Abstract
Foods implicated in human campylobacteriosis include raw or undercooked poultry and raw dairy products. Because Campylobacter spp. are the most frequently reported cause of bacterial infection in the European Union and because conventional methods are cumbersome, rapid methods for Campylobacter detection and quantification in food are needed. With this study we sought to validate, according to the standard procedure (UNI EN ISO 16140:2003), an alternative to the reference analytical method (UNI EN ISO 10272-1:2006) for official controls of Campylobacter spp. in raw milk and dairy products. Milk samples collected from 16 milk vending machines located throughout the Genoa metropolitan area were analyzed using two different methods, an enzyme-linked fluorescent assay (ELFA) and a real-time PCR assay, and evaluated in parallel against the reference method. In addition, a total of 460 samples of raw milk collected from milk vending machines were analyzed by ELFA. Results obtained with ELFA showed it was compliant with UNI EN ISO 10272-1:2006 criteria and that the immunoassay had 100% sensitivity, specificity, and accuracy. Regarding samples of milk vending machines, 5.0% (23/460) tested positive at ELFA screening and were subsequently confirmed as C. jejuni. Validation according to UNI EN ISO 16140:2003 of the ELFA method suggests it may be a useful alternative to conventional methods for detecting Campylobacter spp. in official controls.
Key words: Campylobacter spp, ELFA, RT-PCR, Validation, Dairy products, ISO 16140:2003
Introduction
Campylobacter spp. are recognized as the bacterial agent responsible for campylobacteriosis, a zoonotic disease. According to the latest report of the European Food Safety Authority (EFSA), campylobacteriosis is the most frequently reported human gastrointestinal bacterial infection in the European Union (EU): in 2016 about 246,000 human cases were reported, incrementing of 6.1% the cases than 2015. Furthermore, 461 foodborne outbreaks were reported in Europe in 2016 with 4606 outbreak-related cases (EFSA and ECDC, 2017).
Campylobacters are transmitted via the oral-fecal route, mainly by the ingestion of contaminated food or water. The species most commonly associated with human infections are Campylobacter jejuni, followed by C. coli, and C. lari, though other species, including the non-thermophilic C. fetus, have occasionally been reported to cause human diseases (EFSA, 2015; Rodrigues et al., 2015).
Recent trends in food consumption indicate an increasing preference for local products (short food supply chain) and a growing demand for fresh or minimally processed food products as well as meals ready-to-eat (EFSA Journal 2012). The increasing incidence of foodborne bacterial infections is thought to be related to these trends (EFSA Journal, 2015). Foods implicated in campylobacteriosis include raw or undercooked poultry and raw dairy products. In this respect, at EU level 1896 sampled units were analyzed to detect Campylobacter spp. in milk and milk products (including cheeses) in 2016 with the occurrence around 1%. In the outbreaks associated with the consumption of milk, cheese and dairy products Campylobacter was the causative agent with strong-evidence (22.2%) (EFSA and ECDC, 2017).
The risk of infection is high due to the minimum infecting dose of 500 colony-forming units (CFU)/sample (Stern and Robach, 2003). While pasteurization inactivates Campylobacter spp. in few seconds, the bacterium is quite resistant to lower temperatures, indeed they are able to survive for several weeks in refrigerated milk and water (Haughton et al., 2012).
The reference analytical method for the detection of Campylobacter spp. in dairy products is UNI EN ISO 10272-1:2006, a classical four-step plate isolation procedure that entails sample preparation, isolation, confirmatory testing (Morphology/Motility, Oxidase, Microaerobic growth at 25ºC, Aerobic growth at 41,5ºC), and bacterial identification (Catalase, Hippurate, Indoxyl acetate, Nalidixic acid, Cephalothin). Conventional detection methods are slow and may lack in accuracy because of the complex growth requirements of Campylobacter (Melero et al., 2011) involving prolonged incubation time and selective enrichment to reduce the development of background flora. Moreover, test sensitivity, especially in milk samples, is reduced due to improper sample handling, transport, and storage conditions, which inevitably affect strain viability. Alternative detection procedures such as enzyme-linked immunoassay and/or molecular tests are needed to detect foodborne pathogens more rapidly and accurately and ensure food safety. As stated by EC Regulation No 2073/2005, the use of alternative analytical methods may be authorized if they are validated according to UNI EN ISO 16140:2003 or other internationally accepted protocols. ISO 16140:2003 establishes the general principle and technical characteristics for the validation of alternative methods for the microbiological analysis of food, animal feeding stuff, and environmental and animal specimens. The two step-process entails: validation of alternative methods for use in official controls; and international approval of the results obtained by the alternative method. Diverse real-time PCR assays have been developed to detect Campylobacter spp. in chicken carcasses (Botteldoorn et al., 2008), fresh chicken meat and by-products (Saiyudthong et al., 2015), and foods (Vencia et al., 2014). Although an alternative AOAC-certified procedure to detect Campylobacter from chicken carcass and carcass sponges from turkey is available, currently there are few alternative methods for the detection of Campylobacter spp. in dairy products. Such alternative methods are mainly PCR based methods with high sensitivity and low specificity; this cause increase of analysis time.
