Abstract
Objectives
To develop a model using radiomic features extracted from MR images to distinguish radiation necrosis from tumor progression in brain metastases after Gamma knife radiosurgery.
Methods
We retrospectively identified 87 patients with pathologically confirmed necrosis (24 lesions) or progression (73 lesions), and calculated 285 radiomic features from 4 MR sequences (T1, T1 post-contrast, T2, and fluid-attenuated inversion recovery) obtained at 2 follow-up time points per lesion per patient. Reproducibility of each feature between the two time points was calculated within each group to identify a subset of features with distinct reproducible values between two groups. Changes in radiomic features from one time point to the next (delta radiomics) were used to build a model to classify necrosis and progression lesions.
Results
A combination of 5 radiomic features from both T1 post-contrast and T2 MR images were found to be useful in distinguishing necrosis from progression lesions. Delta radiomic features with a RUSBoost ensemble classifier had an overall predictive accuracy of 73.2% and an area under the curve value of 0.73 in leave-one-out cross-validation.
Conclusions
Delta radiomic features extracted from MR images have potential for distinguishing radiation necrosis from tumor progression after radiosurgery for brain metastases.
Keywords: Delta radiomic features, MRI, radiation necrosis, brain metastases, Gamma knife radiosurgery
Introduction
Brain metastases are the most common type of intracranial tumors, occurring in 9%–17% of patients with cancer [1]. Aggressive treatment of brain metastases with stereotactic radiosurgery has improved the median survival time for patients with brain metastases [2; 3], but approximately 10% of patients receiving radiosurgery for their brain tumor develop radionecrosis [4; 5]. Necrosis typically manifests at 6–9 months after radiosurgery with edema and severe neuropsychological disturbances; these symptoms, and the appearance of radiation necrosis on magnetic resonance imaging (MRI), closely mimic tumor progression. Because tumor progression can be treated but radiation necrosis is irreversible, the ability to distinguish the two after stereotactic radiosurgery (e.g., Gamma Knife) for brain metastases is clinically important.
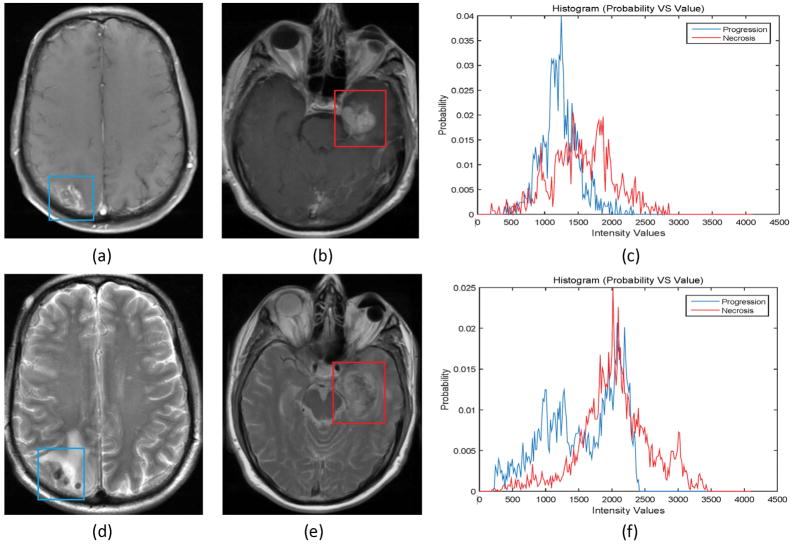
At present, confirmation of necrosis versus progression mainly relies on surgical resection, an invasive approach with risk of surgery induced neurologic deficits and other operative complications such as wound infection. Additionally late stage cancer patients often carry medical comorbidities which can compound surgical risk. On the other hand, part of routine follow-up care after Gamma Knife treatment involves undergoing high-resolution MRI at each visit, which can include T1 weighted (T1) scans, T1 weighted post-contrast (T1c) scans, T2 weighted (T2) scans, fluid-attenuated inversion recovery (FLAIR) scans, and diffusion-weighted MR scans, among others. However, to date none of these scan types can reliably distinguish radiation necrosis from tumor progression based on lesion appearance (Figure 1). Other special imaging approaches [6; 7] may be useful to distinguish radiation necrosis from tumor progression, but they are not generally available.
Figure 1.
Progressive lesions (left) are quite similar in appearance to necrotic lesions (middle) on both T1 weighted post-contrast magnetic resonance (MR) images (a and b) and T2 weighted MR images (d and e). Differences in intensity values within the lesions are shown at right (blue indicates progressive lesion, red necrosis).
