Abstract
Myeloid-derived suppressor cells (MDSCs) have strong immunosuppressive functions and contribute to the formation of the tumor microenvironment. Long non-coding (Lnc) RNAs are highly important factors associated with tumors and may be used as markers for tumor diagnosis, which is valuable for targeted therapy. LncRNA MALAT1 is expressed in various tissues and plays a critical role in cell proliferation, including tumorigenesis and metastasis. However, the role of MALAT1 in MDSCs is unclear. In this study, we observed an increased proportion of MDSCs and elevated levels of the related molecule arginase-1 (ARG-1) in peripheral blood mononuclear cells (PBMCs) obtained from lung cancer patients. The proportion of CD8+ cytotoxic T lymphocyte (CTL) was significantly decreased in PBMCs from lung cancer patients. Moreover, the proportion of CTL cells was negatively correlated with the proportion of MDSCs. Furthermore, MALAT1 levels were decreased in PBMCs from lung cancer patients. The relative expression of MALAT1 was moderate negatively correlated with the proportion of MDSCs. In vitro results indicate that the knockdown of MALAT1 significantly increased the proportion of MDSCs. Our data provide the first evidence that lncRNA MALAT1 negatively regulates MDSCs and is decreased in PBMCs from lung cancer patients.
Keywords: MDSCs, long non-coding RNA, MALAT1, lung cancer
Introduction
Myeloid-derived suppressor cells (MDSCs) are precursors of macrophages, dendritic cells and granulocytes, and they exert immunosuppressive functions. Many studies have shown that MDSCs can suppress immune responses in tumors and other diseases. In humans, MDSCs generally coexpress the genes CD11b and CD33 and lack the MHCII protein HLA-DR; thus, these cells are currently defined as CD33+CD11b+HLA-DR-1-3. Recent comprehensive reviews have shown that MDSCs play important roles in tumor immune escape, tumor development and progression. These cells can play a role in negatively regulating immunity by secreting functional molecules such as arginase-1 (ARG-1), reactive oxygen species (ROS), and inducible nitric oxide synthase (iNOS)4. It is widely known that high expression of ARG-1 by MDSCs inhibits T cell activation, which affects cell immune responses. Recent reviews have established a direct role for ARG-1 in the inhibition of T cell function5, 6.
It has become increasingly clear that long non-coding (lnc) RNAs play crucial roles in malignant tumorigenesis through a diverse range of mechanisms7, 8. LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear enrichment autosomal transcript 2 (NEAT2), is widely expressed in mammals9, 10. Abnormal expression of MALAT1 has been observed in tumor tissues and is associated with many important biological processes, including proliferation, apoptosis, invasion and metastasis of tumor cells11-14. To understand this tumor related molecule, our research focused on how MALAT1 is expressed in peripheral blood mononuclear cells (PBMCs) from lung cancer patients and how MALAT1 impacts MDSCs.
In this study, we detected the proportion of MDSCs in lung cancer patients, the level of ARG-1 in MDSCs, and the percentage of CD8+CTL cells, and we later analyzed the relationship of MALAT1 with MDSCs in PBMCs and further explored the regulatory function of MALAT1 on MDSCs induction in vitro. Our findings provide an experimental basis and new insights to elucidate the role of MALAT1 and MDSCs in the immune function of patients with lung cancer.
Materials and methods
Patients and health volunteers
Thirty patients newly diagnosed with lung cancer were chosen from the Affiliated People's Hospital, Jiangsu University. Among these 30 cases, 15 were female, and the other 15 were male, with an average age of 55±15 years being determined. All lung cancer patients were diagnosed with primary lung cancer and fulfilled the American-European Consensus Group Criteria. Another group of thirty healthy volunteers were also chosen from the Affiliated People's Hospital, Jiangsu University. Among these 30 cases, 16 were female, 14 were male, and the average age was 45±15 years. None of the healthy volunteers had any history of malignant tumors or autoimmune diseases. The study was approved by the Institutional Ethical Board, and each blood sample was collected with prior patient consent.
