Abstract
Purpose: Early diagnosis of cancer enables extended survival and reduced symptoms. To this end, a “three-in-one” nanohybrid of MOF@AuNP@GO is designed as synergistic nanoquencher to develop a novel fluorescence biosensor for rapid and sensitive detection of cancer-related biomarkers.
Methods: The ssDNA absorption affinities and fluorescence quenching abilities of the MOF@AuNP@GO were evaluated using FAM-labeled single-stranded DNA (ssDNA). Then, two specific dye-labeled ssDNA and aptamer probes were designed for the recognition of p53 gene and prostate specific antigen (PSA), respectively. Fluorescence spectra were recorded and ratiometric signal processing was performed.
Results: The designed nanohybrids exhibit enhanced ssDNA binding affinities and fluorescence quenching abilities, which significantly decrease the background signal and increase the signal-to-noise (S/N) ratio, thus lowering the detection limit (LOD). Accordingly, with ratiometric measurement, this developed nanosensor can sensitively measure p53 gene and PSA with LODs of 0.005 nM and 0.01 ng mL-1, respectively. Besides, this method also displays excellent performances with respect to universality, multiplexed detection, specificity, and practicality in human serum.
Conclusion: The designed MOF@AuNP@GO-based fluorescence biosensor can serve as a promising platform for washing-free, rapid and sensitive measurement of cancer biomarkers, making this method well-suited for point-of-care (POC) diagnosis.
Keywords: nanohybrid, nanoquencher, fluorescence, no-wash biosensor, ratiometric, cancer biomarker
Introduction
Early diagnosis of cancer can significantly improve the chances of successful early treatment, and provide good outcomes for cancer patients, including extended survival and reduced symptoms.1-4 Among the existing strategies that have been employed for early detection of cancer-related biomarkers, fluorescence-based biosensors with high sensitivity and multiplex detection capacity have been extensively explored and proposed.5-7 Among the developed biosensors, nanoquencher-based fluorescence biosensors have been widely applied for measuring various tumor biomarkers, such as nucleic acids and proteins, because of their strong fluorescence quenching and no wash workflow.8-11 Currently, the most commonly used nanoquenchers include gold nanoparticles (AuNPs),12, 13 carbon nanotubes (CNTs),14, 15 graphene oxide (GO),16, 17 MoS2 nanosheets,18, 19 and g-C3N4 nanosheets.20-22 These nanoquenchers exhibit high binding affinity to dye-labeled single-stranded DNA (ssDNA) or aptamer molecules to bring dye molecules and nanoquenchers into close vicinity, thereby causing strong fluorescence quenching of dye molecules through energy transfer and electron transfer processes. Despite the great progress made in this field, there still remain several challenges, including suboptimal sensitivity and relatively low reliability with single-intensity signal output.
Metal-organic frameworks (MOFs) are highly ordered crystalline materials with fascinating structures and intriguing features of flexible porosity, high surface area, and uniform open cavities.23, 24 MOFs have been explored for gas storage, separations, catalysis, nanoscale reactors, as well as bioimaging and drug delivery.25-30 Recently, MOFs have also been used as nanoquenchers for rational design of fluorescence biosensors.31, 32 Similar to traditional nanoquenchers, MOFs can directly adsorb dye-labeled ssDNA molecules through electrostatic, π-π stacking, or hydrogen bond interactions. The attached dye molecules encounter strong fluorescence quenching based on the photoinduced electron-transfer (PET) process.33 Several MOF-based nanoquenchers, such as zeolitic imidazolate framework (ZIF)-8,34 H2dtoaCu,35, 36 UiO-66-NH2,37 MIL-101,38 and MIL-88B,39 have been designed and fabricated to develop fluorescence biosensors for target DNA detection. In these developed MOFs, the ligands with a rich π-electron system were chosen to allow for binding between dye-labeled DNA probes and the metal ions, whereas these metals were designed as coordination centers and exhibited intrinsic fluorescence quenching ability via PET processes. Nevertheless, these reported MOF-based nanoquenchers presented insufficient fluorescence quenching efficiency towards dye molecules that resulted in relatively low detection sensitivity. As signal-to-noise (S/N) ratio determines the detection sensitivity of fluorescence biosensors, how to design new MOF nanostructures with higher fluorescence quenching ability to decrease background noise signal and augment the S/N ratio is thus of significance to increase detection sensitivity.
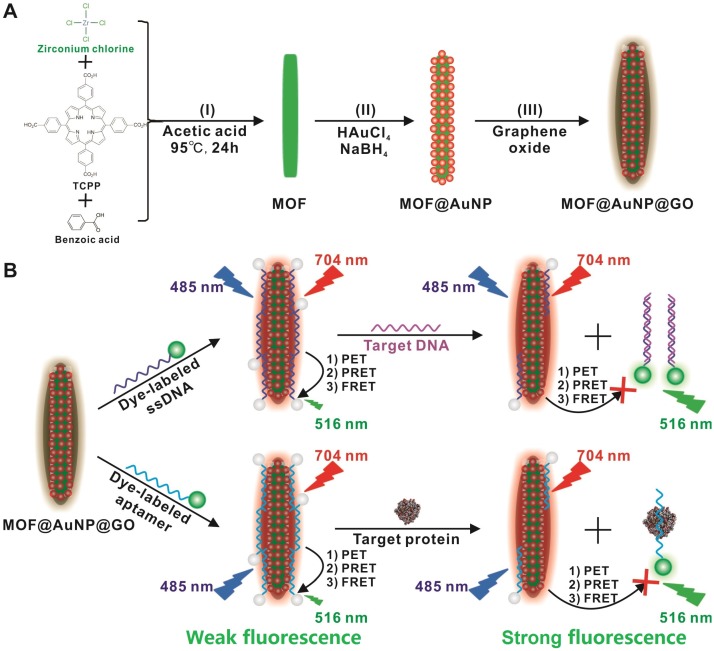
Compared with single-component nanomaterials, hybrid nanomaterials provide more advantages of enhanced physicochemical and optical properties that can serve as promising platforms for improved biosensing, imaging, and therapeutic applications.40 In this work, we designed and applied a “three-in-one” nanohybrid system as a synergistic nanoquencher to develop a highly-sensitive fluorescence biosensor for cancer biomarker detection (Scheme 1). The proposed nanohybrid, namely, MOF@AuNP@GO, consists of three common quenching nanomaterials, including zirconium (Zr)-based MOF nanostructures for quenching via PET process35-37, 41, 42 and meanwhile offering a reference signal to realize ratiometric measurements, gold nanoparticles (AuNPs) for quenching by plasmonic resonance energy transfer (PRET), and graphene oxide (GO) for quenching based on fluorescence resonance energy transfer (FRET) and endowing higher colloidal stability, respectively. The designed hybrid MOF@AuNP@GO nanocomposites are constructed using Zr-based MOF as a template for in situ growth of AuNPs to form the MOF@AuNP nanostructure, and then an ultrathin GO layer is coated onto the MOF@AuNP surface to produce MOF@AuNP@GO via strong π-π stacking and hydrogen bond interactions. The as-prepared MOF@AuNP@GO demonstrates significantly increased binding affinity and quenching ability for dye-labeled ssDNAs or aptamers based on π-π stacking, van der Waals force, or hydrogen bonding, which can largely shorten the overall detection time and reduce the background noise for improved detection sensitivity. Also, the MOF@AuNP@GO can provide a stable NIR emission from tetrakis(4-carboxyphenyl)porphyrin (TCPP) ligand that is unchanged with altering target concentrations, which can act as a reference signal to the sensing signal from the dye molecules to allow for ratiometric measurement. With ratiometric strategies, the biosensor sensitivity and reliability are further improved owing to the increased S/N ratio and decreased susceptibility against analyte-independent interference factors, including inhomogeneous excitation and emission, background light scattering, as well as photobleaching.43-47 In brief, the proposed MOF@AuNP@GO nanohybrid can serve as a promising alternative for the development of nanoquencher-based fluorescence biosensors with high sensitivity and reliability.