An alternative to relying entirely on lesion appearance on various types of scans is to use the radiomic features of the MR image data. Radiomic features are quantitative descriptors that reflect textural variations in image intensity, shape, size or volume to offer information on tumor phenotype [8–13]. Here, we explored the possibility of using radiomic features extracted from MR images for predictive modeling to distinguish radiation necrosis from tumor progression after Gamma Knife radiosurgery for brain metastases.
Materials and Methods
Patient Data
This retrospective analysis of data from MR images was approved by the institutional review board, and the requirement for informed consent was waived. Eligibility criteria were as follows. (a) treatment with Gamma Knife radiosurgery for brain metastases with subsequent pathologically confirmed tumor progression or radiation necrosis via histologic resection or imaging during follow-up from August 2009 through August 2016; (b) availability of at least two sets of MR scans obtained at two separate follow-up times after radiosurgery but before confirmation; and (c) an identical sequence protocol for all MR scans, including T1, T1c, T2, and FLAIR. Patients were excluded if the MRI data were of poor quality because of motion artifacts or poor contrast injection. All MR images were acquired with a 1.5-T MRI system (Signa HDxt; GE Healthcare, Barrington, IL) during routine clinical visits. All images were axial scans with field of view of 22 cm, slice thickness of 5 mm, and slice spacing of 6.5 mm. We identified 87 patients who met these criteria; 1 or 2 lesions were identified per patient, for a total of 73 tumor progression lesions and 24 radiation necrosis lesions. The outcome of the tumor resection was used to label the lesion as necrosis or tumor progression. Table 1 shows the demographic summary of patients enrolled into this study.
Table 1.
The demographic summary of patients enrolled into this study. All data in the table are out of 84 patients, with 3 patients excluded due to the lack of documented radiation data.
| All patients (n = 84) | Necrosis Patients (n= 19) | Non-necrosis Patients (n=65) | |
|---|---|---|---|
| Gender | |||
| Male | 46 | 7 | 39 |
| Female | 38 | 12 | 26 |
| Number Alive | 12 | 3 | 9 |
| Age at SRS (years) | 28–79 | 37–71 | 28–79 |
| Primary Histology | |||
| Lung | 21 | 5 | 16 |
| Melanoma | 42 | 8 | 34 |
| Renal | 5 | 2 | 3 |
| Breast | 10 | 3 | 7 |
| Other | 6 | 1 | 5 |
| Prior WBRT | 22 | 7 | 15 |
| Chemotherapy prior to SRS | 61 | 13 | 48 |
| Chemotherapy after SRS | 37 | 11 | 26 |
| Interval between SRS and resection (days) | 267 (48–1307) | 258 (102–1307) | 26 (48–920) |
| Median (range) | |||
| SRS Radiation Dose (Gy) | 20 (13–24)* | 18 (14–22) | 20 (13–24) |
| Median (range) | |||
| Prior SRS | 9 | 1 | 8 |
Out of 81 patients, with 6 patients excluded due to the lack of documented radiation dose.
Prediction using Radiomic Features
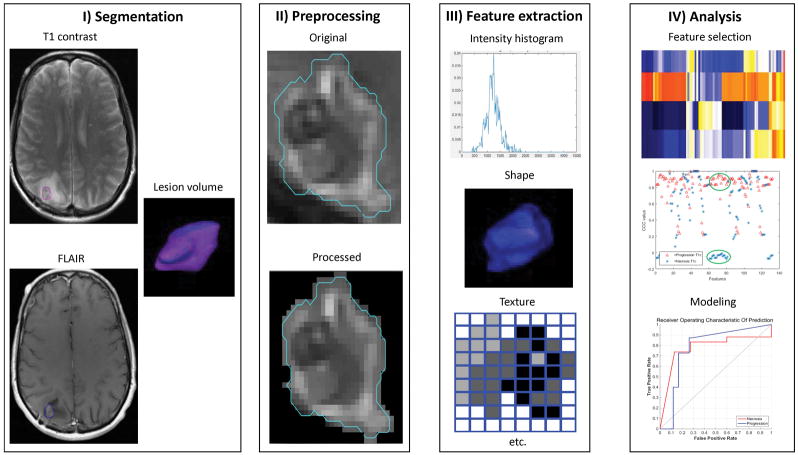
The overall workflow of this study is depicted in Figure 2; each step in the process is described further below.
Figure 2.
A generic framework for using radiomic features to create a predictive model. Steps include lesion segmentation, image preprocessing and feature extraction, and feature selection, analysis, and modeling.
Lesion segmentation
Each lesion was delineated on each type of MRI sequence (T1, T1c, T2, and FLAIR) for each patient by using the VelocityAI software (version 3.0.1; Varian Medical Systems, Atlanta, GA). A radiation oncologist contoured the regions of interest manually on the T1c images because the lesions were easier to identify after contrast injection than on the other scan types. The T1c contour was then rigidly mapped to the other scan sequences (T1, T2, and FLAIR) for each patient at each time point by using the Mattes mutual information metric [14] in the Velocity AI. The radiation oncologist then reviewed the contours on the T1, T2, and FLAIR scans to ensure correct mapping and modified them if necessary.