Peripheral blood mononuclear cell isolation
For each subject, 2 ml peripheral blood treated with EDTA-K2 anticoagulant was centrifuged at 450×g for 5 min at room temperature, and the upper plasma was later collected. Blood cells were diluted with 2 ml PBS buffer, added slowly to the liquid surface of 2 ml Ficoll-Hypaque separation solution, and centrifuged at 450×g at room temperature for 20 min. Finally, the PBMC component was removed carefully and washed with PBS buffer.
RNA isolation and qRT-PCR
Total RNA was extracted with Trizol reagent (Invitrogen, California, USA), and cDNA was synthesized with random primers and a Reverse Transcription Kit (Toyobo, Osaka, Japan). The relative expression of MALAT1 was measured by qRT-PCR using BioRad SYBR Green Super Mix (Bio-Rad, Hercules, USA). Primer sequences were as follows: MALAT1, sense, 5'-CTTTTCCCCCACATTTCCAA-3', antisense, 5'-CTCGTGGCTCAAGTGAGGTG-3'; β-actin, sense, 5'-CACGAAACTACCTTCAACTCC-3', antisense, 5'-CATACTCCTGCTTGCTGATC-3'. The relative expression of MALAT1 was expressed as the ratio of MALAT1 to β-actin transcript. Data were analyzed by BioRad CFX Manager software.
Flow cytometry analysis
Resuspended PBMCs (1×106) were cultured in 1 ml 10% fetal bovine serum RPMI 1640 medium in the presence of 50 ng/ml PMA (Sigma-Aldrich, California, USA) and 1 μg/ml of ionomycin (Sigma-Aldrich). After 1 hour, 1 μg/ml brefeldin-A (eBioscience, San Diego, USA) was added to the culture for 4 hours at 37℃ and 5% CO2. Stimulated PBMCs were stained with PE-Cy5-conjugated anti-CD3 mAb and FITC-conjugated anti-CD8 mAb (eBioscience), then fixed, permeabilized and stained with anti-IFN-γ mAb (eBioscience) with an Intracellular Staining Kit (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. Th1 were defined as CD3+CD8-IFN-γ+, and CTL were defined as CD3+ CD8+ IFN-γ+.
Resuspended PBMCs (1×106) were stained with APC-conjugated anti-HLA-DR mAb (eBioscience), PE-Cy5-conjugated anti-CD11b mAb (Biolegend), FITC-conjugated anti-CD33 mAb (eBioscience), and PE-conjugated anti-ARG-1 mAb (Biolegend); MDSCs were defined as CD33+CD11b+HLA-DR-. All data were acquired with a BD FASCalibur flow cytometer and subsequently analyzed with FlowJo software.
Small interfering RNA (siRNA) knockdown and MDSC induction in vitro
Small interfering RNA (Ribobio, Guangzhou, China) was designed against the sequence of MALAT1, and a nonspecific scrambled siRNA was used as a negative control. Following the manufacturer's protocol, 1×106 PBMCs were transfected with 50 nmol/L MALAT1 siRNA or control for 24 hours, and 10 nmol/ml IL-1β and 10 nmol/ml GM-CSF were added to establish MDSC culture. Cells were cultured at 37℃ for 6 days with cytokine replacement every 2-3 days. Since certain markers are downregulated after several days of culture, recent reviews have quantified and described MDSCs using the myeloid marker CD33+15-17. Cultured cells were collected and analyzed by flow cytometry to determine the proportion of CD33+MDSCs.
Statistical analysis
Data were analyzed with GraphPad Prism (5.0) software. Student's unpaired t test was applied for comparisons. Correlation between variables was quantified by Pearson's correlation coefficients. All bars in graphs are expressed as the mean ± SD. Differences were considered to be statistically significant at p<0.05.