Scheme 1.
(A) Schematic representation for MOF@AuNP@GO synthesis, and (B) Schematic diagrams for fluorescence recovery-based ratiometric no-wash biosensor for target DNA and protein detection by using MOF@AuNP@GO as synergistic nanoquencher.
Results and Discussions
Synthesis and characterization of MOF@AuNP@GO
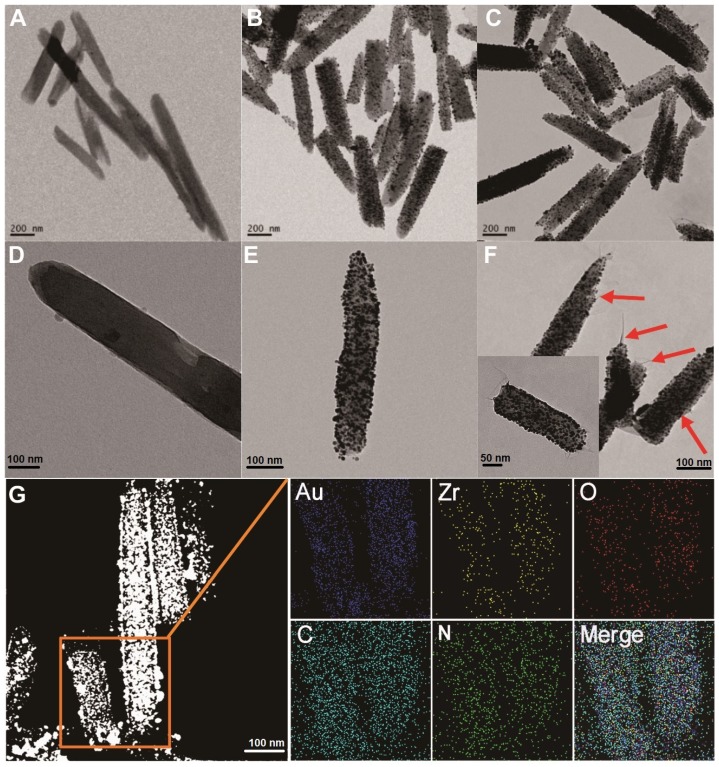
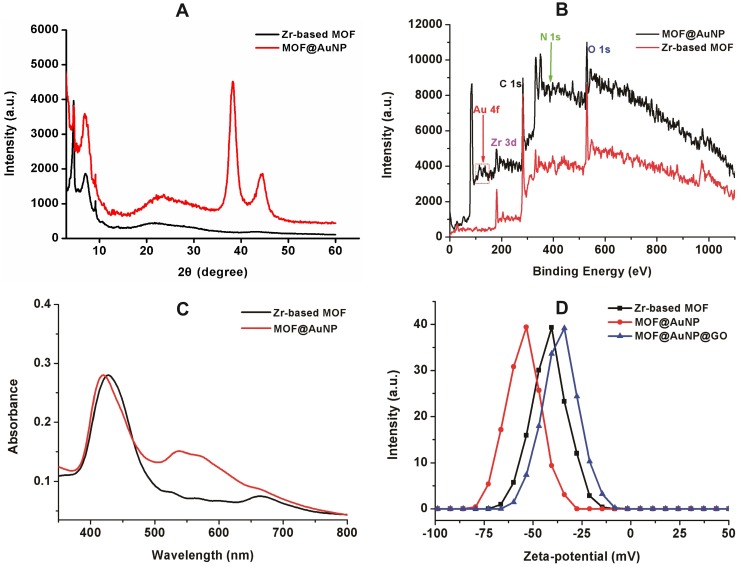
To prepare the MOF@AuNP@GO nanohybrid, Zr-based MOF nanostructures were first synthesized by using TCPP as a ligand, and Zr4+ as a metal coordination center and open site due to the strong coordination between Zr4+ cations and COO- anions. As shown in Figure 1A and 1D, the as-synthesized Zr-based MOF nanoarchitectures exhibit a rod-like single crystal structure, with a length of approximately 500 nm. The powder XRD characterization of Zr-based MOF presents three typical diffraction peaks at 4.8°, 7.1°, and 9.8° (Figure 2A), which is in accordance with the previous reports.48 Besides, the XPS spectrum for Zr-based MOFs indicates the presence of C, N, O, and Zr (Figure 2B). Collectively, these results confirm successful preparation of the Zr-based MOF nanostructures.
Figure 1.
TEM imaging analysis of Zr-based MOF, MOF@AuNP, and MOF@AuNP@GO. (A and D) TEM images of Zr-based MOF, (B and E) TEM images of MOF@AuNP, and (C and F) TEM images of MOF@AuNP@GO, in which the GO layer with filaments was indicated by the red arrows. (G) EDS mapping analysis of the MOF@AuNP@GO nanohybrid.
Figure 2.
Characterization of MOF-based nanostructures. (A) Powder XRD patterns of Zr-based MOF and MOF@AuNP, (B) XPS survey spectrum of Zr-based MOF and MOF@AuNP, (C) UV-Vis absorption spectra of Zr-based MOF and MOF@AuNP, and (D) Zeta-potential analysis of Zr-based MOF, MOF@AuNP and MOF@AuNP@GO.
Then, the Zr-based MOF nonostructures were applied as a template for in situ growth of AuNPs to form MOF@AuNP nanostructures by the classical NaBH4 reduction, with solution color changing from green to red brown. UV-Vis absorption spectra show a shift of the S and Q-band peaks (Figure 2C and Figure S1), further confirming the formation of Au(III) TCPP.24 TEM images show that Zr-based MOFs contain a high density of round Au cross sections, and the AuNP density increases as the amount of HAuCl4 increases (Figure S2). TEM images of the as-prepared MOF@AuNP nanocomposites in Figure 1B and 1E show a high density of uniform AuNPs with average size of ∼4.5 ± 1.1 nm in the Zr-based MOF nanocrystals. As shown in Figure 2D, zeta-potential analysis reveals a decrease of potential from -40.5 mV (MOF) to -55.4 mV (MOF@AuNP). Additionally, the XRD data of the resulting MOF@AuNP nanostructures in Figure 2A indicate the retention of Zr-based MOF structures and appearance of two typical gold peaks, reflecting the formation of crystalline AuNPs, which is further confirmed by XPS spectrum (Figure 2B and Figure S3). These above findings demonstrate the successful nucleation and growth of AuNPs in the Zr-based MOFs, and the synthetic MOF@AuNP nanostructures maintain the crystallinity and structural integrity of original Zr-based MOFs. Later, an ultrathin GO layer was coated onto the surface of MOF@AuNP to prepare the MOF@AuNP@GO nanohybrid based on strong π-π stacking and hydrogen bond interactions. An obvious GO layer with filaments coated on individual MOF@AuNP surface can be clearly seen in the TEM images, as indicated by the red arrows (Figure 1C and 1F), whereas no any thin GO layer was observed on bare MOF@AuNP (Figure 1E). Zeta-potential analysis of the resulting MOF@AuNP@GO showed a significant increase from -55.4 to -30.2 mV after GO coating (Figure 2D). In addition, energy dispersive spectroscopy (EDS) mapping analysis demonstrates the presence of Zr, Au, N, C, and O in the synthetic MOF@AuNP@GO nanocomposite (Figure 1G). Thus, these above results indicate the successful preparation of the designed MOF@AuNP@GO nanohybrid.
