Abstract
Lung cancer is the most frequent cancer type and the leading cause of tumor-associated deaths worldwide. TP53 is an important tumor suppressor gene and is frequently inactivated in lung cancer. E3 ligases targeting p53, such as MDM2, are involved in the development of lung cancer. The E3 ligase HUWE1, which targets many tumor-associated proteins including p53, has been reported to be highly expressed in lung cancer; however, its role in lung tumorigenesis is unclear.
Methods: The expression of HUWE1 and p53 in lung cancer cells was modulated and the phenotypes were assessed by performing soft agar colony forming assays, cell cycle analysis, BrdU incorporation assays, and xenograft tumor growth assays. The effect on tumorigenesis in genetically-engineered mice was also analyzed. The mechanism through which HUWE1 sustained lung cancer cell malignancy was confirmed by western blotting. HUWE1 expression in clinical lung cancer was identified by immunohistochemistry and validated by analyzing lung adenocarcinoma and lung squamous carcinoma samples from the Cancer Genome Atlas (TCGA) database. Finally, we assessed the association between HUWE1 expression and patient outcome using online survival analysis software including survival information from the caBIG, GEO, and TCGA database.
Results: Inactivation of HUWE1 in a human lung cancer cell line inhibited proliferation, colony-forming capacity, and tumorigenicity. Mechanistically, this phenotype was driven by increased p53, which was due to attenuated proteasomal degradation by HUWE1. Up-regulation of p53 inhibited cancer cell malignancy, mainly through the induction of p21 expression and the down-regulation of HIF1α. Huwe1 deletion completely abolished the development of EGFRVIII-induced lung cancer in Huwe1 conditional knockout mice. Furthermore, survival analysis of lung cancer patients showed that increased HUWE1 expression is significantly associated with worse prognosis.
Conclusion: Our data suggest that HUWE1 plays a critical role in lung cancer and that the HUWE1-p53 axis might be a potential target for lung cancer therapy.
Keywords: HUWE1, p53, lung cancer, mouse model
Introduction
Globally, lung cancer is the leading cause of cancer-related death, contributing to 27% of such incidences 1, 2. Furthermore, 80% of lung cancers consist of non-small cell lung cancer (NSCLC), for which only 17.4% patients survive longer than 5 years, despite tremendous efforts to uncover the associated mechanisms and studies to develop new therapies 3, 4. Tumor development involves many cancer-associated genes that act as oncogenes or tumor suppressors. E3 ligases take part in this process by directing specific degradation of these proteins though ubiquitination 5. The dysregulation of E3 ligases frequently results in chemotherapy resistance and poor prognosis 6. E3 ligases have emerged as important targets for tumor therapy. For example, thalidomide and its derivatives, lenalidomide and pomalidomide, inhibit the E3 ligase activity of CRL4CRBN and have been approved by the FDA to treat multiple myeloma 7. Numerous studies have demonstrated that E3 ligases also play important roles in lung cancer. Genetic alterations in c-Cbl, which targets EGFR for proteasomal degradation, were found in lung cancers, and the overexpression of its mutant was determined to promote cellular proliferation and migration 8. In addition, lower expression of the E3 ligase CCNB1IP1 often implies poor survival for lung cancer 9.
TP53 is a classic tumor suppressor gene, encoding a protein that inhibits oncogenesis by inducing growth arrest, apoptosis, senescence, and angiogenesis inhibition. Its inactivation is a frequent event in lung cancer and this is classified as a driver mutation 10. Up-regulation or activation of p53 is beneficial for lung tumor treatment 11. Thus, p53 and its upstream effectors such as E3 ligases have become attractive targets for new targeted therapies for lung cancers 11. MDM2 is believed to be a predominant E3 ligase that regulates p53, and its increased expression has been observed in lung cancer. Inactivation of MDM2 results in p53 accumulation and the induction of cell cycle arrest and apoptosis in lung cancer cell lines 12. Several MDM2 inhibitors have been designed for lung cancer therapy, such as Nutlin-3a, RG7112, and MI-773 13.
HUWE1 (HECT, UBA, and WWE domain containing 1) is another well-known E3 ligase that targets p53 and many other substrates involved in tumorigenesis. However, the role of HUWE1 in cancer is still controversial based on in vitro and in vivo data. In a colon cancer model induced by APC deletion, Huwe1 knockout promotes cancer development 14; however, Huwe1 inactivation was shown to inhibit the proliferation of the colon cancer cell line Ls174T and its tumorigenicity in immunocompromised mice 15. In a mouse skin cancer model, Huwe1 inhibits tumor development by inducing the degradation of the Myc/Miz complex 16. Our previous studies demonstrated that Huwe1 is essential for the transformation of ovarian epithelial cells and for the development of ovarian cancer by ubiquitinating histone H1.3 for subsequent proteasomal degradation 17. These data imply that the role of Huwe1 in cancer might depend on the type of disease. Although HUWE1 was reported to be overexpressed in lung cancer 9, 18 and to mediate HGF-induced cell migration and invasion by targeting TIAM1 for proteasomal degradation in the H1299 lung cancer cell line 19, the role of HUWE1 and the HUWE1-p53 axis in lung cancer initiation and development have not been investigated. In this study, we found that inactivation of HUWE1 inhibited the proliferation, colony-forming activity, and tumorigenicity of the A549 human lung cancer cell line and abolished the development of lung cancer induced by overexpression of EGFRVIII in mice. p53 accumulation upon HUWE1 deletion was found to be one of the main mechanisms association with the inhibition of tumor growth and angiogenesis.
Methods
Animals
The Huwe1 conditional knockout mouse was described in a previous study 20. The HUWE1 gene is located on chromosome X, and thus male mice have only one allele; therefore, here, the genotype of wild-type mice was referred to as Huwe1+/y and that of conditional knockout mice was Huwe1 L/y, where y denotes the Y chromosome. Six-week-old BALB/c nude mice were purchased from Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). The animal protocol was approved by the Institutional Review Board at Kunming Institute of Zoology, Chinese Academy of Sciences.
Cell culture
The A549 human lung adenocarcinoma cell line was purchased from China Infrastructure of Cell Line Resources and tested via short tandem repeat (STR) profiling by the provider. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Life Technologies, USA) medium supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies, USA) and maintained in a humidified incubator with 5% CO2 at 37 °C.
Plasmid construction and lentiviral preparation
The lentiviral vector pTomo-EGFRVIII-IRES-Cre was constructed based on the backbone of the pTomo vector. pTomo was a gift from Inder Verma (Addgene plasmid #26291) 21. The genetic element EGFRVIII-IRES-Cre was cloned into pTomo between the XbaI and SalI restriction sites. For the HUWE1 and TP53 shRNA plasmids, the target sequences were cloned into a Tet-pLKO-puro vector obtained from ADDGENE according to the manufacturer's protocol (http://www.addgene.org/21915/). The following target sequences were used: HUWE1, 5´CGACGAGAACTAGCACAG-AAT3´ 22 and TP53: 5´CGGCGCACAGAGGAAGAGAAT3´ 23. The DNA vectors were co-transfected into HEK293T cells with the helper plasmids pCMVΔ8.9 and pMD2.G at a ratio of 10:5:2 for lentiviral packaging as described previously 17.