Results
Proportion of MDSCs is increased in lung cancer patients
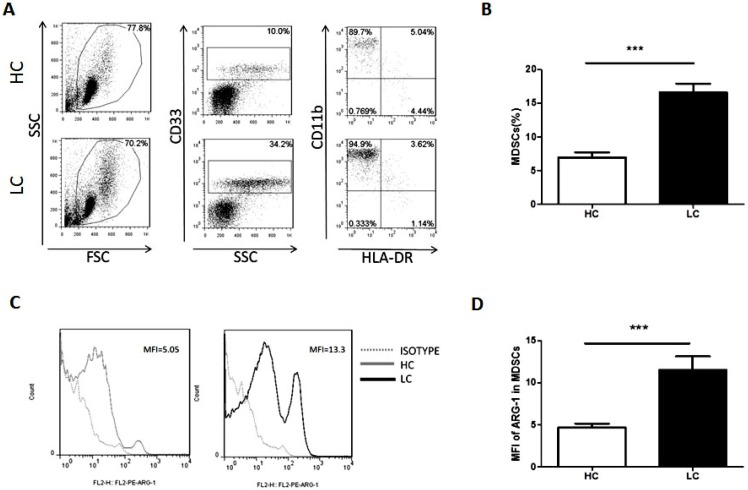
The proportion of MDSCs in PBMCs and the levels of ARG-1 in MDSCs were measured and analyzed by flow cytometry. Compared with healthy volunteers, patients with lung cancer had a significantly increased proportion of MDSCs (Fig. 1A and B). Moreover, the mean fluorescence intensity of ARG-1 in MDSCs was also significantly increased in the lung cancer group (Fig. 1C and D). These results further confirm that MDSCs and the functional factor ARG-1 are increased in lung cancer patients.
Figure 1.
Proportion of MDSCs is increased in lung cancer patients. Peripheral blood mononuclear cells (PBMCs) from lung cancer patients and healthy volunteers were stained with surface antibodies and analyzed by flow cytometry. MDSCs were defined as CD33+CD11b+HLA-DR-. A. Representative flow cytometry dot plots from lung cancer patients and healthy controls are shown. Values correspond to the proportion of MDSCs in PBMCs. B. Proportion of MDSCs in PBMCs from lung cancer patients and healthy controls (n=30). *** p<0.001. C. Representative flow cytometry analysis of ARG-1 in peripheral blood MDSCs from a lung cancer patient and a healthy control. Left panel and right panel show the histogram of mean fluorescence intensity of ARG-1 in MDSCs from a representative HC (the Gray solid line filled) and a representative LC patient (the black solid line filled); the Gray dotted line represents isotype control. D. Levels of ARG-1 in peripheral blood MDSCs from lung cancer patients and healthy controls (n=30). *** p<0.001.
Association between MDSCs and CTL in patients with lung cancer
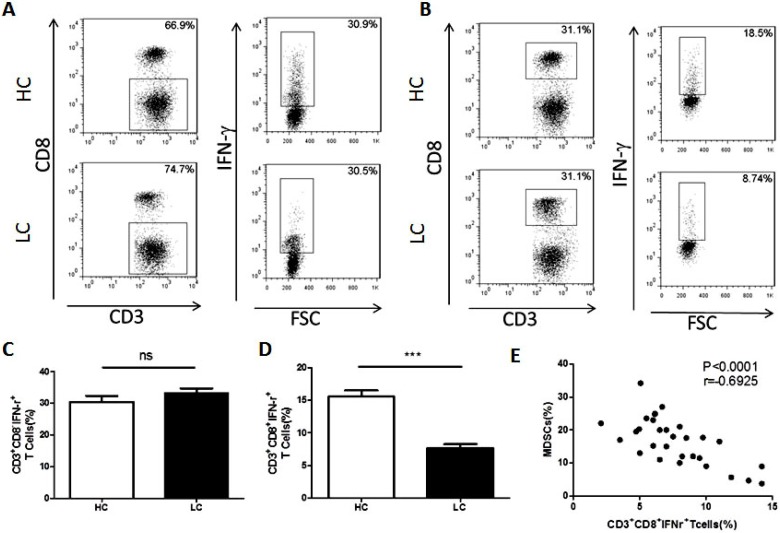
It is widely acknowledged that MDSCs suppress anti-tumor immunity by inhibiting T cells. To examine the association between MDSCs and T cells, the proportion of CD4+Th1 and CD8+CTL cells in PBMCs were compared between healthy volunteers and lung cancer patients. The results showed that the proportion of Th1 cells is unchanged between the lung cancer patients and healthy volunteers (Fig. 2A and C). However, compared to healthy volunteers, the proportion of CTL was significantly decreased in lung cancer patients (Fig. 2B and D). In addition, the proportion of CTL cells was negatively correlated with the percentage of MDSCs in lung cancer PBMCs (Fig. 2E).