Preprocessing and feature calculation
Image preprocessing and radiomic feature extraction and calculation were done with Imaging Biomarker Explorer (IBEX) (http://bit.ly/IBEX_MDAnderson), an open-source software tool based on Matlab and C/C++ [15]. Image preprocessing to reduce uncertainty in feature analysis was done as follows [16]. First, an edge-preserving smoothing filter was applied to the tumor volume before the feature calculations [17] to preserve meaningful edge information while smoothing out undesirable imaging noise. Next, different thresholds were applied to the lesion volume on each scan type to exclude possible brain tissue and to define the final lesion volume. Based on our experience, we set the following low thresholds for image intensity values: 120 for T1; 200 for T1c; 150 for T2; and 50 for FLAIR.
A total of 285 texture features were calculated for each contoured lesion. This feature calculation was done for all four scan types at the two time points, resulting in a total of 2280 radiomic features per lesion. Textural features were organized into six categories according to the feature calculation method: direct intensity and intensity histogram [18; 19]; gray level co-occurrence matrix (COM) [20]; gray level run length matrix (RLM) [21]; geometric shape [22; 23]; neighborhood gray-tone difference matrix (NGTDM) [24]; and (vi) histogram of oriented gradients (HOG) [25; 26] (Table 2). All features were calculated in the 3-dimensional (3D) volume. Some feature values in the COM, RLM, and NGTDM categories were also calculated within 2-dimensional (2D) slices of the lesion volume and then averaged over all slices; these averaged features were called 2.5D features to distinguish them from those calculated from the 3D volume.
Table 2.
Radiomic features used in this study.
| Direct intensity and intensity histogram [112]* | Gray level co-occurrence matrix [132] | Gray level run length matrix [11] | |
|---|---|---|---|
| Energy | Inter-quartile range | Auto correlation | Gray level non-uniformity |
| Global entropy | Kurtosis | Cluster prominence | High gray level run emphasis |
| Global max | Mean absolute deviation | Cluster shade | Low gray level run emphasis |
| Global mean | Median absolute deviation | Cluster tendency | Long run emphasis |
| Global median | Percentile | Contrast | Long run high gray level emphasis |
| Global min | Percentile area | Correlation | |
| Global standard deviation | Quantile | Difference entropy | Long run low gray level emphasis |
| Global uniformity | Range | Dissimilarity | |
| Local entropy max | Skewness | Energy | Short run emphasis |
| Local entropy mean | Gaussian fit amplitude | Entropy | Short run high gray level |
| Local entropy median | Gaussian fit area | Homogeneity | emphasis |
| Local entropy min | Gaussian fit mean | Information measure correlation | Short run low gray level emphasis |
| Local entropy standard deviation | Gaussian fit standard deviation | Inverse different moment norm | |
| Histogram area | Inverse different norm | Run length non-uniformity | |
| Local range max | Local standard deviation median | Inverse variance | Run percentage |
| Local range mean | Local standard deviation min | Max probability | |
| Local range median | Local standard deviation standard | Sum average | |
| Local range min | deviation | Sum entropy | |
| Local range standard deviation | Root mean square | Sum variance | |
| Local standard deviation max | Variance | Variance | |
| Local standard deviation mean | |||
|
| |||
| Geometric shape [14] | Neighborhood gray-tone difference matrix [10] | Histogram of oriented gradients [6] | |
|
| |||
| Compactness | Roundness | Busyness | Inter-Quartile Range |
| Convex | Spherical disproportion | Coarseness | Kurtosis |
| Convex hull volume | Sphericity | Complexity | Mean absolute deviation |
| Mass | Surface area | Contrast | Median absolute deviation |
| Max 3D-diameter | Surface area density | Texture strength | Range |
| Mean breadth | Orientation | Skewness | |
Numbers of features selected from each category are shown in brackets (total=285 features). Shading indicates radiomic features selected for feature modeling by using concordance correlation coefficients (total=43 features).
We also calculated “delta” radiomic features, representing the change in features from one time point to the next, for feature modeling. These delta values were calculated for each MRI sequence, for a total of 1140 delta radiomic features per lesion. Because the time separation between the two sets of MRI scans, ΔT, was different for different patients (range, 9–119 days), these delta radiomic features were not directly comparable, and so we normalized the delta feature value to have a time separation of 30 days for all patients as follow:
where δF is the delta feature value before normalization and δFn is the delta feature value after normalization.
