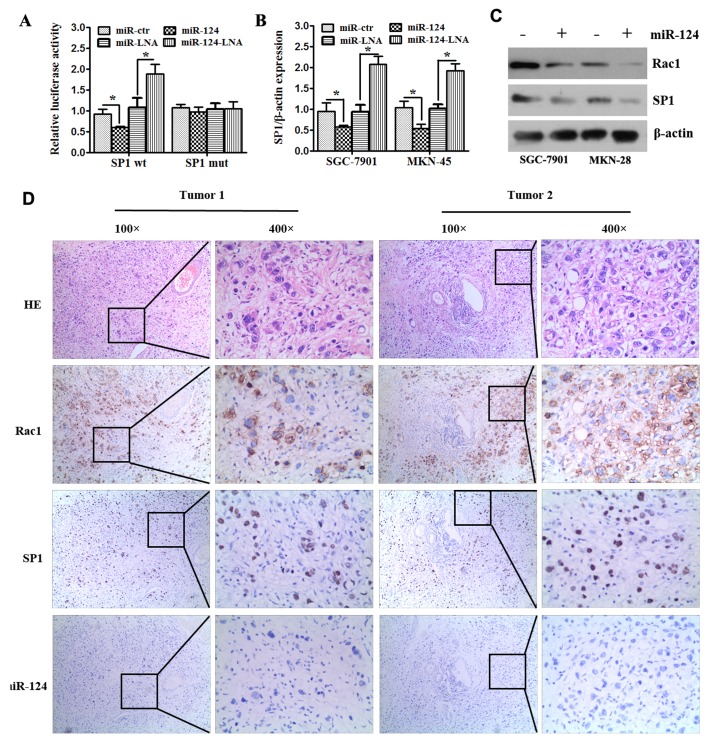
To investigate the molecular mechanism of miR-124-3p in gastric cancer, the TargetScan algorithm (http://www.targetscan.org/vert_71/) was used to search for putative protein-coding gene targets of miR-124, which indicated that SP1 and Rac1 are putative targets of miR-124-3p. Rac1 has previously been identified as a target of miR-124 (17). To ascertain whether SP1 is a direct target of miR-124-3p, luciferase reporter assays were conducted. The full-length SP1 3′-UTR was cloned downstream of the firefly luciferase gene and cotransfected with miR-124-3p mimics, miR-124 inhibitor or their negative controls, and luciferase activity was measured at 48 h post-transfection. SGC-7901 cells cotransfected with SP1 reporter constructs and miR-124-3p exhibited an approximate 44% reduction in luciferase activity with respect to those cotransfected with the scrambled oligonucleotide control (P<0.05). Conversely, miR-124-LNA increased luciferase responsiveness compared with its control (P<0.05). In turn, mutation of the putative miR-124-3p sites in the 3′-UTR of SP1 reduced luciferase responsiveness to miR-124 (Fig. 3A).
Figure 3.
SP1 and Rac1 are direct targets of miR-124-3p. (A) Luciferase assay of SGC-7901 and MKN-28 cells cotransfected with miR-124 mimics, miR-124 inhibitor or corresponding controls and a luciferase reporter containing SP1 3′-untranslated region (SP1-wt) or mutant constructs in which the first four nucleotides of the miR-124 binding site were mutated (SP1-mut). (B) SGC-7901 and MKN-28 cells were transfected with miR-124 mimics, miR-124 inhibitor or controls; miR-124 overexpression inhibited the mRNA expression of SP1. All data are presented as means ± SEM. *P<0.05. (C) SGC-7901 and MKN-28 cells were transfected with miR-124 or scramble; miR-124 overexpression inhibited the protein expression of SP1 and Rac1. (D) Gastric cancer specimens were analyzed by immunohistochemistry and in situ hybridization staining. Representative images of miR-124-3p, SP1 and Rac1 expression and H&E staining are shown (magnification, ×100, ×400). SP1, specificity protein 1; Rac1, Ras-related C3 botulinum toxin substrate 1; miR, microRNA; -wt, wild type; -mut; mutant; -LNA, inhibitor; -ctr, control; H&E, hematoxylin and eosin.