ELFA method was investigated in different study for detection of several foodborne pathogens. ELFA method is reported as alternative procedure to detect Salmonella spp. in raw chicken meat (Rohonczy et al., 2014), Listeria monocytogenes in various food samples (Ueda and Kuwabara, 2010), Escherichia coli O157 in mincemeat (Stefan et al., 2007).
The aim of this study was to evaluate an alternative method to UNI EN ISO 10272-1:2006 that could improve the performance of Campylobacter spp. diagnostic protocols by reducing analysis time. Afterward the method with high performances was validated, according to the standard procedure (UNI EN ISO 16140:2003).
Materials and Methods
Study design
Two different methods were evaluated in parallel against the reference method (UNI EN ISO 10272-1:2006), an enzyme-linked fluorescent assay (ELFA) and a real-time PCR assay, in milk samples collected from 16 milk vending machines throughout the Genoa metropolitan area. Three milk samples testing negative for Campylobacter were spiked with a 50 CFU/mL suspension of Campylobacter jejuni (ATCC 29428). Each sample was divided into 11 aliquots and stored at 4±2°C. An aliquot of each sample was used for pH control; the others were kept refrigerated: T0 (baseline), T1 (1 h), T6 (6 h), T24 (24 h), T30 (30 h), T48 (48 h), T144 (6 days), T192 (8 days), T240 (10 days), and T312 (13 days). Refrigeration temperature and times were based on published data (Jinlin et al., 2011). The samples were processed with enrichment as it follows: 25 mL of milk was placed in Campylobacter enrichment broth (CEB) (Biolife, Milan, Italy, cod. 401286B) supplemented with 12.5 mL of horse blood lysate (Biolife, cod. 90HLX100) and 2.5 mL of Campylobacter Bolton selective supplement (Microbiol, Uta-Cagliari, Italy, cod. 76147), and incubated under microaerophilic conditions at 37±1°C for 4-6 h and then at 41.5±1°C for 44 ±4 h using jars with atmosphere generation system (CampyGen Oxoid™, Oxoid Ltd, Basingstoke, UK, cod. CN0035A). The samples were analyzed using the three analytical methods to detect Campylobacter spp. (Table 1).
Table 1.
Details of the aliquots analyzed and analytical methods applied (h: hours of refrigeration): 3 milk samples spiked with Campylobacter jejuni.
| Aliquots for each sample | Analytical methods |
|---|---|
| pH control | |
| T0 (baseline) | |
| T1 (1h) | |
| T6 (6h) | |
| T24 (24h) | ISO (UNI EN ISO 10272-1:2006) Reference Method |
| T30 (30h) | Real-Time PCR |
| T48 (48h) | Enzyme-Linked Fluorescent Assay (ELFA) |
| T144 (6 days) | |
| T192 (8 days) | |
| T240 (10 days) | |
| T312 (13 days) |
ISO (UNI EN ISO 10272-1:2006) reference method
The reference analytical method represents a classical four-step procedure for Campylobacter spp. detection and enumeration in food and animal feeding stuffs: 1) sample preparation, 2) enrichment, 3) isolation, and 4) confirmatory testing. In order to detect the presence of colonies likely referable to Campylobacter spp., enrichment is followed by seeding the sample on plates of modified charcoal cefoperazonedeoxycholate agar (mCCDA) and Campy Food Agar (CFA) incubated in microaerobic conditions at 41.5±1°C. Suspected colonies are selected from each plate, seeded in Columbia agar, and incubated in microaerophilic conditions at 41.5±1°C. These pure cultures are then used to confirm the presence of Campylobacter spp. by the following assays: a) typical bacteria morphology and motility test; b) incubation at 25±1°C in microaerobiosis for about 44±4 h; c) incubation at 41.5±1°C in aerobiosis for approximately 44±4 hs; d) oxidase test. The presence of Campylobacter spp. is confirmed based on: positive oxidase reaction, typical motility and morphology, absence of growth at 25°C in microaerobiosis and at 41.5°C in aerobiosis.