Feature selection
Not all radiomic features are necessarily useful for distinguishing necrosis from progression; in fact, many features are noisy and may lead to overfitting or misclassification in feature modeling. The extracted features also presumably included substantial redundancy. Therefore, it is important to identify a subset of useful and unique features for feature modeling. Previous studies have reported that quantitative analysis of tumor tissues is more reproducible than quantification of necrotic tissues [27], which led us to use concordance correlation coefficients (CCCs) to quantify the reproducibility of radiomic features for feature selection [16; 22; 28]. The CCC value ranges from −1 to 1, with 1 representing a high reproducibility, 0 no reproducibility, and −1 inverse reproducibility. We calculated the CCC values for each radiomic feature between the two time points for the radiation necrosis group and tumor progression group. Radiomic features with a CCC value > 0.7 for tumor progression and at the same time a CCC value between −0.1 and 0.1 for necrosis were considered potentially useful for distinguishing necrosis from progression.
Feature modeling
We used two sets of features for modeling—the radiomic features at the second time point and the delta features (change from the first to the second time point). The second time point scans were obtained close to the time of pathologic confirmation, and features from those scans may be more consistent with the actual outcomes than features obtained at the first time point. In contrast, the delta radiomic features reflected difference in features from one time point to the next for both the necrotic and progression groups. Delta radiomic features could be distinguishable based on previous reproducibility studies [27]. Radiomic features or delta radiomic features were input into five types of classifiers for modeling: decision trees; discriminant analysis; support vector machines; nearest neighbor classifiers and ensemble classifiers [29] by using the classification learner in Matlab (version 2015b, Mathworks, Natick, MA). The modeling was done on the basis of the selected radiomic features in the previous step using the CCC values, to predict radiation necrosis or tumor progression. A heuristic approach was used by testing the possibility of all combinations of selected features for feature modeling. We used leave-one-out cross validation to validate the predictive models. The performance of the predictive models was evaluated by analysis of the area under the curve (AUC) of the receiver operating characteristic (ROC) and with confusion matrices, which are tables used to describe the performance of the prediction model on a set of test data for which the true values are known. Finally, we compared the prediction performance of the radiomic features at the second time point with that of the delta radiomic features.
Results
Feature Selection
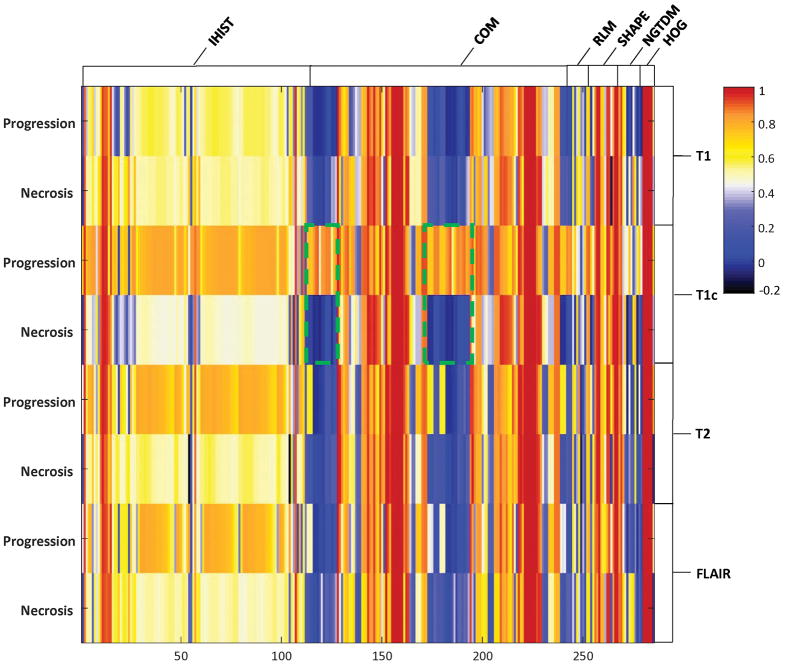
Comparisons of reproducibility for all radiomic features between the necrotic lesions and the progressive lesions are shown as a heat map in Figure 3. Those features showing distinctly different CCCs for necrosis (−0.1 < CCC < 0.1) versus progression (CCC > 0.7) are highlighted for the COM features. Reproducibility of these features was found to be fairly high for the progressive lesions but not for the necrotic lesions. Features considered potentially distinguishable between these two groups were then selected for subsequent modeling. We found that most radiomic features extracted from the T1, T2, and FLAIR sequences did not show sufficient separation between these two groups except the HOG skewness feature extracted from the T2. From the T1c scans, we were able to select 42 radiomic features using the criterion defined by the CCC values, and most of them are COM features (Table 2).
Figure 3.