To confirm SP1 and Rac1 as targets of miR-124, miR-124-3p mimics, miR-124 inhibitor or their scramble controls were transfected into SGC-7901 and MKN-28 cells, and RT-qPCR and western blot analysis were performed to detect the expression of SP1 and Rac1. Following transfection with miR-124 mimics, the results indicated a significant reduction in the mRNA levels of SP1 in SGC-7901 and MKN-28 cells (P<0.05; Fig. 3B), and a marked reduction in the protein levels of SP1 and Rac1 in SGC-7901 and MKN-28 cells (Fig. 3C). By contrast, following transfection with miR-124 inhibitor, the results indicated a significant increase in the mRNA levels of SP1 in SGC-7901 and MKN-28 cells (P<0.05; Fig. 3B). Collectively these results indicate that miR-124-3p downregulates SP1, and likely Rac1, by targeting their mRNA 3′UTR. Additionally, when investigating the clinical relevance of miR-124-3p and its target genes SP1 and Rac1 in gastric cancer tissues by IHC, it was observed that miR-124-3p expression was negatively associated with SP1 and Rac1 expression in randomly selected gastric cancer sections (Fig. 3D).
miR-124-3p represses carcinogenesis in gastric cancer by targeting SP1 and Rac1
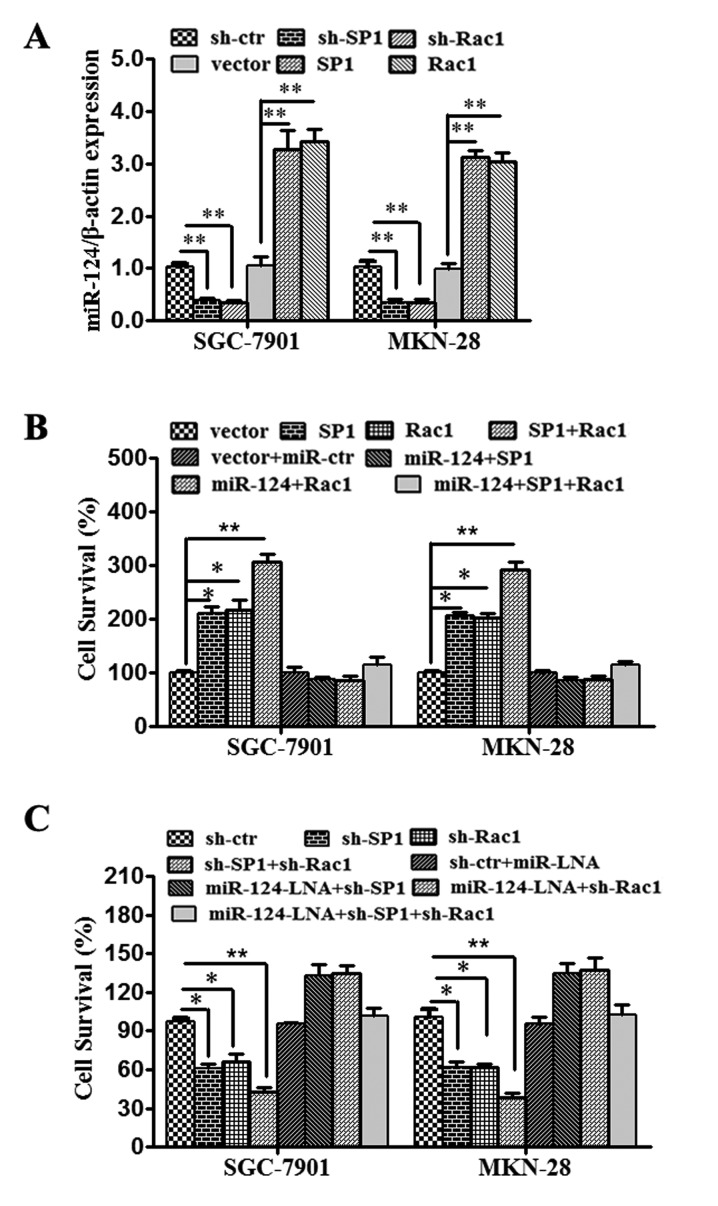
When investigating the functional effect of miR-124-3p and SP1/Rac1 on gastric cancer cells, the previous assays indicated that upregulation of miR-124-3p inhibited the viability of SGC-7901 and MKN-28 cells through functional downregulation of SP1 or/and Rac1 expression. Knockdown and rescue experiments were subsequently performed, in which SGC-7901 and MKN-28 cells were transfected with sh-ctr, sh-SP1, sh-Rac1, vector, SP1 vector or Rac1 vector, and the effects on miR-124 expression confirmed by RT-qPCR (Fig. 4A). As depicted in Fig. 4B, the rate of cell survival was considerably lower in cells transfected with miR-124-3p mimics and SP1 and/or Rac1 vector compared with respective controls. By contrast, downregulation of miR-124-3p by miR-124-LNA or upregulation of SP1 and/or Rac1 by overexpression vectors significantly increased cell survival rate compared with the controls (Fig. 4B and C; P<0.05). These results indicated that transfection of miR-124-3p and knockdown of SP1 and/or Rac1 significantly suppressed the growth of gastric cancer cells in vitro.