Characterization of fluorescence quenching property of MOF@AuNP@GO
The fluorescence quenching ability of the prepared MOF@AuNP@GO was investigated using FAM-labeled ssDNA as a fluorescence probe. To better validate the synergistic quenching effects, MOF alone, MOF@GO, and MOF@AuNP were prepared and used for comparison. Fluorescence quenching capacities of these four MOF-based nanostructures were evaluated by simply incubating FAM-labeled ssDNA (1 nM) with these four nanoquenchers at diverse concentrations, respectively, in which the FAM-labeled ssDNAs can tightly bind onto the nanoquencher surface to cause strong quenching of dye fluorescence. Quenching efficiency (QE) was applied to estimate the quenching capacities, which is calculated based on the following equation: QE (%) = (F0 - F)/F0 × 100%, where F0 and F correspond to the fluorescence intensity of FAM-labeled ssDNA in the absence and presence of nanoquenchers, respectively. Figure S4 indicated the similar fluorescence quenching phenomena in these four MOF-based nanoquenchers, where the QE greatly increased with increasing nanoquencher amount. In following, the value of QE50 was adopted to compare the fluorescence quenching ability, where QE50 is the required nanoquencher amount with fluorescence QE at 50%. As shown in Figure S4, the MOF@AuNP@GO nanohybrid shows the lowest used amount at 1.5 μL for QE50 compared to the other three nanomaterials (MOF at 9.2 μL, MOF@GO at 4.9 μL, and MOF@AuNP at 2.7 μL, respectively), which indicates that the MOF@AuNP@GO exhibits the strongest quenching ability. Moreover, the maximum QE was evaluated, and results show that the Zr-based MOF alone exhibits a high QE of 81.08 ± 3.07%. The high QE was derived from the combined quenching of TCPP ligand, Zr ions, and the MOF structure itself, which is consistent with the previous work.37 Higher QEs were obtained at 89.29 ± 1.86% for MOF@GO, 92.87 ± 2.51% for MOF@AuNP, and 98.84 ± 2.81% for MOF@AuNP@GO, respectively. These results show that the presence of AuNP or GO can largely augment the fluorescence quenching ability of MOF. Especially, the coexistence of AuNP and GO in the MOF nanostructure can result in almost complete quenching with the QE nearly 100%, further confirming superior binding abilities of MOF@AuNP@GO for ssDNA and improved quenching abilities towards dye fluorescence. The Stern-Volmer equation is further applied to describe the fluorescence quenching properties of these four nanoquenchers. By plotting the curve of F0/F values against nanoquencher concentrations, the Stern-Volmer relationship was acquired with the following equation: F0/F =1 + KSV × [Q], where F0 and F represent the fluorescence intensity of FAM-labeled ssDNA in the absence and presence of nanoquenchers, respectively; KSV is the Stern-Volmer quenching constant; and [Q] is the nanoquencher concentration. As shown in Figure S5, an excellent linear relationship between nanoquencher concentrations and F0/F values was obtained for all four MOF-based nanoquenchers. The KSV value from the curve slope can act as an indicator of fluorescence quenching efficiency, and results in Figure S5 showed that among these four nanoquenchers, the MOF@AuNP@GO nanohybrid possessed the largest KSV value, suggesting the strongest fluorescence quenching ability relative to the other three MOF-based nanomaterials. These results illustrate the MOF@AuNP@GO nanocomposites exhibit higher quenching ability than the other three MOF-based nanoquenchers, further demonstrating the enhanced fluorescence quenching effects in the MOF@AuNP@GO nanohybrid. The enhanced quenching capacities led to reduced background fluorescence signal and increased S/N ratio to achieve highly sensitive detection. In brief, the proposed MOF@AuNP@GO is well-suited as a synergistic nanoquencher to increase detection sensitivity of traditional MOF-based fluorescence biosensors.
Development of no-wash fluorescence biosensor
In general, there are two strategies that can be used to construct no-wash fluorescence biosensors: one depends on target-induced fluorescence recovery, and the other rests on target-induced fluorescence retention. For the former, the presence of target can induce the specific hybridization between target DNA and dye-labeled ssDNA pre-adsorbed on nanoquencher to form double-strand DNA (dsDNA) that can detach from the nanoquencher surface to trigger fluorescence recovery of dye molecules due to the lower affinity of dsDNA for nanoquencher than that of ssDNA. For the latter, the target DNA first hybridizes with dye-labeled ssDNA to generate dsDNA, and thus the dye fluorescence can be effectively retained in the presence of nanoquencher. Nonetheless, a growing number of studies have demonstrated higher detection sensitivity was realized by using the fluorescence recovery-based strategy due to the lower background fluorescence signal and higher S/N ratio.3,39 Thus, in this work, the former is selected to realize a highly sensitive no-wash fluorescence biosensor for target detection.
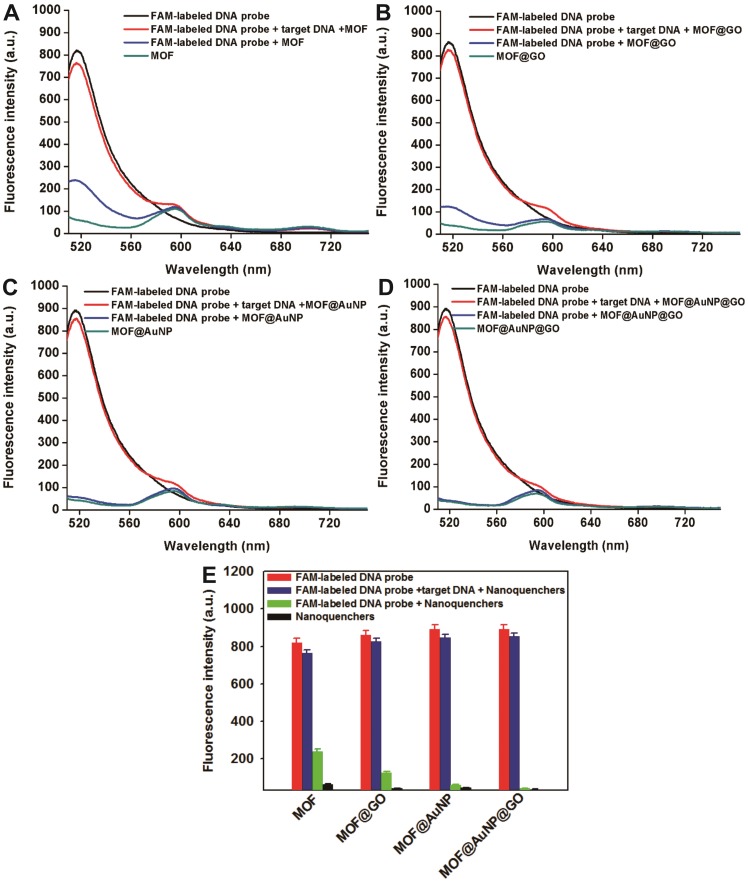
p53 is an important tumor suppressor gene. Its mutation has been observed in various human cancers, and it is considered to be an indicator of tumor progression.49 Thus, p53 gene was chosen as a target DNA in this study. The complementary FAM-labeled ssDNA sequence was designed as a detection probe for p53 gene, and the details of all DNA sequences used can be found in Table S1. The developed MOF@AuNP@GO was applied as a nanoquencher to develop a fluorescence biosensor for p53 gene detection. For comparison, the same procedures were carried out with three other MOF-based nanomaterials, including MOF alone, MOF@GO, and MOF@AuNP, respectively. The fluorescence spectra of FAM-labeled ssDNA probe at different experimental conditions were recorded under excitation at 485 nm. Figure 3D indicates the FAM-labeled ssDNA probe exhibits a strong fluorescence emission at 516 nm. With the addition of MOF@AuNP@GO, the dye fluorescence at 516 nm was remarkably quenched and caused very low fluorescence emission signal, close to that of MOF@AuNP@GO itself. The completely quenched fluorescence emission is due to the strong binding and quenching ability of MOF@AuNP@GO to FAM-labeled ssDNAs. In contrast, with the hybridization of target p53 gene and FAM-labeled ssDNA probe to form dsDNA complex, the pre-quenched fluorescence by MOF@AuNP@GO exhibited an obvious recovery with an efficiency of above 90%. Similar fluorescence quenching and recovery results were observed by using the other three MOF-based nanoquenchers, including MOF alone (Figure 3A), MOF@GO (Figure 3B), and MOF@AuNP (Figure 3C), respectively. These findings demonstrate our designed fluorescence biosensor can accurately detect the presence of target DNA according to the affinity difference of ssDNA and dsDNA with nanoquenchers.