A heat map showing concordance correlation coefficient (CCC) values for each of the radiomic features calculated in the tumor-progression group and those calculated in the necrosis group for the different types of magnetic resonance scans (T1, T1 postcontrast [T1c], T2, and fluid-attenuated inversion recovery [FLAIR]). Distinct CCC values between these two groups were highlighted for the COM features. Abbreviations: IHIST, direct intensity and intensity histogram; COM, gray level co-occurrence matrix; RLM, gray level run length matrix; SHAPE, geometric shape; NGTDM, neighborhood gray-tone difference matrix; HOG, histogram of oriented gradients.
Feature Modeling
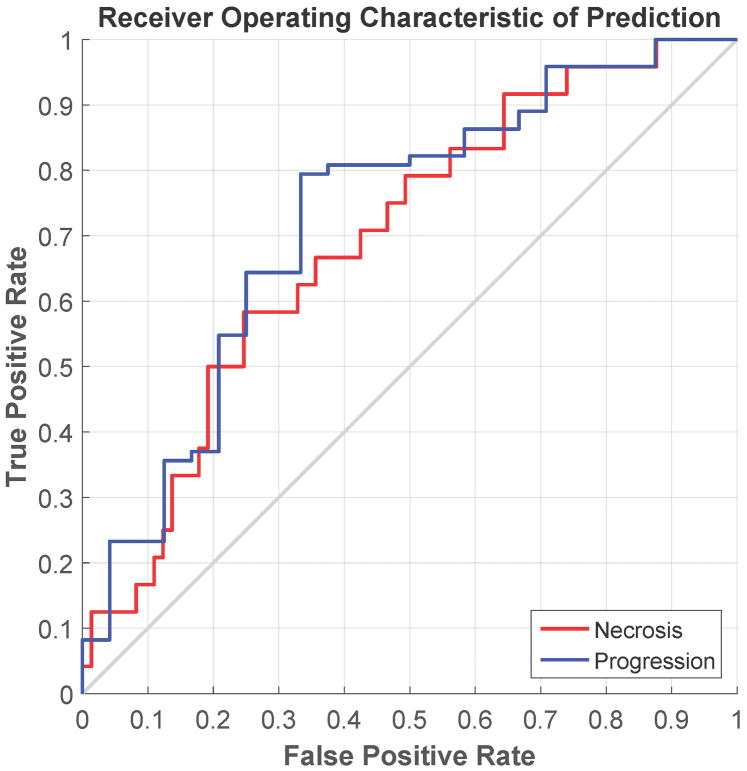
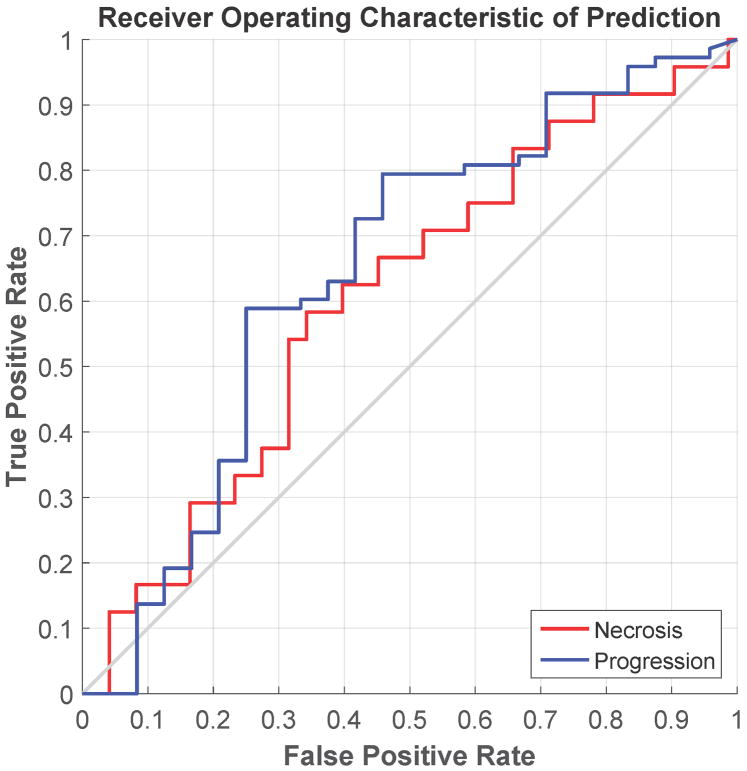
The ROC curves and the confusion matrices for the feature modeling are shown in Figure 4 and Table 3. In feature modeling, because the training data was highly skewed, with the number of the progression lesion much more than that of the necrosis lesion, we found that the best classifier for this situation was the RUSBoost[30], one of the ensemble classifiers in our evaluation. The RUSBoost is a decision tree based classifier. We used the following parameters for RUSBoost classifier in our evaluation: maximum number of splits as 50, number of learners as 150, and learning rate as 0.1. For the delta radiomic features, the best predictive features were the combination of 2 direct intensity features from T1c (energy and variance), 2 2.5D-RLM features from T1c (high gray level run emphasis and short run high gray level emphasis), and the HOG skewness feature from T2. The overall accuracy of the leave-one-out cross validation was 73.2%, with an accuracy for predicting radiation necrosis of 58.3% and an accuracy for predicting tumor progression of 78.1%. The AUC of the ROC curve was 0.73. In contrast, for the radiomic features at the second time point, the overall accuracy of the leave-one-out cross validation for those features was 69.1%, with an accuracy for predicting radiation necrosis of 54.2% and an accuracy for predicting tumor progression of 74.0%. The AUC of the ROC curve was 0.65.