RNA isolation and real-time PCR
Total RNA was isolated using Trizol (Sigma-Aldrich, USA) and DNA contaminants were removed using the TURBO DNA-free TM Kit (Life Technologies, USA). Next, 2 μg of total RNA was reverse transcribed using random primers and the RevertAid First Strand cDNA Synthesis kit (Thermo Scientific, USA). Quantitative PCR was performed in triplicate using the SYBR Green method (Life Technologies, USA) according to the manufacturer's instructions. The cycle threshold (CT) values of the target genes were normalized to those of 18S rRNA. All primers used in this study are shown in Table S1.
Colony formation assay
Colony formation assay in soft agar was performed as described before 17. Briefly, 1.5 × 104 single cells were mixed with 1 mL DMEM supplemented with 10% FBS and 0.3% soft agar (Sigma-Aldrich, USA), and then the mixture was added to the top of the base agar containing 0.6% agarose in a 6-well plate. After 5 weeks, 1 mL of PBS containing 4% formaldehyde and 0.005% crystal violet was added to fix and stain the colonies.
Cell cycle analysis
Cells were digested with 0.25% trypsin-EDTA (Life Technologies, USA) and fixed with 70% ethanol for 12 h at 4 °C. Then, the cells were treated with RNase A (QIAGEN, German) and propidium iodide (PI; Sigma-Aldrich, USA) for 0.5 h. Cell cycle analyses were performed by flow cytometry using the BD FACSVantage™ SE System.
HUWE1 gene editing
The guide RNA (gRNA) sequences for HUWE1 were designed using the Target Finder program and cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid (Addgene plasmid #42230) 24. Constructed plasmids were transfected into 293T cells and genomic DNA was extracted 3 days later. PCR was performed to amplify the gRNA targeting region and a T7 endonuclease I assay was used to assess targeting efficiency. The most efficient gRNA-3840 was then transfected into A549 cells and single-cell culture was established using the limiting dilution method. The HUWE1-null clones were screened by DNA sequencing and western blotting. All gRNA sequences are listed in Table S2.
Xenograft mouse models of lung cancer
A549/teton-shHUWE1 cells, established by transduction of A549 with lentiviral particles expressing HUWE1 shRNA, were resuspended at 1 × 107 cells/mL in PBS containing 30% Matrigel (BD Biosciences, USA) and subcutaneously injected into 6-week-old BALB/c nude mice (1 × 106 cells per mouse). When the subcutaneous tumors reached approximately 5 mm in diameter, doxycycline (DOX) was delivered via the drinking water, which contained 5% sucrose. Tumor volume was calculated based on the formula 0.5 × length × width2 25. The data are shown as the means ± standard deviation (SD), and statistical analyses were performed using GraphPad Prism statistical software.
Intranasal inoculation
Mice were subjected to general anesthesia with 2.5% avertin by intraperitoneal injection (15 mL/kg). The mice were placed with their head forward in a supine position and 2 × 105 lentivirus particles in 70 µL PBS was gradually released into the nostrils with sterile pipette tips. Finally, the mice were allowed to slowly wake up, and tumorigenesis was subsequently observed.
Survival analyses
Survival analyses were performed as described 26. In brief, 1926 lung cancer patients were split into two groups, with the lower tertile expression of the target gene as the cut-off level. The two patient cohorts were compared by a Kaplan-Meier survival plot analysis using the default parameters, and the hazard ratio with 95% confidence intervals and log-rank P value were calculated. The probe set used was 207783_x_at for HUWE1 and 205385_at for MDM2.
Western blotting and co-immunoprecipitation
Tissues or cells were lysed using RIPA buffer 17 supplemented with a 1× protease inhibitor cocktail (Roche, Switzerland) and 0.1 mg/mL phenylmethylsulfonyl fluoride (Sigma-Aldrich, USA) for 30 min on ice. Protein concentration was determined using the BCA protein assay kit (Beyotime, China). Then, 100 μg of protein was subjected to standard SDS-PAGE electrophoresis. For co-immunoprecipitations, 1 × 107 cells were treated with 5 μM MG-132 for 6 h, lysed in lysis buffer [50 mM tris HCl, 150 mM NaCl, 1 mM EDTA, 1% triton X-100, and a 1× protease inhibitor cocktail], and 500 μg of total protein was incubated with 10 μL of p53 antibody and 20 μL protein G agarose (Santa Cruz, USA); co-immunoprecipitation was performed in accordance with the manufacturer's instructions. The antibodies used in the study are shown in Table S3. The use of human cancer tissues was approved by the Institutional Review Board at Kunming Institute of Zoology, Chinese Academy of Sciences.
BrdU incorporation assay
The BrdU incorporation assay was performed as described elsewhere 17. BrdU (Thermo Scientific, USA) was added to the cell medium at a final concentration 10 μM and incubated for 1 h. The labeled cells were fixed with 4% paraformaldehyde, permeabilized with 0.3% triton-X-100, denatured with 2 M HCl, and blocked with 10% goat serum. The cells were stained for 1 h at room temperature with anti-BrdU primary antibody (Abcam, ab6326, 1:250), and then incubated with the CY3-labeled secondary antibody. The data were quantitated by counting the number of BrdU-positive cells in seven random microscopic views for each sample. The data are presented as the means ± SD.
Immunohistochemical staining
Immunohistochemical staining was performed as described previously 17. Information regarding all primary antibodies is provided in Table S3.
Results
HUWE1 expression in lung cancer
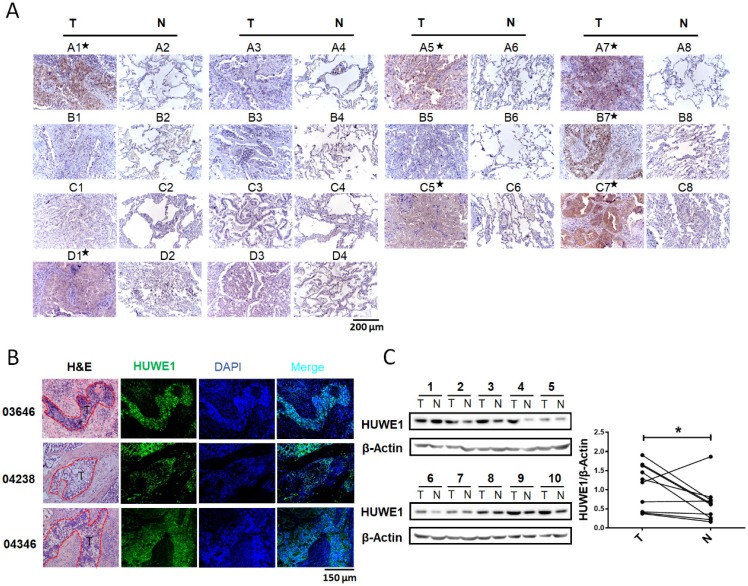
Although an increase in HUWE1 mRNA was reported in lung cancer, HUWE1 expression in lung cancer at the protein level had not been explored. Thus, we performed immunohistochemistry to determine and compare the expression of HUWE1 using a tissue microarray (Shanghai Outdo Biotech Co., ltd) containing 14 pairs of lung cancer and adjacent normal tissues. HUWE1 expression was elevated in cancer tissue in seven of 14 pairs (Figure 1A). To further confirm HUWE1 expression in larger tissue sections, immunohistochemistry was performed on frozen sections and similar staining patterns were observed (Figure 1B). Proteins were also extracted from another 10 pairs of frozen samples for western blot analysis, which confirmed the increase in HUWE1 expression in cancer (Figure 1C). Thus, our experimental results showing protein levels and previous studies exhibiting transcriptional levels all indicate the dysregulation of HUWE1 in lung cancers.