Figure 3.
Fluorescence spectrum assay under different experimental conditions, including FAM-labeled DNA probe, FAM-labeled DNA probe + nanoquenchers, FAM-labeled DNA probe + target p53 gene + nanoquenchers, and nanoquenchers, including MOF alone (A), MOF@GO (B), MOF@AuNP (C), and MOF@AuNP@GO (D). (E) The corresponding fluorescence intensity under different experimental conditions from A to D. The concentrations of FAM-labeled DNA probe and target p53 gene in the final solution are 1 nM and 20 nM, respectively; and the concentrations of four MOF-based nanoquenchers of MOF alone, MOF@GO, MOF@AuNP, and MOF@AuNP@GO in the final solution are 5, 3.75, 2 and 2 µg mL-1, respectively. Fluorescence spectra were collected with the excitation of 485 nm and the emission of 516 nm. Error bars were obtained from three independent repeat experiments.
The quenching and recovery kinetics of various nanoquenchers were investigated by recording time-dependent fluorescence changes of FAM-labeled ssDNA in the absence or presence of target DNA, respectively. As shown in Figure S7, all four nanoquenchers rapidly absorbed FAM-labeled ssDNA, and then reached equilibration within 5 min. In particular, the quenching equilibrium of MOF@AuNP@GO was attained within 1 min, much faster than the other three nanoquenchers, such as 2 min for MOF@GO, and 5 min for MOF alone and for MOF@AuNP (Figure S7A). A possible explanation for the rapid quenching is that the MOF-based nanostructures have conjugated π-system ligands of TCPP and Zr metal center, as well as rich pores on their surface for significantly increased surface areas, attributing to the adsorption of FAM-labeled ssDNA via strong π-stacking interactions between the TCPP ligand and the unpaired nucleobases. More importantly, the presence of AuNP or GO largely improves the fluorescence quenching ability compared with bare MOF nanostructures. On the other hand, upon the addition of target DNA, the specific hybridization of target DNA and FAM-labeled ssDNA can trigger the rapid release of dsDNA from the nanoquencher surface to induce the fluorescence recovery, and the release and recovery processes could be rapidly completed within 5 min for all four MOF-based nanoquenchers (Figure S7B). This is because the formed dsDNA complex with a rigid structure can effectively shield the nucleobases within the negatively charged dsDNA phosphate backbone, which leads to reduced binding affinity between nanoquenchers and dsDNA, thereby causing rapid release of dsDNA from the MOF-based nanoquencher surface. In brief, the fast responses of quenching and recovery strongly supported the requirements for rapid detection. Thus, to maximize the quenching and recovery efficiencies for allowing higher reliability and reproducibility of our designed fluorescence biosensors, 10 min was selected as the overall detection time for all four nanoquenchers in subsequent experiments, including 5 min for quenching and 5 min for target-induced fluorescence recovery, respectively.
Analytical performance of no-wash fluorescence biosensor
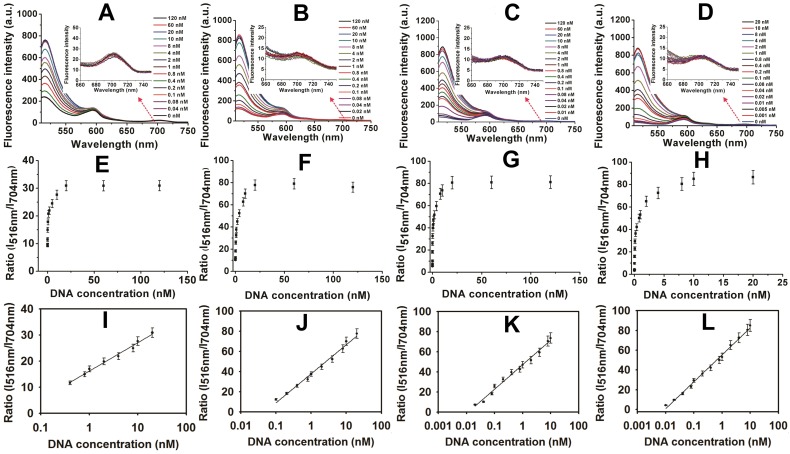
Encouraged by these results, the MOF@AuNP@GO-based fluorescence biosensor was applied to quantify target p53 gene concentrations. The standard curve was established by simply mixing different concentrations of target DNAs with the complex of FAM-labeled ssDNA probe and MOF@AuNP@GO. For comparison, similar detection procedures were performed by using MOF alone, MOF@GO, and MOF@AuNP as controls. The concentration-dependent fluorescence signal changes were collected at different target concentrations ranging from 0 to 120 nM. Figure 4A-D showed the corresponding fluorescence spectra of these biosensors at different target concentrations. For these four MOF-based nanoquenchers, a similar fluorescence signal change trend corresponding to target concentration was observed, in which the fluorescence emission intensity at 516 nm gradually increased with increasing target concentrations. Interestingly, an unchanged fluorescence emission at 704 nm was observed from TCPP ligand that could work as a reference fluorescence signal for ratiometric measurement (Figure S6), further improving the detection sensitivity and reliability of our proposed fluorescence biosensor due to the increased S/N ratio.
Figure 4.
Fluorescence spectra and the corresponding standard curves of four MOF-based nanoquenchers in the presence of different concentrations of target p53 gene. Fluorescence spectra of MOF alone (A), MOF@GO (B), MOF@AuNP (C), and MOF@AuNP@GO (D), the corresponding relationships between the ratio of I516nm/I704nm and target concentrations from A to D, and the corresponding calibration curves at low target concentrations (I, J, K, and L) from E to H, respectively. All experiments were carried out at 37 ℃ in 10 mM Tris-HCl buffered (pH 7.4, containing 100 mM NaCl, 5 mM KCl and 5 mM MgCl2) with concentrations of MOF, MOF@GO, MOF@AuNP, and MOF@AuNP@GO at 5, 3.75, 2 and 2 µg mL-1 in the final solution, respectively. Fluorescence spectra were recorded with the excitation of 485 nm. Error bars were obtained from three repeats.
By using the ratio of I516nm/I704nm as signal readout, the corresponding variations of ratiometric fluorescence signal vs. target DNA concentration were recorded in Figure 4E-H, and an excellent linear relationship was achieved between the ratio of I516nm/I704nm and target concentrations (Figure 4I-L), respectively. According to these developed standard curves, the corresponding linear detection ranges and LODs were obtained and summarized in Table S3. Compared with MOF alone, the increased analytical sensitivities were observed by MOF@GO, MOF@AuNP, and MOF@AuNP@GO (Table S3), which was mainly attributed to lower background signal and higher S/N ratio. Here, the MOF@AuNP@GO nanohybrid exhibited the lowest detection limit (LOD) of 5 pM (S/N = 3), which was approximately a 40, 16, and 4-fold improvement over those of MOF alone (LOD: 0.2 nM), MOF@GO (LOD: 80 pM), and MOF@AuNP (LOD: 20 pM), respectively. Besides p53 gene, the ratiometric fluorescence detection of another important cancer suppressor gene, BRCA1, that is associated with breast cancer, was performed by using MOF@AuNP@GO as a nanoquencher,50 and the experimental details can be found in Supporting Information. Results in Figure S8 and Figure S9 indicated that similar to p53 gene, our developed fluorescence biosensor can be used for sensitive analysis of BRCA1 gene, and the LOD also reached 1 pM (S/N = 3), with a linear detection range from 0.001 to 5 nM. These results demonstrated the generality of our designed fluorescence nanosensor. Additionally, our developed fluorescence quenching-based biosensor was superior or comparable to most of the previously reported fluorescence biosensors based on MOF and other common nanoquenchers in terms of sensitivity and detection time (Table S3). Furthermore, the practicality of our method in biological samples was evaluated by using p53-spiked serum samples at the final concentrations of 0, 0.1, 1, 5, and 10 nM, respectively. Later, these spiked samples were diluted with the reaction buffer for subsequent analysis. Results in Table S4 showed high recoveries of 92% to 103.4% with low relative standard deviation of less than 11% (Table S4), demonstrating the potential of accurately measuring p53 gene in real samples. Such excellent sensing performances make the proposed biosensor well-suited for rapid and sensitive detection of target DNA even in real biological samples.