Figure 2.
Correlation between the proportion of MDSCs and CTL cells in PBMCs from lung cancer patients. A. Representative flow cytometry analysis of Th1 cells in peripheral blood from a lung cancer patient and a healthy control. B. Representative flow cytometry analysis of CTL cells in peripheral blood from a lung cancer patient and a healthy control. C. Proportion of Th1 cells in PBMCs from lung cancer patients and healthy controls (n=30), ns means no significant difference. D. Proportion of CTL cells in PBMCs from lung cancer patients and healthy controls, *** p<0.001. E. Correlation between the proportion of MDSCs and CTL cells in PBMCs from lung cancer patients (n=30). Each data point represents an individual subject, p<0.0001, r=-0.6925.
Association between MALAT1 and MDSCs in patients with lung cancer
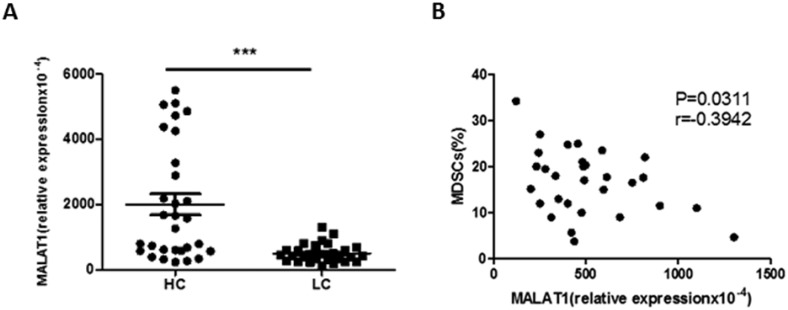
The expression of lncRNA MALAT1 in PBMCs from lung cancer patients was measured by qRT-PCR. Compared with healthy volunteers, the relative expression of MALAT1 in PBMCs was significantly decreased in lung cancer patients (Fig. 3A). Furthermore, there was a moderately negative correlation between the expression of MALAT1 and the proportion of MDSCs in PBMCs from patients with lung cancer (Fig. 3B).
Figure 3.
Correlation between the level of MALAT1 and the proportion of MDSCs in lung cancer patients. A. The level of MALAT1 in PBMCs of lung cancer patients was determined by qRT-PCR. Each data point represents an individual subject and horizontal lines indicate the mean, *** p<0.001. B. Correlation between the level of MALAT1 and the proportion of MDSCs in PBMCs from LC patients. n=30, p=0.0311, r=-0.3942.
Knockdown of MALAT1 increased the proportion of MDSCs in vitro
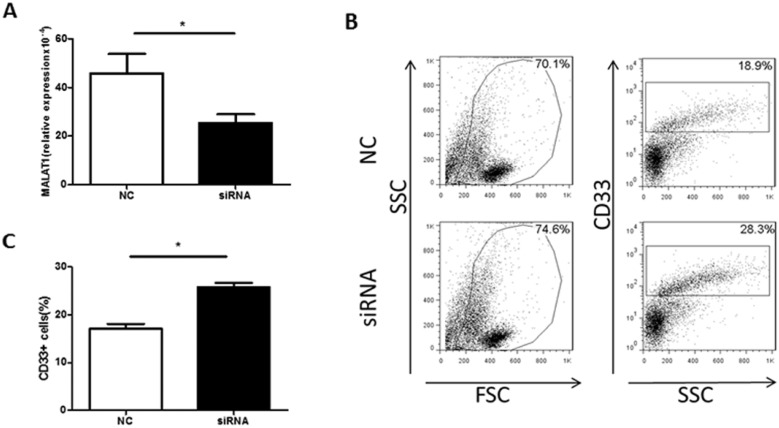
To confirm a direct effect of MALAT1 on MDSCs, a MDSC-inducing culture system was established. PBMCs were transfected with MALAT1 siRNA, which reduced MALAT1 expression (Fig. 4A). Moreover, the MALAT1 siRNA treatment significantly increased the proportion of MDSCs in the PBMC cell culture system (Fig. 4B and C). These data indicate that MALAT1 negatively regulates MDSCs in vitro.