Figure 1.
HUWE1 expression in lung cancer. (A) HUWE1 staining in a tissue microarray containing 14 pairs of lung adenocarcinoma (T) and adjacent normal tissues (N). The samples with significantly elevated HUWE1 expression are marked by asterisks. (B) HUWE1 staining in frozen sections from non-small cell lung cancers. T, tumor; 03646, 04238 and 04346 indicate the numbers of the tumor sample from different patients. Samples 03646 and 04346 were squamous cell carcinoma and 04238 was adenocarcinoma. (C) Immunoblot showing the detection of HUWE1 in the lysates of paired tumor (T) and adjacent normal tissues (N). The tumor type of 1-6 was lung adenocarcinoma, whereas others were lung squamous cell carcinoma. β-actin was used as a loading control. The results were quantified using ImageJ, and intensity values were normalized to the β-actin band (*, P < 0.05, paired t-test).
HUWE1 inactivation inhibits lung cancer cell proliferation and tumor growth
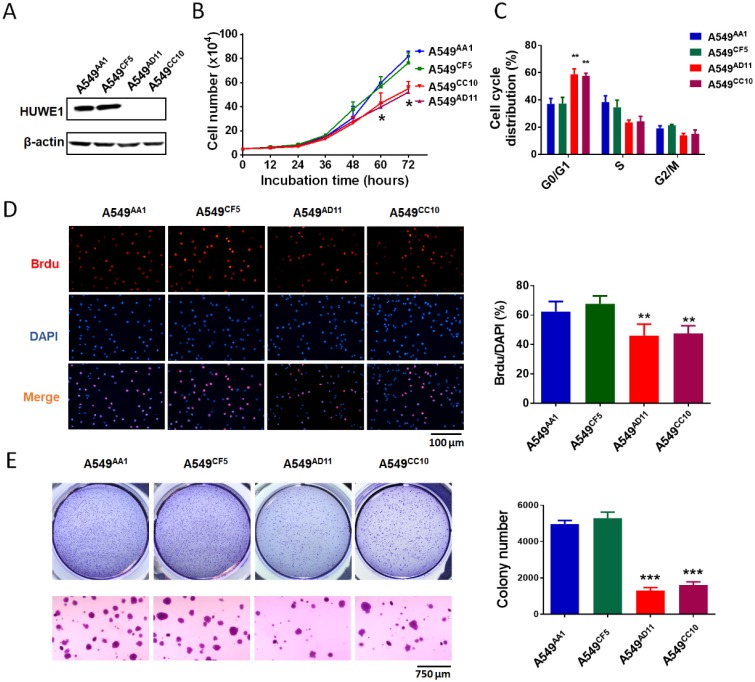
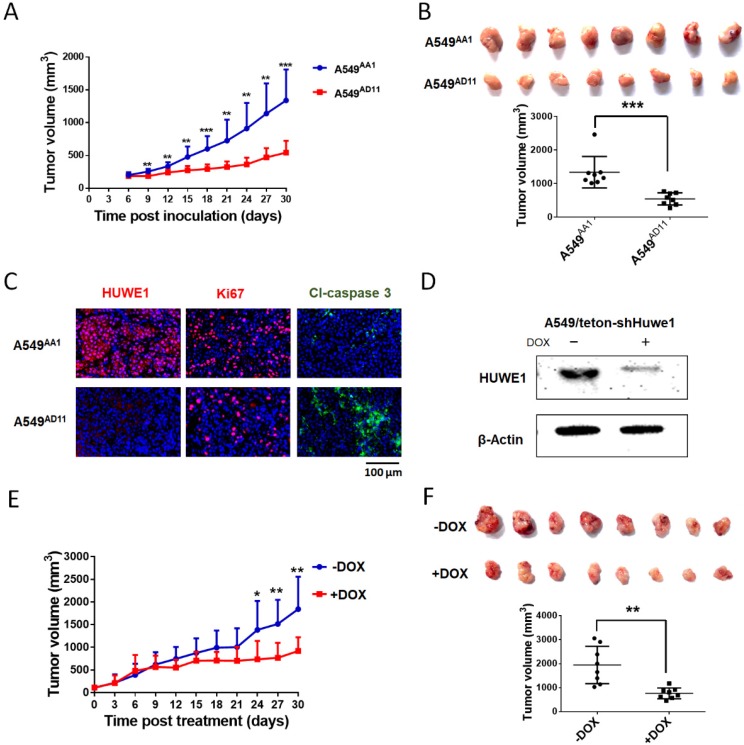
To study the role of HUWE1 in lung cancer, we performed genome editing on the HUWE1 gene in a widely-used lung cancer cell line, A549, using CRISPR/cas9 (Figure S1). Single-cell clones were picked and expanded, and HUWE1 inactivation was confirmed by DNA sequencing and western blotting. HUWE1 intact clones A549AA1 and A549CF5 and HUWE1-null clones A549AD11 and A549CC10 were used for the following experiment (Figure S2 and Figure 2A). As shown in the cell growth curve, the proliferation was significantly inhibited by HUWE1 deletion (Figure 2B). Both an increased proportion of cells in the G1 phase and decreased BrdU-incorporation indicated G1 arrest upon HUWE1 inactivation (Figure 2C-D and Figure S3). Moreover, HUWE1-null clones also showed significantly decreased colony formation capacity (Figure 2E). No obvious cell death was observed in HUWE1-null cells, and this conclusion was also verified by the apoptosis marker cleaved caspase-3 staining (Figure S4). Although it was reported that some substrates of HUWE1 are involved in DNA damage, results from the comet assay did not appear to detect remarkable changes upon HUWE1 deletion (Figure S5). To determine if HUWE1 deletion inhibits tumor growth, HUWE1-intact clones A549AA1 and HUWE1-null clones A549AD11 were injected subcutaneously into immunocompromised mice. A549AD11 xenograft tumors grew significantly slower than formed by A549AA1 cells (Figure 3A-B). Mice were sacrificed after 30 days, and immunohistochemical staining was performed. As expected, HUWE1 was completely absent in tumors derived from A549AD11 cells and expression of the proliferation marker Ki-67 was significantly reduced (Figure 3C). Interestingly, an increase in cellular apoptosis was observed in tumors, which was not observed in vitro (Figure 3C).