Figure 4.
Receiver operating characteristic curve showing the performance of two predictive models in leave-one-out cross-validation. (a) Predictive model using delta radiomic features; (b) predictive model using the radiomic features extracted from the scan obtained at the second time point.
Table 3.
Confusion matrix of the predictive model built from delta radiomic features or the radiomic features at the second time point in leave-one-out cross-validation.
| True Class | |||
|---|---|---|---|
| Delta radiomic features | Necrosis | Progression | |
| Predict | Necrosis | 14 (58.3%) | 10 (41.7%) |
| Progression | 16 (21.9%) | 57 (78.1%) | |
| Radiomic features at second time point | |||
| Predict | Necrosis | 13 (54.2%) | 11 (45.8%) |
| Progression | 19 (26.0%) | 54 (74.0%) | |
Discussion
We developed a predictive model based on radiomic features extracted from MR images to distinguish radiation necrosis from tumor progression after Gamma Knife radiosurgery for brain metastases. Previous studies on feature reproducibility for necrotic tissue [27] led us to propose using CCC values for feature selection, both to reduce redundancy and identify features useful for predictive modeling. We found this feature selection approach to be effective, and believe that our findings will be a useful addition to radiomics research on identifying features that are useful for outcome modeling for clinical applications.
The major finding of this study was that the delta radiomic features extracted from T1c MR images had great potential for distinguishing radiation necrosis from tumor progression for patients treated with Gamma Knife radiosurgery. Because of the unbalanced of the training data and the relatively small amount of the training data, it was difficult to conclude whether a predictive model could be developed to reliably predict the outcome. However, we did find that radiomic features extracted from T1c were more valuable than those extracted from other MR sequences in this study (T1, T2, and FLAIR). In future studies of using imaging features to distinguish radiation necrosis from tumor progression, the features identified in this study should be investigated first for their predictive value.
To the best of our knowledge, we are the first to use MR-based delta radiomic features to distinguish radiation necrosis from tumor progression after Gamma Knife radiosurgery for brain metastatic lesions. Distinguishing between these two conditions is challenging and is the focus of considerable research in the field of neuro-oncology. However, many published studies seek to develop new imaging approaches to improve diagnosis, such as MRI spectroscopy and perfusion MRI [6], dynamic contrast-enhanced MRI [31], and positron emission tomography imaging [7], among others. Although these studies are important, these approaches are generally expensive and require considerable development before clinical use. Some investigators have used radiomic features to distinguish radiation necrosis from tumor progression [10; 12; 32–34], but those studies used radiomic features obtained at a single time point instead of using changes in radiomic features over time to model or classify features. Because radiomic features seem to keep changing over time, determining the best time to extract features for modeling is difficult, and models built on these features may not be sufficiently robust to account for variations over time. Our study found that delta radiomic features had higher predictive value than did radiomic features extracted from a single time point. Our approach may also be useful as a noninvasive way of determining the status of an enlarging lesion after radiosurgery as well as aiding the choice of therapy once a lesion has been detected (e.g., surgical resection versus conservative medical management).