Real-Time PCR
DNA was extracted in 1 mL of the enrichment broth using a QIAamp Mini kit® (Qiagen®, Milan, Italy, cat. 51306) following the manufacturer’s recommendations. For the real-time PCR assay, amplification of a specific gene of C. jejuni gene encoding hippuricase was performed using the primers (F:5’CGGGATAGTTATAGTATTGAAGTT ATTGG3’;R:5’GAAGGAGCATAATAGGA TCTTG3’) (Zhang et al., 2013) and two different amplification mixes based on SYBR Green® (RealMasterMix Fast SYBR™ 2X, 5Prime, ThermoFisher Scientific, Rodano, Italy) and EvaGreen® (SsoFast™ EvaGreen® Supermix, Bio-Rad Laboratories, Milan, Italy), respectively; two DNA intercalators widely used in molecular biology as an alternative to the classical system of the probe (Razzuoli et al., 2011) were applied. Amplification was carried out on a CFX96™ real-time PCR system (BioRad) using the temperature profile described in Razzuoli et al., 2013.
Enzyme-linked fluorescent assay (ELFA)
ELFA is an immunological analytical method that detects antigens and measures the concentration of antibodies in blood plasma. Among the different immunoassay methods, ELFA refers to heterogeneous phase systems in which the antibodies or antigens are adsorbed or bound to a solid substrate (O'Keeffe et al., 2000). The ELFA based on commercial instrument is entirely automated and it is associated ready to use kits. The total enrichment time is 44-52 h and the time of run is 70 minutes (bioMérieux, France). In the present study, the ELFA MiniVIDAS® Campylobacter kit (ELFA CAM) (bioMèrieux, Marcy l’Etoile, France, cat. 30111) was used according to the manufacturer’s instructions, with slight modifications: Bolton broth was prepared according to UNI EN ISO 10272-1:2006 for the enrichment step instead of the suggested enrichment broth.
Statistical analysis
Each test was performed in triplicate. The results obtained by the culture method are expressed qualitatively as presence/absence. For the molecular methods, the microbial challenge tests were normalized by adding 4 ng of DNA to each amplification reaction. Results are expressed as the mean Ct amplification and Delta Ct (Ct at time 0 - Ct refrigeration time) (Talaat et al., 2002). The results obtained with the ELFA method are expressed in TV (relative fluorescence values of the sample/relative fluorescence values of positive control). Samples with TV >0.10 were considered as positive. Within the same group (method detecting presence/absence of Campylobacter), the differences between the means were evaluated using ANOVA (GraphPad Prism 5.03, GraphPad Software, San Diego, CA, USA). The threshold of significance was set at 0.05 (P<0.05).
Validation protocol
Validation according to the standard procedure (UNI EN ISO 16140:2003) of a qualitative method alternative to a reference method is allowed. Two alternative methods to the reference analytical method (UNI EN ISO 10272-1:2006) for the detection of Campylobacter in milk were compared to determine: limit of detection (LOD), relative sensitivity, accuracy, specificity, relative detection level, inclusiveness, and exclusivity. In order to assess the LOD, serial dilutions were prepared from a suspension of C. jejuni (ATCC 29428) 0.5 McFarland (corresponding to 1.5*108 CFU/mL) in physiological solution. A total of 13 serial dilutions were prepared at concentrations of: 1.5*107, 1.5*106, 1.5*105, 1.5*104, 1.5*103, 150, 15, 10, 5, 2, 1, 0.5, and 0.25 CFU/mL. A raw milk sample testing negative for Campylobacter according to the classical method was used to determine the LOD. Each sample was contaminated with 1 mL of a bacterial suspension previously prepared and tested by both the ELFA and the reference analytical method. Method sensitivity, specificity, and accuracy were tested in total of 104 samples of milk and dairy products: 70 milk samples and 34 cheese samples, 45 of which were artificially contaminated with C. jejuni. Comparison tests were performed on these 45 samples:10 samples spiked with a concentration equal to the LOD,13 at LOD*10, 15 at LOD*20, and 7 at LOD*100. In addition, the relative detection level was tested in 18 raw milk samples: 6 spiked with a concentration equal to the LOD, 6 at LOD*3, and 6 testing negatives for Campylobacter (Table 2). In compliance with UNI EN ISO 16140:2003, each LOD was compared using Fisher’s exact test (PrismGraphPad 5.03).