Figure 4.
miR-124-3p represses carcinogenesis in gastric cancer by targeting SP1 and Rac1. (A) SGC-7901 and MKN-28 cells were respectively transfected with sh-ctr, sh-SP1, sh-Rac1, vector, SP1 vector or Rac1 vector; all the transfections were successful. (B) SGC-7901 and MKN-28 cells were respectively transfected with vector, SP1, Rac1, Sp1+Rac1, vector+miR-ctr, miR-124+SP1, miR-124+Rac1 or miR-124+SP1+Rac1, and the viability of the cells was analyzed. (C) SGC-7901 and MKN-28 cells were respectively transfected with sh-ctr, sh-SP1, sh-Rac1, sh-Sp1+sh-Rac1, sh-ctr+miR-LNA, miR-124-LNA+sh-SP1, miR-124-LNA+sh-Rac1 or miR-124-LNA+sh-SP1+sh-Rac1, and the viability of the cells was analyzed. All data are presented as means ± SEM. **P<0.01, *P<0.05. SP1, specificity protein 1; Rac1, Ras-related C3 botulinum toxin substrate 1; miR, microRNA; sh-, short hairpin; -LNA, inhibitor; -ctr, control.
Discussion
miR-124-3p commonly serves integral roles in cell growth, apoptosis, invasion and metastasis, and serves as a tumor suppressor in various cancers (5,8,9). In the present study, the downregulation of miR-124-3p was indicated to be a frequent event in gastric cancer. Furthermore, low-level expression of miR-124-3p was significantly associated with clinical stage and lymph node metastases. Accordingly, Kaplan-Meier survival analysis indicated that patients whose primary tumors exhibited low-level expression of miR-124-3p had shorter OS and RFS rates. In previous studies by our group, it was observed that miRs-200b, −200c and −26a may be prognostic indicators in gastric cancer (14,18). Here, miR-124-3p was indicated as a useful prognostic marker for predicting survival and relapse in patients with gastric cancer.
While the initial results indicated that miR-124-3p acted as a tumor suppressor in gastric cancer, the underlying molecular mechanisms were unknown. Therefore, Rac1 and SP1 were assessed are potential downstream target genes of miR-124. miR-124-3p was determined to bind to complementary sites in the 3′-UTRs of Rac1 and SP1, and induce a significant reduction in the expression levels of Rac1 and SP1. Rac1 is an established GTPase that belongs to the RAS superfamily of small GTP-binding proteins (19). Previous studies have reported that Rac1 expression was frequently increased in colon, breast and gastric cancers, medulloblastoma and lung cancer (20–24). Results also support that Rac1 is implicated in cytoskeletal rearrangements and in the regulation of multiple cancer-associated cellular phenotypes, including cell growth, invasion and metastasis, as well as angiogenesis (25–27). Vader et al (28) demonstrated that silencing of Rac1 in vascular endothelial cells inhibited vascular endothelial growth factor-mediated tube formation as well as endothelial cell migration, invasion and proliferation in vitro. In addition, Yoshida et al (25) observed that selective inhibition of Rac1 activity by the inhibitor NSC23766 suppressed cell growth and induced apoptosis in different breast cancer cell lines without being toxic to normal mammary epithelial cells. SP1, a zinc finger transcription factor, has been reported as upregulated in cervical, lung, gastric, oral squamous and colon cancers, and has been implicated in cell proliferation, migration, apoptosis and angiogenesis through regulation of a variety of cancer-associated genes (29–33). In the present study, Rac1 and SP1 were upregulated and negatively associated with miR-124-3p levels in gastric cancer tissues. Furthermore, Rac1 and SP1 overexpression rescued the inhibitory effect of miR-124 on cell survival. Collectively these results demonstrated that miR-124-3p may inhibit gastric cancer growth by targeting Rac1 and SP1.
Overall, the present study identified the potential role of miR-124-3p in gastric cancer progression and as an independent predictor of OS and RFS. The data further indicated that miR-124-3p suppressed gastric cancer growth by regulating Rac1 and SP1. These findings suggest miR-124-3p may be employed as a novel prognostic biomarker, as an indicator of treatment strategy and/or as a potential therapeutic target in gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and Technology Department of Hunan Province, China (grant no. 2018JJ6071), the Education Department of Hunan Province, China (grant no. 16C1426) and the Science and Technology Department of Hengyang, China (grant no. 2015KJ16).