Our study had some limitations. First, the progression group had two times more data samples than the necrosis group. This imbalance greatly skewed the outcome of feature modeling for most classifiers limiting the predictive capability and accuracy. The limited numbers of lesions (and patients) reflected the nature of the treatment. We plan to add more data samples to our model when new patient data become available. Second, although patients were asked to return for scheduled follow-up visits, the follow-up interval varied among patients, and thus the time separation for calculating the delta radiomic features was different among patients. In an attempt to mitigate this limitation, we scaled the time interval to 30 days for all patients, under the assumption that if the radiomic features changed, that change would be linear during that period. This may be an oversimplification; although the assumption is reasonable, it may not be consistent with the actual changes in features, which are unknown at this time. Including data samples at additional time points could potentially address this problem. Third, uncertainties were present in our analysis that could have affected the outcomes. The boundaries of the actual lesion were not known. Lesion segmentation was based on the best judgment of the radiation oncologists and could have varied among individuals. Unlike CT images, MR image values are not calibrated, and thus the same tissue could appear different on different MRI systems. Although some have proposed ways of normalizing MR intensity values [35], the efficacy of normalization is not known. Hence rather than using normalization, we chose threshold values for intensity of the lesion volume based on our experience, acknowledging that contrast-enhanced MR scans might not be suitable for intensity normalization. Finally, implementation of current radiomic analysis at clinical routine is still difficult because it involves complex computational steps with frequent human interactions. In addition, radionomic features differentiating tumor progression and necrosis might also be dependent of the tumor origin and biology. Currently, a distinctive analysis of radiomic features in correlation to their tumor origin is not available. This will be an important topic of our future studies.
In conclusion, we developed a prediction tool using changes in radiomic features extracted from MR scans to distinguish radiation necrosis from tumor progression after brain radiosurgery for metastatic lesions. We found that combination of several delta radiomic features from both T1-weighted post-contrast and T2-weighted MR scans gave rise to the best prediction performance in a RUSBoost ensemble model. This tool may aid decision-making regarding the choice of surgical resection versus conservative medical management for patients suspected of having progression or necrosis after Gamma Knife radiosurgery for brain metastases.
Key points.
Some radiomic features showed better reproducibility for progressive lesions than necrotic ones
Delta radiomic features can help to distinguish radiation necrosis from tumor progression
Delta radiomic features had better predictive value than did traditional radiomic features
Acknowledgments
The authors would like to thank Christine Wogan for reviewing this manuscript. This work was funded in part by Cancer Center Support (Core) Grant CA016672 from the National Cancer Institute, National Institutes of Health, to The University of Texas MD Anderson Cancer Center.
Abbreviation and acronyms
- AUC
Area under the curve
- CCC
Concordance correlation coefficient
- COM
Co-occurrence matrix
- FLAIR
Fluid-attenuated inversion recovery
- HOG
Histogram of oriented gradients
- IBEX
Imaging Biomarker Explorer
- MRI
Magnetic resonance imaging
- NGTDM
Neighborhood gray-tone difference matrix
- RLM
Run length matrix
- ROC
Receiver operating characteristic
- T1c
T1 weighted post-contrast
Footnotes
Meeting presentations: This work was presented in part at the 2016 American Association of Physicists in Medicine Annual Meeting.
References
- 1.Nayak L, Lee EQ, Wen PY. Epidemiology of brain metastases. Current Oncology Reports. 2012;14:48–54. doi: 10.1007/s11912-011-0203-y. [DOI] [PubMed] [Google Scholar]
- 2.Kondziolka D, Martin JJ, Flickinger JC, et al. Long-term survivors after gamma knife radiosurgery for brain metastases. Cancer. 2005;104:2784–2791. doi: 10.1002/cncr.21545. [DOI] [PubMed] [Google Scholar]
- 3.Elaimy AL, Mackay AR, Lamoreaux WT, et al. Clinical outcomes of stereotactic radiosurgery in the treatment of patients with metastatic brain tumors. World Neurosurg. 2011;75:673–683. doi: 10.1016/j.wneu.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Minniti G, Clarke E, Lanzetta G, et al. Stereotactic radiosurgery for brain metastases: analysis of outcome and risk of brain radionecrosis. Radiation Oncology. 2011;6:48. doi: 10.1186/1748-717X-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw E, Scott C, Souhami L, et al. Single dose radiosurgical treatment of recurrent previously irradiated primary brain tumors and brain metastases: Final report of RTOG protocol 90-05. International Journal of Radiation Oncology Biology Physics. 2000;47:291–298. doi: 10.1016/s0360-3016(99)00507-6. [DOI] [PubMed] [Google Scholar]