Figure 2.
HUWE1 deletion impairs cell proliferation and colony formation. (A) Immunoblot showing the deletion of the HUWE1 in A549 cells; β-actin was used as a loading control. (B) Growth curves for HUWE1-wild type cells and HUWE1-null cells. The data are presented as the means ± SD (*, P < 0.05, unpaired t-test). (C) Cell cycle distribution analysis for HUWE1-wild type cells and HUWE1-null cells. The data are presented as the means ± SD of three independent experiments shown in Figure S3 (**, P < 0.01, unpaired t-test). (D) BrdU incorporation assay was used to evaluate the DNA synthesis and proliferation rates of the indicated cell lines. The data are presented as the means ± SD (**, P < 0.01, unpaired t-test). (E) Soft agar colony formation assays using the indicated cell clones. Quantification of colony numbers is shown at the right. The data are presented as the means ± SD (***, P < 0.001, unpaired t-test).
Figure 3.
HUWE1 inactivation inhibits the tumorigenicity of lung cancer cells. (A) The HUWE1-intact clone A549AA1 and HUWE1-null clone A549AD11 were injected subcutaneously into BALB/c nude mice. Tumor sizes were measured every 3 days. The results are presented as the mean volume ± SD (**, P < 0.01; ***, P < 0.001, unpaired t-test). (B) Tumors were dissected 30 days after the injection of tumor cells. The quantification of tumor volumes is shown below. Each dot represents one tumor volume in a single mouse. The data are presented as the means ± SD (***, P < 0.001, unpaired t-test). (C) HUWE1, Ki67, and cleaved (Cl)-caspase 3 staining in indicated cancer tissues. (D) Western blot showing the expression of HUWE1 in the lysates of A549/teton-shHUWE1 cells with or without doxycycline (DOX). β-actin was used as a loading control. (E) Growth curve of the tumors formed by A549/teton-shHUWE1 cells in BALB/c nude mice. Doxycycline (DOX) or sucrose was added to the drinking water of mice 7 days after the injection of tumor cells. Tumor sizes were then measured every 3 days. The results are presented as the mean volume ± SD (*, P < 0.05; **, P < 0.01, unpaired t-test). (F) Tumors were dissected at the end of experiments. Quantification of tumor volumes is shown below. Each dot represents one tumor volume in a single mouse. The data are presented as the means ± SD (**, P < 0.01, unpaired t-test).
To further confirm the role of HUWE1 in lung cancer cells, A549 cells were infected with lentivirus expressing tetracycline-inducible HUWE1 shRNA (A549/teton-shHUWE1). HUWE1 knockdown was confirmed by western blotting (Figure 3D). As shown in HUWE1-null cells, knockdown of this gene impaired proliferation and colony-forming activity in lung cancer cells but had no effect on apoptosis. (Figure S6). A549/teton-shHUWE1 cells were subcutaneously injected into immunocompromised mice and HUWE1 knockdown was induced by doxycycline 7 days after injection when tumor sizes reached approximately 5 mm in diameter. Treatment with doxycycline inhibited tumor growth and decreased tumor size (Figure 3E). When mice were sacrificed on day 32 after Dox treatment, the mice that received Dox treatment had significantly diminished tumor burden compared to that in the control group (Figure 3F). Immunohistochemical staining indicated a decrease in HUWE1 protein expression in Dox-treated tumors (Figure S7). HUWE1 knockdown also decreased the number of proliferating cells and increased the expression of the apoptosis marker cleaved-caspase 3 (Figure S7).
p53 is a key mediator of the phenotype induced by HUWE1 inhibition
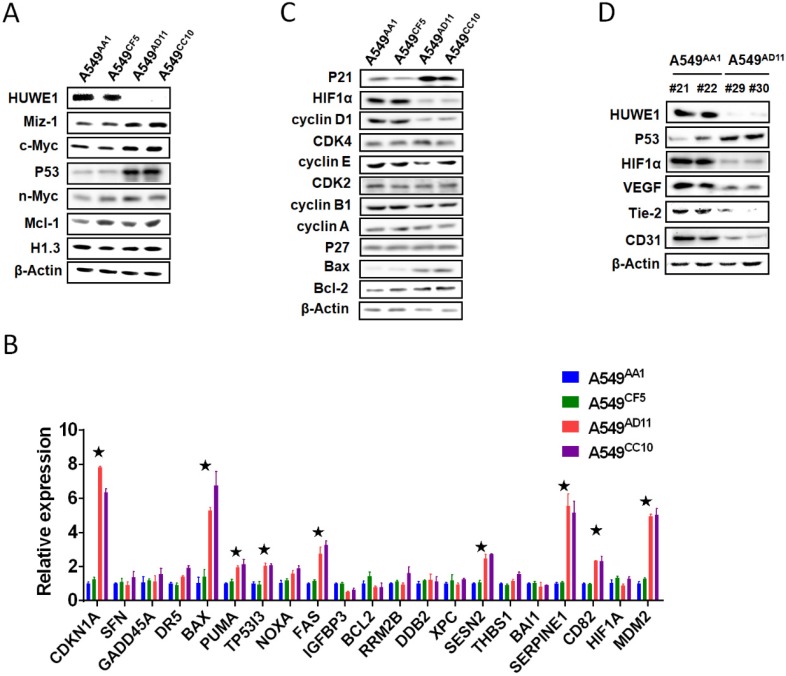
To elucidate the molecular mechanisms through which HUWE1 inactivation inhibits tumor growth, the substrates of HUWE1 were detected by western blotting. p53 was significantly accumulated, whereas some other substrates such as Miz-1 and c-MYC increased slightly (Figure 4A). No significant change was found regarding the expression of histone H1.3, which was reported to be a substrate of HUWE1 and to mediate the role of HUWE1 in ovarian cancer (Figure 4A). We then determined the expression of p53-regulated genes by real-time PCR. Consistent with our expectations, many genes encoding proteins such as the cell-cycle inhibitor CDKN1A, the apoptosis promoters PUMA, TP53I3, BAX, and FAX, the DNA damage response-associated SESN2, and the angiogenesis inhibitors SERPINE1 and CD82 were up-regulated upon HUWE1 inactivation (fold change > 2; Figure 4B). The MDM2 gene encoding an E3 ligase that ubiquitinates p53 for proteasome-mediated degradation also increased (Figure 4B), which might be due to feedback regulation 27. These data demonstrated the increase in p53 expression and activation of the p53 pathway. p53 is a well-known tumor suppressor and thus we speculated that its increase might explain the inhibition of tumor growth by HUWE1 inactivation in lung cancer.