Table 2.
Details of validation protocol; determination of limit of detection (LOD), relative sensitivity, accuracy, specificity, relative detection level. All samples were tested by ELFA and reference analytical method.
| LOD | 13 serial dilutions | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CFU/mL | 1.5*107 | 1.5*106 | 1.5*105 | 1.5*104 | 1.5*103 | 150 | 15 | 10 | 5 | 2 | 1 | 0.25 |
| 1 mL bacterial suspension + milk sample | ||||||||||||
| Sensitivity specificity accuracy (tot 104 samples) | 10 samples | 13 samples | 15 samples | 7 samples | 59 samples | |||||||
| Spiked level | equal to the LOD | LOD*10 | LOD*20 | LOD*100 | negative | |||||||
| Relative detection level (tot 18 samples) | 6 samples | 6 samples | 6 samples | - | - | |||||||
| Spiked level | equal to the LOD | LOD*3 | negative | - | - | |||||||
Naturally contaminated sample analysis
Between 1 January 2014 and 31 December 2016, the ELFA method was used in our laboratory to analyze raw milk samples collected from milk vending machines. A total of 460 samples were analyzed; all positive samples were confirmed by the reference analytical method.
Results and Discussion
The results of the molecular biology tests support the hypothesis that both fluorophores correctly detected the presence of bacterial genomes (T0 to T312). EvaGreen showed greater sensitivity than SYBRGreen; a difference of 3±1.09 in Ct indicated an equal DNA concentration in all samples (Figure 1). No differences in Ct with respect to refrigeration time were observed (Figure 2). This is consistent with previously published data and was probably due to the higher affinity of EvaGreen for dsDNA, which yields higher fluorescence signals (Razzuoli et al., 2011). Moreover, Campylobacter spp. DNA could still be detected at T312 (13 days of refrigeration) but there was no colony growth on the plate, demonstrating that Campylobacter were no longer viable (Figure 3). ELFA detected the presence of Campylobacter spp. between T0 and T240. The fluorescence signal levels indicated a significant decrease (P<0.001) from T0 to T144; at T48 of refrigeration, the fluorescence signal decreased significantly (P<0.001) through to T312 (Figure 4).
Figure 1.
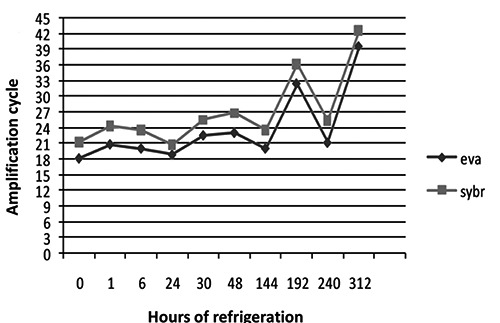
Comparison between fluorophores. Data are expressed as arithmetic means of the amplification cycles (Ct) detected by the two fluorophores tested at different hours of refrigeration: EvaGreen showed greater sensitivity (eva: EvaGreen; sybr: SYBRGreen).
Figure 2.
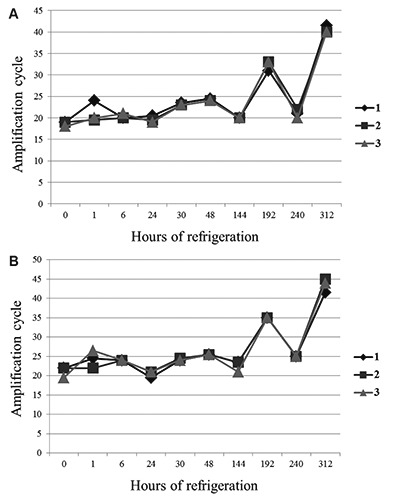
Performance of 3 dropped samples. The increase in amplification cycle (Ct) indicates a decrease in Campylobacter concentration. Data are expressed as the arithmetic means of the Ct of the 3 samples. A) Test using EvaGreen. B) Test using SYBRGreen.
Figure 3.