Figure 4.
Knockdown of MALAT1 expression increases the proportion of MDSCs in vitro. A. The level of MALAT1 in PBMCs from healthy volunteers was determined by qRT-PCR, * p<0.05. B. Representative flow cytometry plots of MDSCs after MALAT1 siRNA or negative control transfection. C. Comparison of the proportion of CD33+MDSCs in PBMCs after MALAT1 siRNA and negative control transfection, * p<0.05.
Discussion
MDSCs play important roles in tumor immune escape, tumor development, and tumor progression. Many studies have reported that MDSCs can exhibit immunosuppressive effects on T cells through a variety of mechanisms, such as secretion of ARG-1, ROS, and iNOS18. ARG-1 breaks down arginine, which results in the inhibition of T cell activation. Nagaraj et al. reported that MDSCs had elevated levels of cyclooxygenase (COX-2) and prostaglandin E2 (PGE2), which repressed the quantity and function of CD8+T cells19. Some studies found markedly increased numbers of MDSCs and high levels of arginase in tumor tissues from lung cancer, and miR-9 regulated MDSCs differentiation by targeting the runt-related transcription factor 120. The above research suggests that there are many ways to inhibit MDSCs. Zou and colleagues confirmed that MDSCs play multiple roles within the tumor microenvironment, such as inhibiting effector cells while also activating tumor cells to promote tumor development21. Several studies have shown that the quantity of MDSCs in tumor patients is significantly higher than in healthy people and is closely correlated with disease progression. Our results show that the proportion of MDSCs in PBMCs from lung cancer patients was significantly higher than from healthy controls. Therefore, it can be concluded that MDSCs exhibit major immunosuppressive function within the tumor microenvironment.
It is well-known that MDSCs are divided into two groups: granulocyte MDSCs (G-MDSCs) and mononuclear MDSCs (M-MDSCs). While G-MDSCs produce high levels of ROS and low levels of NO, M-MDSCs conversely produce low ROS and high NO. Both G-MDSCs and M-MDSCs secrete ARG-122. Our experiments analyzed the average fluorescence intensity of ARG-1 in MDSCs by flow cytometry. We observed that ARG-1 levels in patients with lung cancer were significantly higher than those of healthy controls. Recent data suggest that there is a close association between the availability of arginine and the regulation of T-cell proliferation2, 23. Moreover, COX-2 inhibitors downregulate the expression of ARG-1 by MDSCs, enhancing antitumor T-cell responses and increasing the therapeutic efficacy of immunotherapy23-25. Consistent with previous reviews, our results indicate that in lung cancer patients, elevated expression of ARG-1 in MDSCs increases immunological inhibition of T cells.
CD4+T cells and CD8+T cells play important roles in the body's response to tumors. CD4+T cells assist CD8+T cells, NK cells or macrophages to kill tumors by secreting Th1 cytokines. CD8+T cells can directly kill tumor cells by secreting perforin and granular enzymes. Our study showed that there was no obvious alteration of Th1 cells in PBMCs isolated from lung cancer patients compared to healthy controls, but CTL cells in cancer patients were significantly less abundant than in the controls. These results suggest that CTL cells may play a major role in adaptive immune responses to lung cancer. In addition, we observed a negative correlation between the proportions of CTLs and MDSCs in lung cancer. Therefore, we speculate that MDSCs primarily promote tumor development by inhibiting CTL production, thereby exerting immunosuppressive functions in the development of lung cancer.