Figure 4.
p53 is the key mediator of HUWE1 in lung cancer. (A) The main substrates of HUWE1 were detected by western blotting in HUWE1-wild type and HUWE1-null cells; β-actin was used as a loading control. (B) Total RNA was isolated from the indicated cells. Real-time quantitative RT-PCR was used to determine the expression of genes transcriptionally regulated by p53 in HUWE1-wild type cells and HUWE1-null cells. The genes with fold change > 2 are marked by asterisks. The data are presented as the means ± SD. (C) Immunoblotting was used to determine the expression of cell-cycle associated proteins in the lysates of HUWE1-wild type and HUWE1-null cells; β-actin was used as a loading control. (D) Proteins were extracted from the tumors formed by the HUWE1-intact clone A549AA1 and the HUWE1-null clone A549AD11 in BALB/c nude mice, as described in Figure 3A. Markers of angiogenesis in the lysates were detected by western blotting; #21, #22, #29, and #30 indicate the serial numbers of mice. β-actin was used as a loading control.
Considering that decreased cell proliferation was one of the main phenotypes associated with HUWE1 inactivation, the expression of cell-cycle associated proteins was assessed. Similar to real-time PCR data, p21 remarkably increased upon HUWE1 deletion, which could be attributed to p53 upregulation. Cyclin D1, the target protein of p21, also decreased in HUWE1-null cells (Figure 4C). It has been reported that the cyclinD1-CDK4 complex promotes passage through the G1 phase by inhibiting the retinoblastoma protein 28. We assessed expression of core c-Myc downstream targets including cyclin A, cyclin E, and CDK4, and found no significant changes when HUWE1 was inactivated (Figure 4C).
As the significant difference in apoptosis between in vivo and in vitro conditions was speculated to be potentially caused by hypoxemia, HIF1α, which is also regulated by p53 on the protein level and plays an important role in enhancing tumor growth by promoting angiogenesis and regulating cellular metabolism to overcome hypoxia, was also assessed by western blotting; this protein was found to be down-regulated with HUWE1 ablation (Figure 4C). To determine if HUWE1 affects tumorigenesis by maintaining HIF1α expression, markers of angiogenesis including VEGF, Tie2, and CD31 were detected in the lysates of subcutaneous tumors formed by A549 cells. Results showed that all of these proteins were strikingly down-regulated with HUWE1 inactivation (Figure 4D and Figure S8).
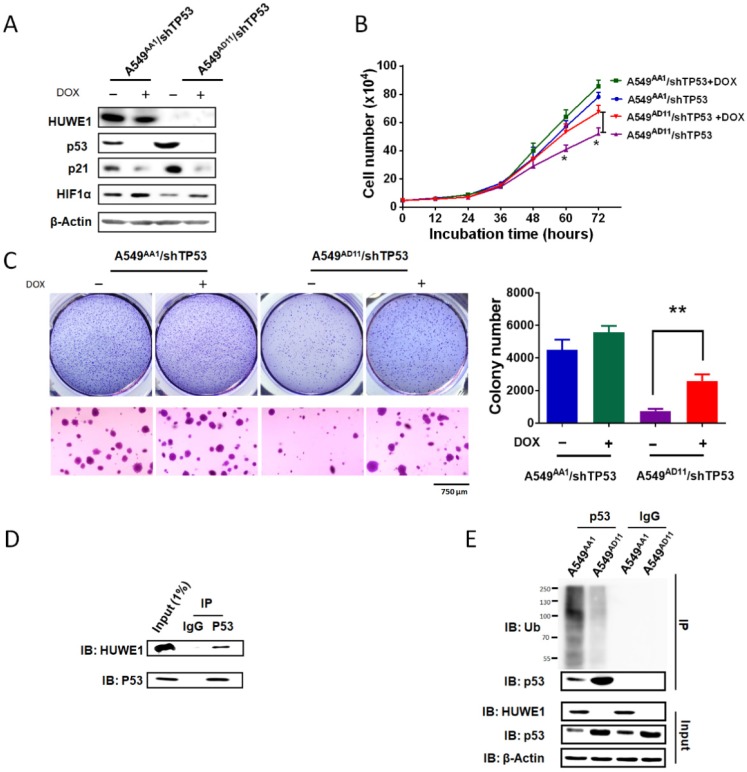
To confirm the role of p53 in mediating the phenotype associated with HUWE1 down-regulation, TP53 was silenced in HUWE1-null cells. The up-regulation of p21 and down-regulation of HIF1α mediated by HUWE1 deletion were partially rescued by p53 silencing (Figure 5A). Phenotypically, the effects of HUWE1 ablation on cell proliferation and colony formation were also partially rescued (Figure 5B-C). However, reduced p53 expression did not restore SERPINE1 and CD82 expression on the mRNA level, although they were up-regulated upon HUWE1 inactivation (Figure S9). We speculated that they could be regulated by HUWE1 in a p53-independent manner. Taken together, the aforementioned data demonstrate that p53 is one of the key mediators of HUWE1 in lung cancer.
Figure 5.
p53 silencing partially rescues the effects of HUWE1 deletion. (A) Western blotting for the indicated proteins in teton-shTP53 lentivirus-infected HUWE1-wild type cells and HUWE1-null cells; TP53 knockdown was induced by doxycycline (DOX). β-actin was used as a loading control. (B) Growth curves for the indicated cells. The data are presented as the means ± SD (*, P < 0.05, unpaired t-test). (C) Soft agar colony formation assay of the indicated cells. Quantification of colony numbers is shown. The data are presented as the means ± SD (**, P < 0.01, unpaired t-test). (D) Proteins were extracted from the HUWE1-intact clone A549AA1. Co-immunoprecipitation assays were performed to assess the interaction between HUWE1 and p53 using the p53 antibody. Rabbit IgG was used as a negative control. (E) In vivo ubiquitination assays to evaluate the amount of ubiquitinated p53 in HUWE1 wild type cells and HUWE1-null cells. The cells were treated with 5 μM MG-132 for 6 h, then lysed, and subjected to immunoprecipitation with the p53 antibody. Ubiquitinated p53 was detected by immunoblotting with an anti-ubiquitin antibody.
Ubiquitin-dependent p53 degradation mediated by HUWE1 was reported in previous studies 29. Here we determined the associated mechanism in lung cancer cells. Co-immunoprecipitation was performed in A549 cells harboring intact HUWE1. Western blot analysis demonstrated binding between HUWE1 and p53 (Figure 5D). Next, ubiquitination assays were performed in HUWE1-intact and HUWE1-null cells and we found that HUWE1 deletion decreased the level of ubiquitinated p53 (Figure 5E).