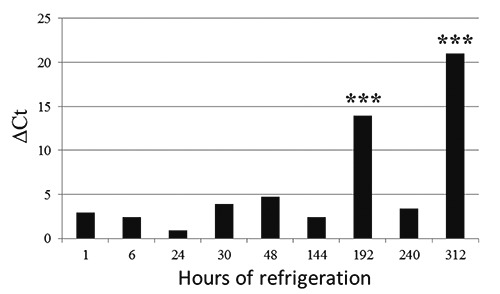
Change in Ct in relation to refrigeration time. ΔCt = difference between TX and T0. ***Statistically significant P<0.0001.
Figure 4.
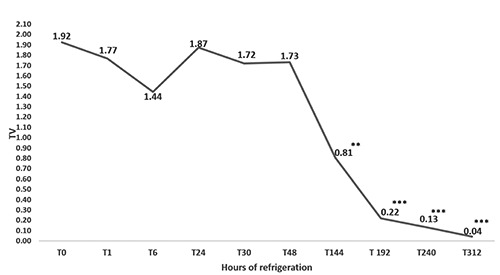
Changes in TV (relative fluorescence values of the sample/relative fluorescence values of positive control) with longer refrigeration time. Statistically significant: *P<0.05; **P<0.001; ***P<0.0001.
Because the real-time PCR assay detected Campylobacter DNA after several days of refrigeration (Figure 3), but the bacteria could not always be isolated on the plate, only the ELFA method was tested to determine whether it was compliant with UNI EN ISO 10272-1:2006. Campylobater were detectable at concentrations from 1.5*107 to 1 CFU/25 mL, demonstrating similar performance for both methods, with a LOD of 1 CFU/25 mL for the ELFA method (Figure 5). There were no differences between the fluorescence values at higher concentrations (from 1.5*107 to 1.5*104); however, the values were drastically decreased between 1500-150 CFU/25 mL (Figure 5). The ELFA method showed 100% sensitivity (IC 95% 0.97-1.00), specificity (IC 95% 0.98-1.00), and accuracy (IC 95% 0.99-1.00). Analysis of the 70 milk samples showed concordant results for 42 samples that tested negative and 28 positive; analysis of the 34 cheese samples showed that 17 tested positive and 17 negative. ELFA and reference method showed the same results for all samples under study. The associated test P-value of Fisher’s exact test was P=1 for all relative detection level levels, suggesting a 100% probability that the positive samples were correctly identified.
Figure 5.
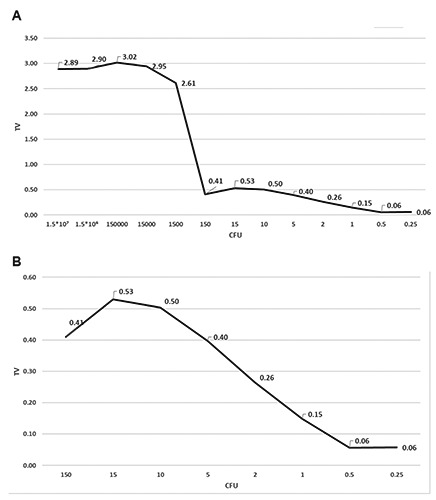
Limit of detection (LOD). Data are expressed in TV (relative fluorescence values of the sample/relative fluorescence values of positive control). A) Campylobater concentrations were detectable from 1.5*107 to 1 CFU/25 g. Concentrations were drastically decreased between 1500-150 CFU/25 mL but were detected until 1 CFU/25 g. B) Concentrations <0.10 TV were considered negative.
Between 1 January 2014 and 31 December 2016, we used the ELFA method for raw milk sample analysis in our food control laboratory. Out of the 460 samples analyzed, 5.0% (23/460) tested positive at ELFA screening and were subsequently confirmed by isolation of the bacterial strain; in all cases C. jejuni was isolated.
Conclusions
Several microorganisms can be present in dairy products (e.g., Campylobacter spp., Salmonella spp., Listeria monocytogenes, verocytotoxin-producing Escherichia coli) coming from animal reservoirs, and which can outcome important sources of foodborne illness.
However, one of the major disadvantages of DNA based techniques is their inability to distinguish between DNA from viable and dead cells (Nocker et al., 2006), which is probably the most important obstacle for implementation of these methods in routine applications.