Availability of data and materials
All data generated or analyzed in this study are included in this article.
Authors' contributions
FL and LX designed the study. FL, HH and JZ performed experiments and collected and analyzed data. FL, LT, XA and ZZ wrote, reviewed and revised manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest regarding the publication of this study.
References
- 1.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 2.Slack FJ, Weidhaas JB. MicroRNA in cancer prognosis. N Engl J Med. 2008;359:2720–2722. doi: 10.1056/NEJMe0808667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng CJ, Slack FJ. The duality of oncomiR addiction in the maintenance and treatment of cancer. Cancer J. 2012;18:232–237. doi: 10.1097/PPO.0b013e318258b75b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Zhou L, Li Y, Lin S, Tomuleasa C. MicroRNA-21 stimulates gastric cancer growth and invasion by inhibiting the tumor suppressor effects of programmed cell death protein 4 and phosphatase and tensin homolog. J BUON. 2014;19:228–236. [PubMed] [Google Scholar]
- 5.Li L, Luo J, Wang B, Wang D, Xie X, Yuan L, Guo J, Xi S, Gao J, Lin X, et al. MicroRNA-124 targets flotillin-1 to regulate proliferation and migration in breast cancer. Mol Cancer. 2013;12:163. doi: 10.1186/1476-4598-12-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park SY, Kim H, Yoon S, Bae JA, Choi SY, Jung YD, Kim KK. KITENIN-targeting microRNA-124 suppresses colorectal cancer cell motility and tumorigenesis. Mol Ther. 2014;22:1653–1664. doi: 10.1038/mt.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang S, Zhao Y, Hu K, Xu C, Wang L, Liu J, Yao A, Zhang H, Cao F. miR-124 exhibits antiproliferative and antiaggressive effects on prostate cancer cells through PACE4 pathway. Prostate. 2014;74:1095–1106. doi: 10.1002/pros.22822. [DOI] [PubMed] [Google Scholar]
- 8.Xie L, Zhang Z, Tan Z, He R, Zeng X, Xie Y, Li S, Tang G, Tang H, He X. MicroRNA-124 inhibits proliferation and induces apoptosis by directly repressing EZH2 in gastric cancer. Mol Cell Biochem. 2014;392:153–159. doi: 10.1007/s11010-014-2028-0. [DOI] [PubMed] [Google Scholar]
- 9.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Wang Q, Zhao Q, Di W. miR-124 inhibits the migration and invasion of ovarian cancer cells by targeting SphK1. J Ovarian Res. 2013;6:84. doi: 10.1186/1757-2215-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An L, Liu Y, Wu A, Guan Y. microRNA-124 inhibits migration and invasion by down-regulating ROCK1 in glioma. PLoS One. 2013;8:e69478. doi: 10.1371/journal.pone.0069478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia J, Wu Z, Yu C, He W, Zheng H, He Y, Jian W, Chen L, Zhang L, Li W. miR-124 inhibits cell proliferation in gastric cancer through down-regulation of SPHK1. J Pathol. 2012;227:470–480. doi: 10.1002/path.4030. [DOI] [PubMed] [Google Scholar]
- 13.Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, et al. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 14.Tang H, Deng M, Tang Y, Xie X, Guo J, Kong Y, Ye F, Su Q, Xie X. miR 200b and miR 200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19:5602–5612. doi: 10.1158/1078-0432.CCR-13-1326. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Liu A, Yu Q, Peng Z, Huang Y, Diao S, Cheng J, Wang W, Hong M. miR-200b inhibits CD133+ glioma cells by targeting the AKT pathway. Oncol Lett. 2017;13:4701–4707. doi: 10.3892/ol.2017.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu CB, Li QL, Hu JF, Zhang Q, Xie JP, Deng L. miR-124 inhibits growth and invasion of gastric cancer by targeting ROCK1. Asian Pac J Cancer Prev. 2014;15:6543–6546. doi: 10.7314/APJCP.2014.15.16.6543. [DOI] [PubMed] [Google Scholar]