- 6.Chuang MT, Liu YS, Tsai YS, Chen YC, Wang CK. Differentiating Radiation-Induced Necrosis from Recurrent Brain Tumor Using MR Perfusion and Spectroscopy: A Meta-Analysis. PLoS ONE. 2016:11. doi: 10.1371/journal.pone.0141438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai G, Mahadevan A, Hackney D, et al. Diagnostic Accuracy of PET, SPECT, and Arterial Spin-Labeling in Differentiating Tumor Recurrence from Necrosis in Cerebral Metastasis after Stereotactic Radiosurgery. American Journal Of Neuroradiology. 2015;36:2250–2255. doi: 10.3174/ajnr.A4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature Communications. 2014:5. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proceedings of the National Academy of Sciences. 2015;112:E6265–E6273. doi: 10.1073/pnas.1505935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273:168–174. doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itakura H, Achrol AS, Mitchell LA, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Science translational medicine. 2015;7:303ra138–303ra138. doi: 10.1126/scitranslmed.aaa7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto S, Han W, Kim Y, et al. Breast cancer: radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology. 2015;275:384–392. doi: 10.1148/radiol.15142698. [DOI] [PubMed] [Google Scholar]
- 14.Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W. PET-CT image registration in the chest using free-form deformations. IEEE Transactions On Medical Imaging. 2003;22:120–128. doi: 10.1109/TMI.2003.809072. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: An open infrastructure software platform to facilitate collaborative work in radiomics. Medical Physics. 2015;42:1341–1353. doi: 10.1118/1.4908210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Zhang L, Fave XJ, et al. Uncertainty analysis of quantitative imaging features extracted from contrast-enhanced CT in lung tumors. Comput Med Imaging Graph. 2016;48:1–8. doi: 10.1016/j.compmedimag.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burger W, Burge MJ. Edge-Preserving Smoothing FiltersPrinciples of Digital Image Processing. Springer; 2013. pp. 119–167. [Google Scholar]
- 18.Cunliffe AR, Armato SG, III, Fei XM, Tuohy RE, Al-Hallaq HA. Lung texture in serial thoracic CT scans: Registration-based methods to compare anatomically matched regionsa) Medical Physics. 2013;40:061906. doi: 10.1118/1.4805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fried DV, Tucker SL, Zhou S, et al. Prognostic Value and Reproducibility of Pretreatment CT Texture Features in Stage III Non-Small Cell Lung Cancer. International Journal of Radiation Oncology*Biology*Physics. 2014;90:834–842. doi: 10.1016/j.ijrobp.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE T Syst Man Cyb. 1973;3(6):610–621. [Google Scholar]
- 21.Tang X. Texture information in run-length matrices. IEEE Transactions on Image Processing. 1998;7:1602–1609. doi: 10.1109/83.725367. [DOI] [PubMed] [Google Scholar]
- 22.Legland D, Kiêu K, Devaux M-F. Computation of Minkowski measures on 2D and 3D binary images. Image Anal Stereol. 2007;26:83–92. [Google Scholar]
- 23.Basu S. Developing predictive models for lung tumor analysis 2012 [Google Scholar]
- 24.Amadasun M, King R. Textural Features Corresponding to Textural Properties. Ieee Transactions on Systems Man and Cybernetics. 1989;19:1264–1274. [Google Scholar]
- 25.Dalal N, Triggs B. Histograms of oriented gradients for human detection. IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR); 2005. pp. 886–893. [Google Scholar]
- 26.Tiwari P, Prasanna P, Wolansky L, et al. Computer-Extracted Texture Features to Distinguish Cerebral Radionecrosis from Recurrent Brain Tumors on Multiparametric MRI: A Feasibility Study. AJNR Am J Neuroradiol. 2016 doi: 10.3174/ajnr.A4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galizia MS, Töre HG, Chalian H, McCarthy R, Salem R, Yaghmai V. MDCT necrosis quantification in the assessment of hepatocellular carcinoma response to yttrium 90 radioembolization therapy: comparison of two-dimensional and volumetric techniques. Academic Radiology. 2012;19:48–54. doi: 10.1016/j.acra.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Lin LIK. A Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- 29.Duda RO, Hart PE, Stork DG. Pattern Classification. John Wiley and Sons, Inc; 2001. [Google Scholar]
- 30.Seiffert C, Khoshgoftaar TM, Van Hulse J, Napolitano A. RUSBoost: A Hybrid Approach to Alleviating Class Imbalance. Ieee Transactions on Systems Man and Cybernetics Part a-Systems and Humans. 2010;40:185–197. [Google Scholar]
- 31.Barajas RF, Chang JS, Segal MR, et al. Differentiation of Recurrent Glioblastoma Multiforme from Radiation Necrosis after External Beam Radiation Therapy with Dynamic Susceptibility-weighted Contrast-enhanced Perfusion MR Imaging. Radiology. 2009;253:486–496. doi: 10.1148/radiol.2532090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kickingereder P, Burth S, Wick A, et al. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology. 2016;280:880–889. doi: 10.1148/radiol.2016160845. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Zhu Y, Burnside ES, et al. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology. 2016:152110. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larroza A, Moratal D, Paredes-Sánchez A, et al. Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. Journal of Magnetic Resonance Imaging. 2015;42:1362–1368. doi: 10.1002/jmri.24913. [DOI] [PubMed] [Google Scholar]
- 35.Shinohara RT, Sweeney EM, Goldsmith J, et al. Statistical normalization techniques for magnetic resonance imaging. NeuroImage: Clinical. 2014;6:9–19. doi: 10.1016/j.nicl.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]