Recent studies have shown that lncRNA MALAT1 is closely related to lung cancer, cervical cancer, colorectal cancer and liver cancer. Garen and Song demonstrated that overexpression of MALAT1 reduces the function of the tumor suppressor PSF26. Schmidt et al. also showed that MALAT1 is abnormally overexpressed in non-small cell lung cancer (NSCLC) tumor cells or tumor tissues27. However, several reports have shown low expression of MALAT1 in the peripheral blood of lung cancer patients28, which is in contrast to the high expression observed in tumor tissues, and the mechanism behind this discrepancy is unclear. Our results showed that the relative expression of MALAT1 in PBMCs of lung cancer patients was significantly lower than that of healthy controls. In addition, two previous experimental studies have demonstrated a negative correlation between the abundance of MDSCs and CTLs in PBMCs from lung cancer patients. As such, we wondered whether MALAT1was also associated with MDSCs and CTLs. Our results showed a moderately negative correlation between the level of MALAT1 and the proportion of MDSCs in PBMCs from patients with lung cancer but with no direct correlation with CTLs. We thus speculated that MALAT1 may have a direct role in regulating the differentiation of MDSCs. We designed a MALAT1 siRNA to transfect PBMCs and established a MDSC-inducing culture system. The proportion of MDSCs was significantly increased after PBMCs were transfected with MALAT1 siRNA. These results show that knockdown of MALAT1 increases the quantity of MDSCs by regulating their differentiation.
Our data suggest that MALAT1 plays a direct role in regulating MDSCs expansion, which provides a new understanding of lung cancer development and offers potential targets for auxiliary diagnosis and treatment in lung cancer.
Acknowledgments
This work was supported by the Jiangsu Province's Key Medical Talents Program (Grant No. ZDRCB2016018), the Summit of the Six Top Talents Program of Jiangsu Province (Grant No. 2015-WSN-116), the Specialized Project for Clinical Medicine of Jiangsu Province (Grant No. BL2014065), the Natural Science Foundation of Jiangsu (Grant No. BK20150533), the China Postdoctoral Science Foundation (Grant Nos. 2016M600382, 2016M590423, 2017T100336), the Jiangsu Postdoctoral Science Foundation (Grant No.1601082B), and the Jiangsu Province “333” Project (Grant No. BRA2017128).
Compliance with ethical standards
The study was approved by the respective Ethics Committee of the Affiliated People's Hospital of Jiangsu University. Written informed consent was obtained from all the subjects in accordance with the Declaration of Helsinki.
Abbreviations
- LncRNA
Long non-coding RNA
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- MDSCs
Myeloid-derived suppressor cells
- Th1
T helper 1
- CTL
Cytotoxic T lymphocyte
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- PBMCs
Peripheral blood mononuclear cells
- Arg1
Arginase 1
- siRNA
Small interfering RNA.