Huwe1 deletion inhibits lung cancer development in mice
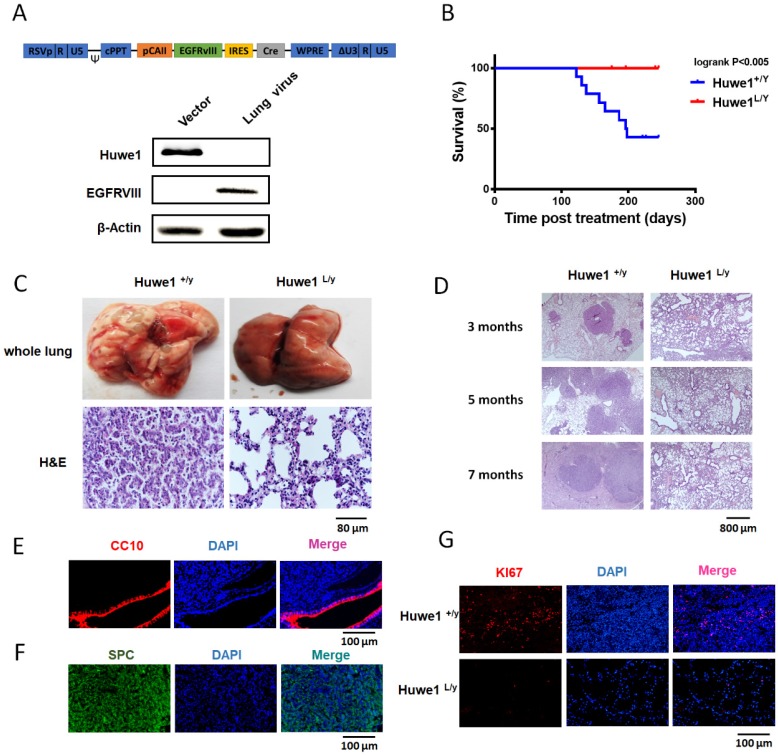
To further elucidate the role of Huwe1 in lung cancer, we studied its effect on lung cancer formation in Huwe1 conditional knockout mice. Alterations in EGFR have been confirmed to be driver mutations in human lung cancer and are widely used in mouse models for lung cancer research 10, 30. Here we constructed a lentiviral vector expressing EGFRVIII and Cre driven by the carbonic anhydrase II (CAII) promoter, which is expressed mainly in alveolar epithelial cells and was previously demonstrated to efficiently drive adenocarcinoma formation in mouse lungs 31. Lentiviral particles were tested for EGFRVIII expression and Huwe1 deletion in vitro (Figure 6A), and these were then delivered via intranasal infection. We determined that 54% of Huwe1+/Y mice developed lung cancer by 8 months after lentivirus infection, whereas all Huwe1L/Y mice were healthy with no obvious signs of lung cancer (Figure 6B).
Figure 6.
Huwe1 deletion inhibits lung cancer development in mice. (A) The schematic of lentiviral vector pTomo-EGFRVIII-IRES-cre. EGFRVIII expression and Cre function were verified in HUWE1L/L mouse ovary epithelial cells by western blotting; β-actin was used as a loading control. (B) Kaplan-Meier survival curve showing Huwe1+/Y and Huwe1L/Y mouse survival after pTomo-EGFRVIII-IRES-cre lentivirus infection; n = 13. (C) Whole lungs collected from Huwe1+/Y and Huwe1L/Y mice and corresponding histopathologic assay (H&E staining). (D) Huwe1+/Y and Huwe1L/Y mice were sacrificed at different stages after lentivirus infection. H&E staining in paraffin sections cut from the tumors showed different lung cancer burdens. (E-G) CC10, SPC, and Ki67 staining in paraffin-embedded tumor sections.
Similar to previous reports of EGFRVIII-induced lung cancer, histological examination of lung cancer in Huwe1+/Y mice showed characteristics of lung adenocarcinoma 32, 33 (Figure 6C-D). This was also confirmed by pulmonary-associated surfactant protein C (SPC)-positive and clara cell-specific protein (CC10)-negative staining (Figure 6E-F). Cellular proliferation was also detected in cancers, and the rate of Ki67 positivity was approximately 10%, which is similar to that in human adenocarcinomas 34 (Figure 6G). In conclusion, these data demonstrated the critical role of Huwe1 in lung cancer development.
High expression of HUWE1 is associated with poor survival in lung cancer patients
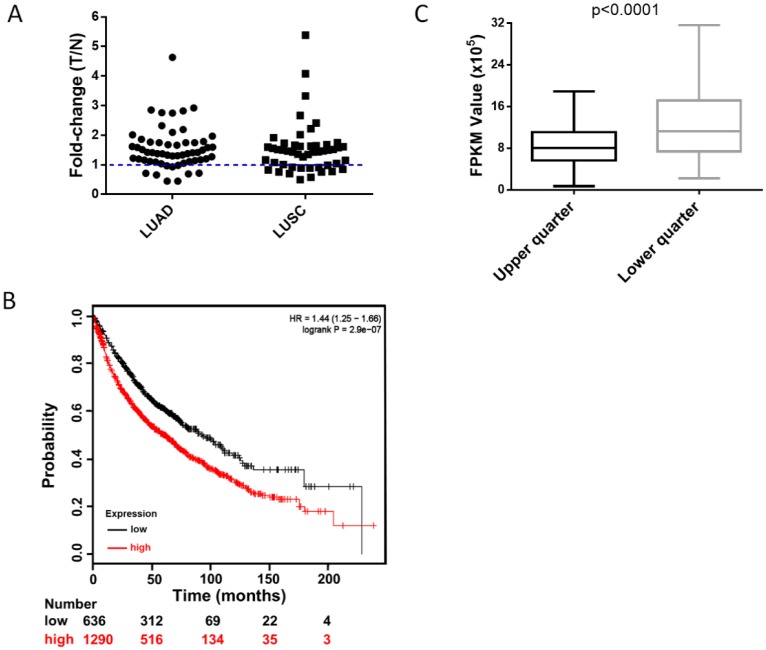
To clarify the clinical relevance of HUWE1, we analyzed the RNA-Seq data of paired lung cancer and para-cancerous tissues from The Cancer Genome Atlas (TCGA) and found that approximately 82% of lung adenocarcinomas and 73% of lung squamous cell carcinomas had elevated HUWE1 expression compared to that in paired normal tissue (fold change > 1, Figure 7A). Unexpectedly, the change in MDM2 was not significant (Figure S10A). Importantly, survival analysis showed that increased HUWE1 expression is significantly associated with poorer prognosis using online survival analysis software including survival information of the caBIG, GEO, and TCGA databases 26 (Figure 7B). However, there was no significant association between MDM2 expression and prognosis (Figure S10B). By analyzing the RNA-Seq data of lung adenocarcinoma and squamous cancer tissues from TCGA, we found that the expression of CDKN1A (the gene encoding p21) in the HUWE1-low expression group was significantly higher than that in the in HUWE1-high expression group (Figure 7C), which was consistent with our experimental data. Meanwhile, MDM2 had no obvious relationship with CDKN1A (Figure S10C). The non-significant correlation between HIF1α and HUWE1 at the transcription level, which was shown in our experiment, was also confirmed by the RNA-Seq data from the TCGA (data not shown). These results suggested that HUWE1 regulates HIF1α expression primarily at the protein level in non-small-cell lung carcinoma.
Figure 7.