- 18.Deng M, Tang HL, Lu XH, Liu MY, Lu XM, Gu YX, Liu JF, He ZM. miR-26a suppresses tumor growth and metastasis by targeting FGF9 in gastric cancer. PLoS One. 2013;8:e72662. doi: 10.1371/journal.pone.0072662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennings RT, Knaus UG. Rho family and Rap GTPase activation assays. Methods Mol Biol. 2014;1124:79–88. doi: 10.1007/978-1-62703-845-4_6. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H, Dong T, Zhou H, Wang L, Huang A, Feng B, Quan Y, Jin R, Zhang W, Sun J, et al. miR-320a suppresses colorectal cancer progression by targeting Rac1. Carcinogenesis. 2014;35:886–895. doi: 10.1093/carcin/bgt378. [DOI] [PubMed] [Google Scholar]
- 21.Dokmanovic M, Hirsch DS, Shen Y, Wu WJ. Rac1 contributes to trastuzumab resistance of breast cancer cells: Rac1 as a potential therapeutic target for the treatment of trastuzumab-resistant breast cancer. Mol Cancer Ther. 2009;8:1557–1569. doi: 10.1158/1535-7163.MCT-09-0140. [DOI] [PubMed] [Google Scholar]
- 22.Zhan H, Liang H, Liu X, Deng J, Wang B, Hao X. Expression of Rac1, HIF-1α, and VEGF in gastric carcinoma: Correlation with angiogenesis and prognosis. Onkologie. 2013;36:102–107. doi: 10.1159/000348525. [DOI] [PubMed] [Google Scholar]
- 23.Chen B, Gao Y, Jiang T, Ding J, Zeng Y, Xu R, Jiang X. Inhibition of tumor cell migration and invasion through knockdown of Rac1 expression in medulloblastoma cells. Cell Mol Neurobiol. 2011;31:251–257. doi: 10.1007/s10571-010-9615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gastonguay A, Berg T, Hauser AD, Schuld N, Lorimer E, Williams CL. The role of Rac1 in the regulation of NF-κB activity, cell proliferation, and cell migration in non-small cell lung carcinoma. Cancer Biol Ther. 2012;13:647–656. doi: 10.4161/cbt.20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida T, Zhang Y, Rivera Rosado LA, Chen J, Khan T, Moon SY, Zhang B. Blockade of Rac1 activity induces G1 cell cycle arrest or apoptosis in breast cancer cells through downregulation of cyclin D1, survivin, and X-linked inhibitor of apoptosis protein. Mol Cancer Ther. 2010;9:1657–1668. doi: 10.1158/1535-7163.MCT-09-0906. [DOI] [PubMed] [Google Scholar]
- 26.Rathinam R, Berrier A, Alahari SK. Role of Rho GTPases and their regulators in cancer progression. Front Biosci. 2011;16:2561–2571. doi: 10.2741/3872. [DOI] [PubMed] [Google Scholar]
- 27.Bid HK, Roberts RD, Manchanda PK, Houghton PJ. RAC1: An emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol Cancer Ther. 2013;12:1925–1934. doi: 10.1158/1535-7163.MCT-13-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vader P, van der Meel R, Symons MH, Fens MH, Pieters E, Wilschut KJ, Storm G, Jarzabek M, Gallagher WM, Schiffelers RM, et al. Examining the role of Rac1 in tumor angiogenesis and growth: A clinically relevant RNAi-mediated approach. Angiogenesis. 2011;14:457–466. doi: 10.1007/s10456-011-9229-x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Li S, Yan Q, Chen X, Yang Y, Liu X, Wan X. Interferon-β induced microRNA-129-5p down-regulates HPV-18 E6 and E7 viral gene expression by targeting SP1 in cervical cancer cells. PLoS One. 2013;8:e81366. doi: 10.1371/journal.pone.0081366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang WB, Chen PH, Hsu T, Fu TF, Su WC, Liaw H, Chang WC, Hung JJ. Sp1-mediated microRNA-182 expression regulates lung cancer progression. Oncotarget. 2014;5:740–753. doi: 10.18632/oncotarget.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, Zhu W, Shu Y, Liu P. miR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588:1168–1177. doi: 10.1016/j.febslet.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 32.Kim DW, Ko SM, Jeon YJ, Noh YW, Choi NJ, Cho SD, Moon HS, Cho YS, Shin JC, Park SM, et al. Anti-proliferative effect of honokiol in oral squamous cancer through the regulation of specificity protein 1. Int J Oncol. 2013;43:1103–1110. doi: 10.3892/ijo.2013.2028. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Zhang W, Guo Z, Ma F, Wu Y, Bai Y, Gong W, Chen Y, Cheng T, Zhi F, et al. Inhibition of the transcription factor Sp1 suppresses colon cancer stem cell growth and induces apoptosis in vitro and in nude mouse xenografts. Oncol Rep. 2013;30:1782–1792. doi: 10.3892/or.2013.2627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed in this study are included in this article.