Results of the present study suggest that ELFA correctly identified Campylobacter in the artificially contaminated samples; its performance was comparable to that of the reference analytical method (ISO 10272-1:2006) as validated with the standard procedure. Validation of the ELFA method for the detection of Campylobacter spp. in official controls is a fundamental step to reduce the lengthy analysis time and the costs of conventional methods. With the ELFA method, we were able to shorten analysis time for the detection of Campylobacter spp., which was particularly advantageous for the analysis of suspected samples and it may benefit the entire food trade.
Funding Statement
Funding: none.
References
- Botteldoorn N, Van Coillie E, Piessens V, Rasschaert G, Debruyne L, Heyndrickx M, Herman L, Messens W, 2008. Quantification of Campylobacter spp. in chicken carcass rinse by real-time PCR. J Appl Microbiol 105:1909-18. [DOI] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2017. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J 15:5077, 228 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control), 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J 13:4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA Journal, 2012. EU Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2010. EFSA J 10:2597. [Google Scholar]
- Haughton PN, Lyng J, Cronin D, Fanning S, Whyte P, 2012. Effect of crust freezing applied alone and in combination with ultraviolet light on the survival of Campylobacter on raw chicken. Food Microbiol 32:147-52. [DOI] [PubMed] [Google Scholar]
- Melero B, Cocolin L, Rantsiou K, Jaime I, Rovira J, 2011. Comparison between conventional and qPCR methods for enumerating Campylobacter jejuni in a poultry processing plant. Food Microbiol 28:1353-8. [DOI] [PubMed] [Google Scholar]
- Nocker A, Cheung C, Camper A, 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods 67:310-20. [DOI] [PubMed] [Google Scholar]
- O’Keeffe M, 2000. Residue analysis in food. Principles and applications. 1st ed., Harwood Academic Publishers O’Keeffe M, Amsterdam (The Netherlands). [Google Scholar]
- Razzuoli E, Villa R, Amadori M, 2013. IPEC-J2 cells as reporter system of the anti-inflammatory control actions of interferon-alpha. J Interferon Cytokine Res 33:597-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzuoli E, Villa R, Sossi E, Amadori M, 2011. Reverse transcription real-time PCR for detection of porcine interferon α and β genes. Scand J Immunol 74:412-8. [DOI] [PubMed] [Google Scholar]
- Rodrigues RC, Pocheron AL, Hernould M, Haddad N, Tresse O, Cappelier JM, 2015. Description of Campylobacter jejuni Bf, an atypical aero-tolerant strain. Gut Pathog 19:7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohonczy K, Zoller L, Hermann Z, Fodor A, Mráz B, Tabajdi-Pintér V, 2014. Comparison of an automated ELFA and two different real-time PCR techniques for Salmonella detection in poultry samples. Acta Microbiol Immunol Hung 61:261-72. [DOI] [PubMed] [Google Scholar]
- Saiyudthong S, Phusri K, Buates S, 2015. Rapid Detection of Campylobacter jejuni, Campylobacter coli, and Campylobacter lari in Fresh Chicken Meat and By-Products in Bangkok, Thailand, Using Modified Multiplex PCR. J Food Prot 78:1363-9. [DOI] [PubMed] [Google Scholar]
- Stefan A, Scaramagli S, Bergami R, Mazzini C, Barbanera M, Perelle S, Fach P, 2007. Real-time PCR and enzyme-linked fluorescent assay methods for detecting Shiga-toxin-producing Escherichia coli in mincemeat samples. Can J Microbiol 53:337-42. [DOI] [PubMed] [Google Scholar]
- Stern NJ, Robach MC, 2003. Enumeration of Campylobacter spp. in broiler faeces and in corresponding processed carcasses. J Food Prot 66:1557-63. [DOI] [PubMed] [Google Scholar]
- Talaat AM, Howard ST, Hale W, Lyons R, Garner H, Johnston SA, 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda S, Kuwabara Y. Evaluation of an enzyme-linked fluorescent assay for the detection of Listeria monocytogenes from food. Biocontrol Sci 2010;15:91-5. [DOI] [PubMed] [Google Scholar]
- Vencia W, Nogarol C, Bianchi DM, Gallina S, Zuccon F, Adriano D, Gramaglia M, Decastelli L, 2014. Validation according to ISO 16140:2003 of a commercial realtime PCR-based method for detecting Campylobacter jejuni, C. coli, and C. lari in foods. Int J Food Microbiol 177:78-80. [DOI] [PubMed] [Google Scholar]