High expression of HUWE1 is associated with poor survival in lung cancer. (A) The fold-change of HUWE1 mRNA expression in lung cancer (T) and adjacent normal tissue (N). The RNAseq data was downloaded from the TCGA website. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma. (B) Kaplan-Meier plot showing overall survival of non-small-cell lung cancer patients stratified by high or low HUWE1 mRNA expression (lower tertile). (C) Box plot showing the mRNA expression of CDKN1A in the HUWE1-low expression group (lower quarter) and HUWE1-high expression group (upper quarter) (P < 0.001, unpaired t-test).
Discussion
Many HUWE1 substrates have been identified, such as the tumor suppressors BRCA1 35 and p53 29, the oncogenes c-Myc 18 and n-Myc 36, the anti-apoptotic factor Mcl-1 37, the cell cycle protein CDC6 38, the DNA damage repair-related proteins H2AX 39 and TopBP1 40, the DNA polymerase Pol β 41 and Pol λ 42, and key proteins of the Wnt signaling pathway, including β-catenin 43 and Dishevelled 44. By controlling the stability of its substrates, HUWE1 is involved in biological processes such as DNA damage repair, cell proliferation, apoptosis, differentiation, migration, neurodevelopment, and spermatogenesis. HUWE1 overexpression was reported in cancers of the ovary, lung, breast, colon, liver, and pancreas 9, 18. However, as shown in the TCGA database, deletions or mutations in the HUWE1 gene were also found in a significant number of human samples such as lung, esophageal, cervical, melanoma, head and neck. These results suggest that HUWE1 plays a diverse role in different cancer types, or even in different patients with the same type. This was also confirmed by in vivo and in vitro studies 14-16, 29, 37. Previous data showed that HUWE1 performed oncogenic functions mainly through the activation of the proto-oncogene c-Myc 18, 45, or by targeting histone H1.3 for ubiquitination and degradation 17. However, in this study, we found that the oncogenic function of HUWE1 occurs through a different mechanism. p53, rather than H1.3 or c-Myc, was the most significantly increased substrate. In HUWE1-null cells, p21, a well-known transcriptional target of TP53, increased at the mRNA and protein level, and resulted in the G1 arrest of cancer cells. Meanwhile, another p53 target protein HIF1α was down-regulated, which blocked angiogenesis in a xenograft model.
p53 accumulation mediated by HUWE1 inactivation has been frequently reported to induce apoptosis, for example, in U2OS cells in vitro 29 and in mouse pancreatic β-cells in vivo 46. Nonetheless, HUWE1 inactivation in a human lung cancer cell line did not induce cellular apoptosis in vitro. Interestingly, HUWE1 inactivation obviously resulted in apoptosis in xenograft tumors. This might have resulted from the hypoxic environment and the decrease in HIF1α in solid tumors. HIF1α is a recognized oncogene that has been reported to help cancer cells overcome hypoxia through its role in initiating angiogenesis and regulating cellular metabolism. Although an association between HUWE1 and HIF1α has not been reported, another E3 ligase, MDM2, which targets p53, was shown to inhibit HIF1α directly or through degradation of the p53/HIF1α complex 47. In our experiments, the fact that p53 silencing could rescue the expression of HIF1α implied that the decrease in this protein was mediated by p53 instead of through direct degradation by HUWE1.
Although MDM2 has been reported as the primary E3 ligase that is directly involved in p53 regulation, an increasing number of studies suggest that p53 proteasomal degradation is also part of the MDM2-independent pathway 29, 48. p53 was the first identified substrate of HUWE1 29, 49. However, no direct evidence has demonstrated a role for the HUWE1-p53 axis in lung cancer. Based on the analysis of paired lung cancer and adjacent normal tissues from the TCGA database, we found that increased HUWE1 expression was much higher than that of MDM2. Accordingly, samples with increased HUWE1 expression were associated with worse prognosis. Meanwhile, MDM2 expression did not correlate with survival. These data suggested that HUWE1 might play more important roles in lung cancer and could be more clinically relevant than MDM2. Small molecules inhibiting the MDM2-p53 association have been tested for lung cancer therapy in preclinical studies 13. Our results suggest that the HUWE1-p53 axis might represent a better target for lung cancer treatment. Some efforts have been made to screen or design molecules that target HUWE1 for cancer therapy. BI8622 and BI8626 inhibit the proliferation and colony formation of the colon cancer cell line Ls174T by preventing MYC-dependent transactivation 15.
Taken together, HUWE1 plays a critical role in the initiation and development of lung cancer by controlling p53 proteasomal degradation. Hence, the HUWE1-p53 axis is important for mediating the effects of HUWE1 in lung cancer, and thus could represent a potential target for lung cancer treatment.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (NSFC, 81171960), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA 01040403), the Top Talents Program of Yunnan Province, China (2012HA014) to X Zhao and National Natural Science Foundation of China (81773432) to X Yu.
Contributions
D. Yang design and performed all the experiments. Q. Tu, B. Sun and H. Yang prepared lentivirus and performed in vivo experiments. D. Cheng, L. Yan and Y. Cao performed pathology and immunostaining work. H. Dai, Y. Cao, H. Jiang, J. Luo and B. Mao provided material support. X. Zhao, H. Jiang and X. Yu contributed to analysis, interpretation of data and the writing of manuscript, and were responsible for study supervision.
Abbreviations
- BrdU
bromodeoxyuridine
- CC10
clara cell-specific 10-kDa protein
- DAPI
4', 6-diamidino-2-phenylindole, dihydrochloride
- DMEM
Dulbecco's modified Eagle's medium
- DOX
doxycycline
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- gRNA
guide RNA
- H&E
hematoxylin and eosin
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- NSCLC
non-small cell lung cancer
- PBS
phosphate buffered saline
- PI
propidium iodide
- RIPA
radio-immunoprecipitation assay
- SD
standard deviation
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SPC
pulmonary-associated surfactant protein C
- STR
short tandem repeat.
Supplementary Material
Supplementary figures 1-10.
Supplementary table S1.
Supplementary table S2.
Supplementary table S3.