Besides gene biomarkers, protein biomarkers also play an important role in early diagnosis of cancer. For example, prostate specific antigen (PSA) is a glycoprotein specifically secreted by prostatic tissues, which has become a well-defined marker for prostate cancer diagnosis.3 Recently, no-wash fluorescence biosensors have been considered as a promising platform for protein biomarker detection based on the combination of dye-labeled aptamer probe and nanoquenchers. Here, to realize rapid and sensitive detection of PSA, the FAM-labeled aptamer against PSA was designed as a fluorescent probe, and MOF@AuNP@GO was applied as a nanoquencher. Figure S10 shows the fluorescence spectra of FAM-labeled aptamer probe at different experimental conditions with an excitation at 485 nm. A strong fluorescence emission at 516 nm was observed in the FAM-labeled aptamer probe. However, with the addition of MOF@AuNP@GO, the fluorescence emission at 516 nm was nearly completely quenched. Conversely, the presence of target PSA led to an obvious fluorescence recovery owing to the formation of aptamer and target PSA complex that could cause the aptamer probes to be released from the nanoquencher surface. These observations demonstrate the possibility of our developed nanosensor for detecting cancer-related protein biomarkers. The fluorescence spectra of FAM-labeled aptamer probe at different PSA concentrations were depicted in Figure S11A. Results found the fluorescence emission intensity at 516 nm gradually rose with increasing PSA concentrations, whereas an unchanged fluorescence emission at 704 nm could act as a reference fluorescence signal for ratiometric detection. By recording the ratio of I516nm/I704nm, the relationship between ratiometric fluorescence signal and PSA concentration was described in Figure S11B, and a linear detection range from 0.01 to 10 ng/mL was attained, with a LOD of 0.01 ng/mL (Figure S11C), which is better than most other reported nanoquencher-based fluorescence biosensors (Table S5). Moreover, compared to these widely used commercial ELISA kits, our biosensor also exhibits comparable detection sensitivity with simpler operation and shorter analysis time (Table S5). Subsequently, the selectivity of our prepared fluorescent biosensor was further confirmed with other common disease protein biomarkers in serum, such as human serum albumin (HSA), C-reaction protein (CRP), alpha fetoprotein (AFP), carcinoembryonic antigen (CEA), as well as procalcitonin (PCT). Results in Figure S12A shows that compared with the negative control, a specific increase in the ratio of I516nm/I704nm was observed when PSA was presented in serum sample, whereas no obvious changes were seen in the presence of other proteins, indicating high selectivity of our biosensor for PSA against other interfering substances. Owing to the advantages of rapidity, sensitivity and specificity, this proposed biosensor was further extended to clinical detection of PSA in real blood samples. A correlation analysis of our developed nanosensor with a commercial ELISA kit method was performed to evaluate the reliability and acceptability in 30 PSA-spiked independent human serum samples. These serum samples were first diluted with the Tris-HCl reaction buffer for subsequent analysis. As revealed in Figure S12B, the results obtained by the two methods were highly consistent with a linear correlation coefficient (R2) of 0.985 at PSA concentrations ranging from 0 to 120 ng mL-1, indicating the robustness of the designed fluorescence biosensor in the sensitive and reliable detection of cancer-related biomarkers from complex biological samples.
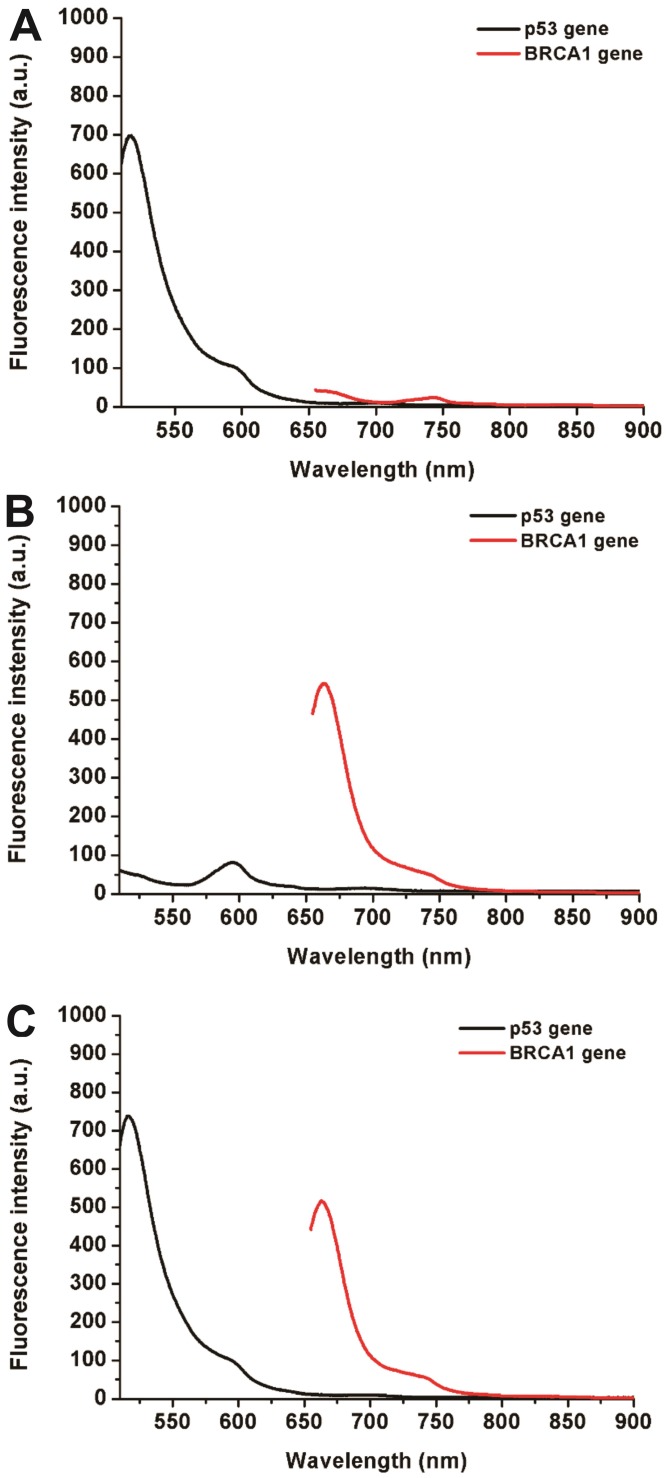
Simultaneous detection of multiple biomarkers in a single clinical sample is critical for reliable cancer diagnosis since it can provide much information from limited volumes. In this work, a multiplexed no-wash fluorescence nanosensor platform based on MOF@AuNP@GO as a nanoquencher was established for simultaneous detection of two cancer-associated gene biomarkers, including p53 and BRCA1 gene, by using FAM-labeled ssDNA probe against p53 and Cy5-labeled ssDNA probe against BRCA1, respectively. Figure 5 showed the corresponding fluorescence spectrum changes for simultaneous detection of p53 and BRCA1 genes. Results indicated that our designed nanosensor provides specific responses to the corresponding target DNA molecules, and exhibits an increased fluorescence signal at the corresponding wavelengths. The presence of p53 gene can only cause a specific fluorescence emission at 516 nm (Figure 5A), whereas the presence of BRCA1 gene can also only induce an obvious fluorescence emission at 662 nm (Figure 5B). Only the coexistence of p53 and BRCA1 gene in the samples can result in two significantly increased fluorescence emissions at 516 and 662 nm (Figure 5C). These results proved that our proposed MOF@AuNP@GO-based fluorescence nanosensor is very suitable for simultaneous detection of multiple target analytes in mixed samples.