References
- 1.Diaz-Montero CM, Finke J, Montero AJ. Myeloid-derived suppressor cells in cancer: therapeutic, predictive, and prognostic implications. Semin Oncol. 2014;41:174–184. doi: 10.1053/j.seminoncol.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Tian J, Wang S. The potential therapeutic role of myeloid-derived suppressor cells in autoimmune arthritis. Semin Arthritis Rheum. 2016;45:490–495. doi: 10.1016/j.semarthrit.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Ma J, Xu H, Wang S. Immunosuppressive role of myeloid-derived suppressor cells and Therapeutic Targeting in lung cancer. J Immunol Res. 2018;2018:6319649. doi: 10.1155/2018/6319649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindau D, Gielen P, Kroesen M, Wesseling P, Adema GJ. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 2013;138:105–115. doi: 10.1111/imm.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez PC, Ochoa AC. Arginine regulation by myeloid derived suppressor cells and tolerance in cancer: mechanisms and therapeutic perspectives. Immunol Rev. 2008;222:180–191. doi: 10.1111/j.1600-065X.2008.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian X, Tian J, Tang X, Ma J, Wang S. Long non-coding RNAs in the regulation of myeloid cells. J Hematol Oncol. 2016;9:99. doi: 10.1186/s13045-016-0333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian X, Ma J, Wang T, Tian J, Zhang Y, Mao L, Xu H, Wang S. LncRNA HOTAIRM1-HOXA1 axis down-regulates the immunosuppressive activity of myeloid-derived suppressor cells in lung cancer. Front Immunol. 2018;9:473. doi: 10.3389/fimmu.2018.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 10.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, Wu JW. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36:3355–3359. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]
- 12.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 13.Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–128. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 14.Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L. et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostlin N, Hofstadter K, Ostermeir AL, Spring B, Leiber A, Haen S, Abele H, Bauer P, Pollheimer J, Hartl D. et al. Granulocytic Myeloid-Derived Suppressor Cells Accumulate in Human Placenta and Polarize toward a Th2 Phenotype. J Immunol. 2016;196:1132–1145. doi: 10.4049/jimmunol.1500340. [DOI] [PubMed] [Google Scholar]
- 16.Lechner MG, Liebertz DJ, Epstein AL. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J Immunol. 2010;185:2273–2284. doi: 10.4049/jimmunol.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lechner MG, Megiel C, Russell SM, Bingham B, Arger N, Woo T, Epstein AL. Functional characterization of human Cd33+ and Cd11b+ myeloid-derived suppressor cell subsets induced from peripheral blood mononuclear cells co-cultured with a diverse set of human tumor cell lines. J Transl Med. 2011;9:90. doi: 10.1186/1479-5876-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schouppe E, Van Overmeire E, Laoui D, Keirsse J, Van Ginderachter JA. Modulation of CD8(+) T-cell activation events by monocytic and granulocytic myeloid-derived suppressor cells. Immunobiology. 2013;218:1385–1391. doi: 10.1016/j.imbio.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Nagaraj S, Nelson A, Youn JI, Cheng P, Quiceno D, Gabrilovich DI. Antigen-specific CD4(+) T cells regulate function of myeloid-derived suppressor cells in cancer via retrograde MHC class II signaling. Cancer Res. 2012;72:928–938. doi: 10.1158/0008-5472.CAN-11-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian J, Rui K, Tang X, Ma J, Wang Y, Tian X, Zhang Y, Xu H, Lu L, Wang S. MicroRNA-9 regulates the differentiation and function of myeloid-derived suppressor cells via targeting Runx1. J Immunol. 2015;195:1301–1311. doi: 10.4049/jimmunol.1500209. [DOI] [PubMed] [Google Scholar]
- 21.Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH, Vatan L, Szeliga W, Mao Y, Thomas DG. et al. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39:611–621. doi: 10.1016/j.immuni.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez PC, Hernandez CP, Quiceno D, Dubinett SM, Zabaleta J, Ochoa JB, Gilbert J, Ochoa AC. Arginase I in myeloid suppressor cells is induced by COX-2 in lung carcinoma. J Exp Med. 2005;202:931–939. doi: 10.1084/jem.20050715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talmadge JE, Hood KC, Zobel LC, Shafer LR, Coles M, Toth B. Chemoprevention by cyclooxygenase-2 inhibition reduces immature myeloid suppressor cell expansion. Int Immunopharmacol. 2007;7:140–151. doi: 10.1016/j.intimp.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 25.Zea AH, Rodriguez PC, Atkins MB, Hernandez C, Signoretti S, Zabaleta J, McDermott D, Quiceno D, Youmans A, O'Neill A. et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65:3044–3048. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 26.Garen A, Song X. Regulatory roles of tumor-suppressor proteins and noncoding RNA in cancer and normal cell functions. Int J Cancer. 2008;122:1687–1689. doi: 10.1002/ijc.23285. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A. et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 28.Guo F, Yu F, Wang J, Li Y, Li Y, Li Z, Zhou Q. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed Rep. 2015;3:309–312. doi: 10.3892/br.2015.422. [DOI] [PMC free article] [PubMed] [Google Scholar]