Supplementary figures 11-24.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR. et al. NCCN guidelines insights: non-small cell lung cancer, version 4.2016. J Natl Compr Canc Netw. 2016;14:255–64. doi: 10.6004/jnccn.2016.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–46. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karve TM, Cheema AK. Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J Amino Acids. 2011;2011:207691. doi: 10.4061/2011/207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–54. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer ES, Bohm K, Lydeard JR, Yang H, Stadler MB, Cavadini S. et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan YHC, Krishnaswamy S, Nandi S, Kanteti R, Vora S, Onel K. et al. CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. Plos One. 2010;5:e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Confalonieri S, Quarto M, Goisis G, Nuciforo P, Donzelli M, Jodice G. et al. Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. Oncogene. 2009;28:2959–68. doi: 10.1038/onc.2009.156. [DOI] [PubMed] [Google Scholar]
- 10.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S. et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–21. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Sun Y. Targeting p53 for novel anticancer therapy. Transl Oncol. 2010;3:1–12. doi: 10.1593/tlo.09250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hai J, Sakashita S, Allo G, Ludkovski O, Ng C, Shepherd FA. et al. Inhibiting MDM2-p53 interaction suppresses tumor growth in patient-derived non-Small cell lung cancer xenograft models. Journal of Thoracic Oncology. 2015;10:1172–80. doi: 10.1097/JTO.0000000000000584. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Zeng SX, Lu H. Targeting p53-MDM2-MDMX loop for cancer therapy. Subcell Biochem. 2014;85:281–319. doi: 10.1007/978-94-017-9211-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myant KB, Cammareri P, Hodder MC, Wills J, Von Kriegsheim A, Gyorffy B. et al. HUWE1 is a critical colonic tumour suppressor gene that prevents MYC signalling, DNA damage accumulation and tumour initiation. EMBO Mol Med. 2017;9:181–97. doi: 10.15252/emmm.201606684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peter S, Bultinck J, Myant K, Jaenicke LA, Walz S, Muller J. et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol Med. 2014;6:1525–41. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue S, Hao Z, Elia AJ, Cescon D, Zhou L, Silvester J. et al. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev. 2013;27:1101–14. doi: 10.1101/gad.214577.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang D, Sun B, Zhang XH, Cheng DM, Yu XP, Yan LZ. et al. Huwe1 sustains normal ovarian epithelial cell transformation and tumor growth through the histone H1.3-H19 cascade. Cancer Res. 2017;77:4773–84. doi: 10.1158/0008-5472.CAN-16-2597. [DOI] [PubMed] [Google Scholar]
- 18.Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R. et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–21. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Vaughan L, Tan CT, Chapman A, Nonaka D, Mack NA, Smith D. et al. HUWE1 ubiquitylates and degrades the RAC activator TIAM1 promoting cell-cell adhesion disassembly, migration, and invasion. Cell Rep. 2015;10:88–102. doi: 10.1016/j.celrep.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, D'Arca D, Lim WK, Brahmachary M, Carro MS, Ludwig T. et al. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell. 2009;17:210–21. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marumoto T, Tashiro A, Friedmann-Morvinski D, Scadeng M, Soda Y, Gage FH. et al. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15:110–6. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan C-H, Lee S-W, Li C-F, Wang J, Yang W-L, Wu C-Y. et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol. 2010;12:457–67. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi DL, Cobrinik D. MDM2 but not MDM4 promotes retinoblastoma cell proliferation through p53-independent regulation of MYCN translation. Oncogene. 2017;36:1760–9. doi: 10.1038/onc.2016.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31:229–34. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- 26.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer (vol 8, e82241, 2013) Plos One. 2014;9:e111842. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 28.Matsushime H, Ewen ME, Strom DK, Kato JY, Hanks SK, Roussel MF. et al. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992;71:323–34. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 29.Chen DL, Kon N, Li MY, Zhang WZ, Qin J, Gu W. ARF-BP1/mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–83. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 30.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19:643–64. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- 31.Xia Y, Yeddula N, Leblanc M, Ke E, Zhang Y, Oldfield E. et al. Reduced cell proliferation by IKK2 depletion in a mouse lung-cancer model. Nat Cell Biol. 2012;14:257–65. doi: 10.1038/ncb2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A. et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2006;103:7817–22. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–8. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 34.Rekhtman N. Neuroendocrine tumors of the lung: an update. Arch Pathol Lab Med. 2010;134:1628–38. doi: 10.5858/2009-0583-RAR.1. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Lu G, Li L, Yi J, Yan K, Wang Y. et al. HUWE1 interacts with BRCA1 and promotes its degradation in the ubiquitin-proteasome pathway. Biochem Biophys Res Commun. 2014;444:549–54. doi: 10.1016/j.bbrc.2014.01.075. [DOI] [PubMed] [Google Scholar]
- 36.Zhao XD, Heng JIT, Guardavaccaro D, Jiang R, Pagano M, Guillemot F. et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–53. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–95. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Hall JR, Kow E, Nevis KR, Lu CK, Luce KS, Zhong Q. et al. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell. 2007;18:3340–50. doi: 10.1091/mbc.E07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atsumi Y, Minakawa Y, Ono M, Dobashi S, Shinohe K, Shinohara A. et al. ATM and SIRT6/SNF2H mediate transient H2AX stabilization when DSBs form by blocking HUWE1 to allow efficient gammaH2AX foci formation. Cell Rep. 2015;13:2728–40. doi: 10.1016/j.celrep.2015.11.054. [DOI] [PubMed] [Google Scholar]
- 40.Herold S, Hock A, Herkert B, Berns K, Mullenders J, Beijersbergen R. et al. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. Embo J. 2008;27:2851–61. doi: 10.1038/emboj.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsons JL, Tait PS, Finch D, Dianova II, Edelmann MJ, Khoronenkova SV. et al. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. Embo J. 2009;28:3207–15. doi: 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markkanen E, van Loon B, Ferrari E, Parsons JL, Dianov GL, Hubscher U. Regulation of oxidative DNA damage repair by DNA polymerase lambda and MutYH by cross-talk of phosphorylation and ubiquitination. Proc Natl Acad Sci U S A. 2012;109:437–42. doi: 10.1073/pnas.1110449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dominguez-Brauer C, Khatun R, Elia AJ, Thu KL, Ramachandran P, Baniasadi SP. et al. E3 ubiquitin ligase Mule targets beta-catenin under conditions of hyperactive Wnt signaling. Proc Natl Acad Sci USA. 2017;114:E1148–E57. doi: 10.1073/pnas.1621355114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Groot RE, Ganji RS, Bernatik O, Lloyd-Lewis B, Seipel K, Sedova K. et al. Huwe1-mediated ubiquitylation of dishevelled defines a negative feedback loop in the Wnt signaling pathway. Sci Signal. 2014;7:ra26. doi: 10.1126/scisignal.2004985. [DOI] [PubMed] [Google Scholar]
- 45.Schaub FX, Cleveland JL. Tipping the MYC-MIZ1 balance: targeting the HUWE1 ubiquitin ligase selectively blocks MYC-activated genes. EMBO Mol Med. 2014;6:1509–11. doi: 10.15252/emmm.201404735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Luk CT, Schroer SA, Smith AM, Li X, Cai EP. et al. Dichotomous role of pancreatic HUWE1/MULE/ARF-BP1 in modulating beta cell apoptosis in mice under physiological and genotoxic conditions. Diabetologia. 2014;57:1889–98. doi: 10.1007/s00125-014-3295-8. [DOI] [PubMed] [Google Scholar]
- 47.Sermeus A, Michiels C. Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis. 2011;2:e164. doi: 10.1038/cddis.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsvetkov P, Reuven N, Shaul Y. Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ. 2010;17:103–8. doi: 10.1038/cdd.2009.67. [DOI] [PubMed] [Google Scholar]
- 49.Gu J, Dubner R, Fornace AJ Jr, Iadarola MJ. UREB1, a tyrosine phosphorylated nuclear protein, inhibits p53 transactivation. Oncogene. 1995;11:2175–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures 1-10.
Supplementary table S1.
Supplementary table S2.
Supplementary table S3.
Supplementary figures 11-24.