Figure 5.
Fluorescence spectra for simultaneous detection of two different DNA targets, including p53 and BRCA1 genes. The concentrations of FAM-labeled p53 probe and Cy5-labeled BRCA1 probe were 1 nM. These two probes were mixed in the presence of different target combinations: (A) p53 gene without BRCA1 gene, (B) BRCA1 gene without p53 gene, and (C) p53 and BRCA1 gene. Fluorescence spectra were collected with the excitation/emission wavelengths of 485 nm/516 nm for FAM, and 633 nm/662 nm for Cy5, respectively.
Moreover, the specificity of our proposed nanosensor was determined by using complementary target p53 gene (T), single-base mismatched target p53 gene (T1), double-base mismatched target p53 gene (T2), triple-base mismatched target p53 gene (T3), and non-complementary target. The ratio of I516nm/I704nm was employed to estimate the target-induced fluorescence signal changes. As shown in Figure S13, only the addition of T, T1, and T2 could cause a distinct increase in the ratio of I516nm/I704nm, whereas a negligible change in the ratio of I516nm/I704nm was observed in the presence of T3 and non-complementary target compared with the control group. These findings suggest the excellent selectivity of our developed no-wash nanosensor using MOF@AuNP@GO against target analytes, even with a single nucleotide mismatch, providing great potential for target quantitative detection in real samples.
Conclusions
In this work, we synthesized a “three-in-one” nanohybrid of MOF@AuNP@GO that exhibited improved absorption affinities to ssDNA molecules and quenching capacities to dye fluorescence. Due to its outstanding discrimination ability against ssDNA and dsDNA, the as-prepared MOF@AuNP@GO with ultrahigh QE was proposed as a synergistic nanoquencher to develop a no-wash fluorescence biosensor for rapid and sensitive detection of cancer-related biomarkers. The designed strategy effectively reduced the background fluorescence signal, and significantly magnified the S/N ratio, further improving detection sensitivity. On the other hand, by using ratiometric fluorescence signal, the sensitivity and reliability were further enhanced. Accordingly, the LODs of our developed nanosensor are as low as 0.005 nM for target DNA, and 0.01 ng/mL for target protein. The LODs are superior or comparable to most of the previously reported fluorescence biosensors based on various nanoquenchers. In addition, the designed nanosensor exhibits excellent performance with respect to universality, multiplex detection, selectivity, as well as practicality. More importantly, the reported fluorescence biosensor does not require the wash step, making this method well-suited for point-of-care (POC) diagnosis. In summary, the designed MOF@AuNP@GO nanohybrid can serve as a promising and competitive alternative for the fabrication of nanoquencher-based biosensing platforms, providing a new perspective for designing nanomaterial-based fluorescence quenchers with high QE.
Experimental Section
Materials
Gold(III) chloride hydrate, N'N-dimethylformamide (DMF), hydrazine monohydrate (NH2NH2·H2O), zirconium(IV) chloride (ZrCl4), sodium borohydride (NaBH4), and benzoic acid were obtained from Sigma-Aldrich. TCPP was purchased from Fisher Scientific. GO was prepared and characterized according to our previous work.51 Commercial ELISA kits for human PSA were obtained from Abnova Corporation. All DNA strands listed in Table S1 (Supporting Information) were synthesized and purified by Integrated DNA Technologies (Coralville, IA, USA). All chemicals were of analytical grade and were used without further purification. Milli-Q water was utilized throughout this work.
Synthesis of Zr-based MOF nanostructures
The Zr-based MOF was prepared through a previously reported method with slight modification.52 Briefly, ZrCl4 (75 mg), TCPP (30 mg), and benzoic acid (1750 mg) were first ultrasonically dissolved in 10 mL DMF solution in a 20 mL Pyrex vial. Then, the resulting mixture was heated to 95 ℃ by oil bath and reacted for 24 h. After slowly cooling down to room temperature, the as-prepared reddish-brown product was washed twice with ethanol and once with water by centrifugation at 4500 rpm for 10 min. The final product, with bright green color, was re-dispersed in aqueous solution. To measure the concentrations of Zr-based MOF crystals in aqueous solution, after drying a certain amount of Zr-based MOF solution at 100 ºC, the weight of dry Zr-based MOF was recorded by an analytical balance (Mettler-Toledo, LLC).
Synthesis of MOF@AuNP@GO nanostructures
To prepare the hybrid GO@AuNP@MOF nanostructures, we first prepared the MOF@AuNP nanostructures through in situ growth of AuNP on the Zr-based MOF surface as previously described.53 Briefly, 80 μL of HAuCl4 (10 mg mL-1) was added into 10 mL aqueous solution containing Zr-based MOF nanostructure at a final concentration of 0.1 mg mL-1. Later, the resultant mixture was stirred, and then 25 μL of freshly prepared NaBH4 solution (3.8 mg mL-1) was rapidly added. Then, with the solution color change from green to red brown, the formed mixture was washed with water twice to remove the unreacted chemicals through centrifugation at 4500 rpm for 10 min. Finally, the obtained MOF@AuNP nanostructures were collected and re-dispersed in water.
After the synthesis of MOF@AuNP, ultrathin GO was coated onto the surface of MOF@AuNP based on a previous report.51 In a typical synthesis procedure of MOF@AuNP@GO, 100 μL of the resulting MOF@AuNP solution (0.1 mg mL-1) was slowly added dropwise into 2 mL of GO solution (0.25 mg mL-1) containing a small amount of NH2NH2·H2O under ultrasound assistance. Subsequently, the mixture continued to react at 60 ºC for 72 h with ultrasonic treatment. Finally, the synthetic MOF@AuNP@GO nanocomposites were washed with water twice to remove excess chemicals, and then re-dispersed in water.
Characterization
Fluorescence spectra were recorded by using a Hitachi F-7000 fluorescence spectrophotometer. The UV-Vis absorption spectra were recorded via a Genesys 10s UV-Vis spectrophotometer (Thermo Scientific). Dynamic light scattering (DLS) measurement was carried out on a scientific nanoparticle analyzer (SZ-100, Horiba). Powder X-ray diffraction (XRD) patterns were observed on a Rigaku Dmax2550PC polycrystal X-ray diffractometer using Cu Kα radiation (Rigaku Co., Tokyo, Japan). Transmission electron microscopy (TEM) images were recorded on a Tecnai TF30 transmission electron microscope (FEI, Hillsboro, OR) equipped with a Gatan Ultrascan 1000 CCD camera (Gatan, Pleasaton, CA). Scanning electron microscope (SEM) imaging was performed on a Hitachi SU-70 Schottky field emission gun scanning electron microscope.
Fluorescence DNA assays
No-wash fluorescent biosensor for target DNA detection was performed according to a previous method.39 Tris-HCl buffered solution (10 mM, pH 7.4) containing 100 mM NaCl, 5 mM MgCl2, and 5 mM KCl was prepared and selected as the reaction buffer solution. The details of the detection assay were as follows: 10 µL of MOF@AuNP@GO (50 µg mL-1) was added into 20 µL of reaction buffer solution containing FAM-labeled DNA probe (1 nM) and incubated at 37 ℃ for 5 min. Next, 20 µL of target DNA with different concentrations ranging from 0 to 120 nM was added into the mixture, which then hybridized with FAM-labeled DNA probe over 5 min. For kinetic studies of the fluorescence quenching and recovery of FAM-labeled DNA probe, the fluorescence spectra of FAM were collected after addition of MOF@AuNP@GO or target DNA, respectively. The excitation/emission for FAM was set at 485 nm/516 nm, respectively. No-wash fluorescent biosensor detection of BRCA1 using Cy5-labeled DNA probe was carried out through a similar procedure, in which the fluorescence spectra were recorded with the excitation at 633 nm and the emission at 662 nm, respectively.
For multiplexed DNA detection based on our developed no-wash biosensor, 20 µL of MOF@AuNP@GO (50 µg mL-1) was added into 10 µL of reaction buffer solution containing FAM-labeled p53 probe (1 nM) and Cy5-labeled BRCA1 probe (1 nM), and incubated at 37 ℃ for 5 min. Subsequently, target p53 gene (20 μL, 2 nM), target BRCA1 gene (20 μL, 1 nM), or the mixture of target p53 gene (10 μL, 4 nM) and target BRCA1 gene (10 μL, 2 nM), were added into the mixed solution, and allowed to hybridize with FAM or Cy5-labeled DNA probes for another 5 min. The fluorescence spectra were collected at the excitation/emission wavelengths of 485 nm/516nm for FAM, and 633/662 nm for Cy5, respectively.
Fluorescence PSA assays
No-wash fluorescence biosensor for PSA detection was conducted as previously described.16 Briefly, FAM-labeled PSA aptamer probe (1 nM) was prepared in 20 mM pH 7.4 Tris-HCl buffered solution containing 100 mM NaCl, 5 mM MgCl2, and 5 mM KCl, and then mixed with MOF@AuNP@GO (50 µg mL-1) at 37 ℃ for 5 min before the addition of PSA. Later, target PSA at concentrations ranging from 0 to 100 ng mL-1 was added into the above mixed solutions. After reaction for another 30 min at 37 ℃, fluorescence spectra were detected and recorded at the excitation/emission wavelengths of 485 nm/516 nm, respectively.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (31760485, 21335004), Major Projects of Natural Science Foundation of Jiangxi province (20161ACB20002), and the Intramural Research Program, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health. X. Huang (201606820007) and Z. He (201606190118) also thank the support from China Scholarship Council (CSC).
Supplementary Material
UV-visible absorption spectrum, TEM imaging, fluorescence spectra, comparison of fluorescence quenching ability, DNA sequences, comparison with existing no-wash fluorescent biosensors and commercial detection methods for DNA and protein detection, and additional results.
References
- 1.Ranjan R, Esimbekova EN, Kratasyuk VA. Rapid biosensing tools for cancer biomarkers. Biosens Bioelectron. 2017;87:918–30. doi: 10.1016/j.bios.2016.09.061. [DOI] [PubMed] [Google Scholar]
- 2.Wu L, Qu X. Cancer biomarker detection: Recent achievements and challenges. Chem Soc Rev. 2015;44:2963–97. doi: 10.1039/c4cs00370e. [DOI] [PubMed] [Google Scholar]
- 3.Huang X, Liu Y, Yung B. et al. Nanotechnology-enhanced no-wash biosensors for in vitro diagnostics of cancer. ACS Nano. 2017;11:5238–93. doi: 10.1021/acsnano.7b02618. [DOI] [PubMed] [Google Scholar]
- 4.Devi RV, Doble M, Verma RS. Nanomaterials for early detection of cancer biomarker with special emphasis on gold nanoparticles in immunoassays/sensors. Biosens Bioelectron. 2015;68:688–98. doi: 10.1016/j.bios.2015.01.066. [DOI] [PubMed] [Google Scholar]
- 5.Chinen AB, Guan CM, Ferrer JR. et al. Nanoparticle probes for the detection of cancer biomarkers, cells, and tissues by fluorescence. Chem Rev. 2015;115:10530–74. doi: 10.1021/acs.chemrev.5b00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yezhelyev MV, Al-Hajj A, Morris C. et al. In situ molecular profiling of breast cancer biomarkers with multicolor quantum dots. Adv Mater. 2007;19:3146–51. [Google Scholar]
- 7.Alptürk O, Rusin O, Fakayode SO. et al. Lanthanide complexes as fluorescent indicators for neutral sugars and cancer biomarkers. Proc Natl Acad Sci U S A. 2006;103:9756–60. doi: 10.1073/pnas.0603758103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Guo Y, Xianyu Y. et al. Nanomaterials for ultrasensitive protein detection. Adv Mater. 2013;25:3802–19. doi: 10.1002/adma.201301334. [DOI] [PubMed] [Google Scholar]
- 9.Jans H, Huo Q. Gold nanoparticle-enabled biological and chemical detection and analysis. Chem Soc Rev. 2012;41:2849–66. doi: 10.1039/c1cs15280g. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Asiri AM, Tang Z. et al. Graphene based materials for biomedical applications. Mater Today. 2013;16:365–73. [Google Scholar]
- 11.Tian F, Lyu J, Shi J. et al. Graphene and graphene-like two-denominational materials based fluorescence resonance energy transfer (FRET) assays for biological applications. Biosens Bioelectron. 2017;89:123–35. doi: 10.1016/j.bios.2016.06.046. [DOI] [PubMed] [Google Scholar]
- 12.Wilson R. The use of gold nanoparticles in diagnostics and detection. Chem Soc Rev. 2008;37:2028–45. doi: 10.1039/b712179m. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Rothberg LJ. DNA sequence detection using selective fluorescence quenching of tagged oligonucleotide probes by gold nanoparticles. Anal Chem. 2004;76:5414–7. doi: 10.1021/ac049173n. [DOI] [PubMed] [Google Scholar]
- 14.Yang R, Jin J, Chen Y. et al. Carbon nanotube-quenched fluorescent oligonucleotides: Probes that fluoresce upon hybridization. J Am Chem Soc. 2008;130:8351–8. doi: 10.1021/ja800604z. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Zhang X, Yang R. et al. Single-walled carbon nanotubes as optical materials for biosensing. Nanoscale. 2011;3:1949–56. doi: 10.1039/c0nr01014f. [DOI] [PubMed] [Google Scholar]
- 16.Lu CH, Yang HH, Zhu CL. et al. A graphene platform for sensing biomolecules. Angew Chem. 2009;121:4879–81. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- 17.Li F, Pei H, Wang L. et al. Nanomaterial-based fluorescent DNA analysis: A comparative study of the quenching effects of graphene oxide, carbon nanotubes, and gold nanoparticles. Adv Funct Mater. 2013;23:4140–8. [Google Scholar]
- 18.Zhu C, Zeng Z, Li H. et al. Single-layer MoS2-based nanoprobes for homogeneous detection of biomolecules. J Am Chem Soc. 2013;135:5998–6001. doi: 10.1021/ja4019572. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y, Shi Y, Yang HY. et al. A novel single-layered MoS2 nanosheet based microfluidic biosensor for ultrasensitive detection of DNA. Nanoscale. 2015;7:2245–9. doi: 10.1039/c4nr07162j. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Wang W, Lei J. et al. Fluorescence quenching of carbon nitride nanosheet through its interaction with DNA for versatile fluorescence sensing. Anal Chem. 2013;85:12182–8. doi: 10.1021/ac403646n. [DOI] [PubMed] [Google Scholar]
- 21.Yang G, Zhu C, Du D. et al. Graphene-like two-dimensional layered nanomaterials: Applications in biosensors and nanomedicine. Nanoscale. 2015;7:14217–31. doi: 10.1039/c5nr03398e. [DOI] [PubMed] [Google Scholar]
- 22.Ping J, Zhou Y, Wu Y. et al. Recent advances in aptasensors based on graphene and graphene-like nanomaterials. Biosens Bioelectron. 2015;64:373–85. doi: 10.1016/j.bios.2014.08.090. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Luque R, Li Y. Controllable design of tunable nanostructures inside metal-organic frameworks. Chem Soc Rev. 2017;46:4614–30. doi: 10.1039/c6cs00537c. [DOI] [PubMed] [Google Scholar]
- 24.Volosskiy B, Niwa K, Chen Y. et al. Metal-organic framework templated synthesis of ultrathin, well-aligned metallic nanowires. ACS Nano. 2015;9:3044–9. doi: 10.1021/nn5072446. [DOI] [PubMed] [Google Scholar]
- 25.Kuppler RJ, Timmons DJ, Fang Q-R. et al. Potential applications of metal-organic frameworks. Coord Chem Rev. 2009;253:3042–66. [Google Scholar]
- 26.Rosi NL, Eckert J, Eddaoudi M. et al. Hydrogen storage in microporous metal-organic frameworks. Science. 2003;300:1127–9. doi: 10.1126/science.1083440. [DOI] [PubMed] [Google Scholar]
- 27.Yang Q, Xu Q, Jiang H-L. Metal-organic frameworks meet metal nanoparticles: Synergistic effect for enhanced catalysis. Chem Soc Rev. 2017;46:4774–808. doi: 10.1039/c6cs00724d. [DOI] [PubMed] [Google Scholar]
- 28.Li J-R, Kuppler RJ, Zhou H-C. Selective gas adsorption and separation in metal-organic frameworks. Chem Soc Rev. 2009;38:1477–504. doi: 10.1039/b802426j. [DOI] [PubMed] [Google Scholar]
- 29.Horcajada P, Chalati T, Serre C. et al. Porous metal-organic-framework nanoscale carriers as a potential platform for drug delivery and imaging. Nat Mater. 2010;9:172–8. doi: 10.1038/nmat2608. [DOI] [PubMed] [Google Scholar]
- 30.Della Rocca J, Liu D, Lin W. Nanoscale metal-organic frameworks for biomedical imaging and drug delivery. Acc Chem Res. 2011;44:957–68. doi: 10.1021/ar200028a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreno LE, Leong K, Farha OK. et al. Metal-organic framework materials as chemical sensors. Chem Rev. 2012;112:1105–25. doi: 10.1021/cr200324t. [DOI] [PubMed] [Google Scholar]
- 32.He C, Liu D, Lin W. Nanomedicine applications of hybrid nanomaterials built from metal-ligand coordination bonds: Nanoscale metal-organic frameworks and nanoscale coordination polymers. Chem Rev. 2015;115:11079–108. doi: 10.1021/acs.chemrev.5b00125. [DOI] [PubMed] [Google Scholar]
- 33.De Silva AP, Gunaratne HN, Gunnlaugsson T. et al. Signaling recognition events with fluorescent sensors and switches. Chem Rev. 1997;97:1515–66. doi: 10.1021/cr960386p. [DOI] [PubMed] [Google Scholar]
- 34.Liu S, Wang L, Tian J. et al. Application of zeolitic imidazolate framework-8 nanoparticles for the fluorescence-enhanced detection of nucleic acids. ChemPhysChem. 2012;77:23–6. [Google Scholar]
- 35.Wei X, Zheng L, Luo F. et al. Fluorescence biosensor for the H5N1 antibody based on a metal-organic framework platform. J Mater Chem B. 2013;1:1812–7. doi: 10.1039/c3tb00501a. [DOI] [PubMed] [Google Scholar]
- 36.Zhu X, Zheng H, Wei X. et al. Metal-organic framework (MOF): A novel sensing platform for biomolecules. Chem Commun. 2013;49:1276–8. doi: 10.1039/c2cc36661d. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H-T, Zhang J-W, Huang G. et al. An amine-functionalized metal-organic framework as a sensing platform for DNA detection. Chem Commun. 2014;50:12069–72. doi: 10.1039/c4cc05571c. [DOI] [PubMed] [Google Scholar]
- 38.Fang JM, Leng F, Zhao XJ. et al. Metal-organic framework MIL-101 as a low background signal platform for label-free DNA detection. Analyst. 2014;139:801–6. doi: 10.1039/c3an01975f. [DOI] [PubMed] [Google Scholar]
- 39.Tian J, Liu Q, Shi J. et al. Rapid, sensitive, and selective fluorescent DNA detection using iron-based metal-organic framework nanorods: Synergies of the metal center and organic linker. Biosens Bioelectron. 2015;71:1–6. doi: 10.1016/j.bios.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Taylor-Pashow KM, Della Rocca J, Huxford RC. et al. Hybrid nanomaterials for biomedical applications. Chem Commun. 2010;46:5832–49. doi: 10.1039/c002073g. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Zheng H, Zhu X. et al. Metal-organic frameworks-based biosensor for sequence-specific recognition of double-stranded DNA. Analyst. 2013;138:3490–3. doi: 10.1039/c3an00426k. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y, Han J, Xue P. et al. Nano metal-organic framework (NOMF)-based strategies for multiplexed microrna detection in solution and living cancer cells. Nanoscale. 2015;7:1753–9. doi: 10.1039/c4nr05447d. [DOI] [PubMed] [Google Scholar]
- 43.Yuan L, Jin F, Zeng Z. et al. Engineering a fret strategy to achieve a ratiometric two-photon fluorescence response with a large emission shift and its application to fluorescence imaging. Chem Sci. 2015;6:2360–5. doi: 10.1039/c4sc03883e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Zhu C, Yang Z. et al. A ratiometric fluorescent probe for rapid detection of hydrogen sulfide in mitochondria. Angew Chem. 2013;125:1732–5. doi: 10.1002/anie.201207701. [DOI] [PubMed] [Google Scholar]
- 45.Lee MH, Kim JS, Sessler JL. Small molecule-based ratiometric fluorescence probes for cations, anions, and biomolecules. Chem Soc Rev. 2015;44:4185–91. doi: 10.1039/c4cs00280f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Z, Baek K-H, Kim HN. et al. Zn2+-triggered amide tautomerization produces a highly Zn2+-selective, cell-permeable, and ratiometric fluorescent sensor. J Am Chem Soc. 2009;132:601–10. doi: 10.1021/ja907334j. [DOI] [PubMed] [Google Scholar]
- 47.Huang X, Song J, Yung BC. et al. Ratiometric optical nanoprobes enable accurate molecular detection and imaging. Chem Soc Rev. 2018 doi: 10.1039/C7CS00612H. DOI: 10.1039/c7cs00612h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang G-Y, Zhuang Y-H, Shan D. et al. Zirconium-based porphyrinic metal-organic framework (PCN-222): Enhanced photoelectrochemical response and its application for label-free phosphoprotein detection. Anal Chem. 2016;88:11207–12. doi: 10.1021/acs.analchem.6b03484. [DOI] [PubMed] [Google Scholar]
- 49.Chang H, Qi C, Yi Q-L. et al. P53 gene deletion detected by fluorescence in situ hybridization is an adverse prognostic factor for patients with multiple myeloma following autologous stem cell transplantation. Blood. 2005;105:358–60. doi: 10.1182/blood-2004-04-1363. [DOI] [PubMed] [Google Scholar]
- 50.Bochar DA, Wang L, Beniya H. et al. BRCA1 is associated with a human SWI/SNF-related complex: Linking chromatin remodeling to breast cancer. Cell. 2000;102:257–65. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 51.Lin L-S, Yang X, Niu G. et al. Dual-enhanced photothermal conversion properties of reduced graphene oxide-coated gold superparticles for light-triggered acoustic and thermal theranostics. Nanoscale. 2016;8:2116–22. doi: 10.1039/c5nr07552a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng D, Gu ZY, Li JR. et al. Zirconium-metalloporphyrin PCN-222: Mesoporous metal-organic frameworks with ultrahigh stability as biomimetic catalysts. Angew Chem. 2012;124:10453–6. doi: 10.1002/anie.201204475. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y, Zhao M, Han S. et al. Growth of au nanoparticles on 2D metalloporphyrinic metal-organic framework nanosheets used as biomimetic catalysts for cascade reactions. Adv Mater. 2017;29:1700102. doi: 10.1002/adma.201700102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
UV-visible absorption spectrum, TEM imaging, fluorescence spectra, comparison of fluorescence quenching ability, DNA sequences, comparison with existing no-wash fluorescent biosensors and commercial detection methods for DNA and protein detection, and additional results.