Abstract
MicroRNA (miRNA) is a small non-coding RNA with an established function to regulate genes at the post-transcriptional level leading to suppression or degradation of its messenger RNA expression (mRNA). Its dysregulation plays a vital role in a variety of biological and pathological processes including cancer. A lot of algorithms have been established to predict the target sites of miRNA, but experimentally identifying and validating its target region is still lacking. Guidance in experimental procedures is really needed to find genuine miRNA targets. Therefore, in this review, we provide an outline on the workflow in predicting and validating the targeted sites of miRNA using several methods as a guideline for the scientists. The final outcome of this type of experiment is essential to explore the major impact of miRNAmRNA interaction involved in the biological processes and to assist miRNA-based drug development in the future.
Key words: microRNA, cancer, algorithm, target identification
Introduction
MicroRNA (miRNA) is a short single-stranded non-coding RNA, approximately 18 to 25 nucleotides in length. It is generated from a primary miRNA transcript (pri-miRNA) which is transcribed from non-protein-coding genes or from the introns of protein- coding genes in the nucleus. Then, pri-miRNA is cleaved by RNAse III enzyme Drosha to form hairpin precursor miRNA (premiRNA) with ~70 nucleotides in length. This is followed by the pre-miRNA is transported into cytoplasm by a Ran-GTP-dependent nuclear export factor, Exportin-5. The pre-miRNA in the cytoplasm is further processed by RNAse III-like nuclease Dicer to generate a mature miRNA strand which is roughly 22 nucleotides in duplex form.1
More than a decade after miRNA was first discovered in 1993, it has started to draw the attention of the science community as it was emerged as an important regulator of gene expression by sharing its extensive sequence complimentarily.2 It was known to control in every aspect in biology such as development, proliferation, differentiation and metabolism. Dysregulation of miRNAs has been reported to lead to many human diseases, including cardiovascular disorders, complex genetic diseases and cancer.3-5
MicroRNA in cancer
It was found that cancer is the most relevant pathology in the world of miRNA-mRNA interactions, where many of the miRNA target sites are clustered in cancer-associated genomic region.6 It can take part either as an oncogene (oncomir), which is upregulated in tumor tissues to promote oncogenesis. On the other hand, it could act as a tumor-suppressor gene (anti-oncomir), which is downregulated in tumor tissues that promotes tumor growth. Dysregulation of these miRNAs could then promote oncogenesis.
MiRNAs have been attested to take part in many types of cancers such as gastric, ovary, lungs, thyroid, colon, prostate and breasts.7-13 Our previous studies on endometrioid endometrial cancer have identified several important transcripts and their related molecular pathways.14,15 The up- or down-regulation of these genes are most likely regulated by miRNAs. The relationship between miRNAs and mRNAs is complex as each miRNA could have multiple target sites on the targeted mRNA and vice versa with each mRNA could be targeted by several miRNAs.16 For example, in breast cancer, miR-29a/-b/-c has been reported to act as an oncogene, but in lung tumor, it turned out to be a tumor-suppressor gene.17,18 While in other studies, loss of miR-23b leads to bladder cancer cell migration and invasion, however in renal cell carcinoma, it reduced invasion by inducing apoptosis.19,20 Hence, miRNA is considered as the master regulator in cellular networks and signaling cascade as it regulates about 60% of the targeted mRNAs.21 To date, about 28,465 miRNAs have been discovered (miRBase Release 21: http://www.mirbase.org, accessed in March 2nd, 2018). To recognize the targeted mRNA, they do not need to completely match as it could identify through a perfect real complement sequence, known as seed region. Usually the seed region located at position 2 to 8 from 5’miRNA, where at this binding site, a stable Watson-Crick base pairs were energetically produced with the targeted mRNAs.21 Then, it could regulate the gene expression either by repressing the mRNA translation or causing mRNA degradation.22,23
The genetic diversity in different types of tumor is associated with different miRNAs in diverse actions and makes the relationship between miRNA and mRNA become more complex. They can participate in various mechanisms and pathways, by targeting key molecules and their networks involved in apoptosis, cell cycle, cell adhesion, cell migration, chromosome stability and DNA repair.24 As the expression of genes regulated by miRNAs is very distinct, thus, it is very essential to validate which target regions of each individual miRNA will give high impact on the gene expression. This is to help to elucidate their biological functions. Table 1 shows several databases available on the reported and experimentally validated miRNA-target interactions at the moment.25-28 However, to study miRNA-target interactions is very challenging as there are many prediction models while only a few have been biologically validated.29 So in this review, we summarize the workflow of molecular network analysis in terms of the prediction methods of miRNAs targets and the experimental approaches that have been described for the identification of their targets. In Figure 1, we proposed a workflow as a guideline in predicting and validating the targeted sites of miRNAs.
Table 1.
Databases of validated miRNA-target interactions.
| Name | URL | Reference |
|---|---|---|
| MirWalk 2.0 | http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2 | (25) |
| MirTarBase | http://mirtarbase.mbc.nctu.edu.tw/php | (26) |
| miRecords | http://c1.accurascience.com/miRecords | (27) |
| miRTarCLIP | http://mirtarclip.mbc.nctu.edu.tw | (28) |
Figure 1.
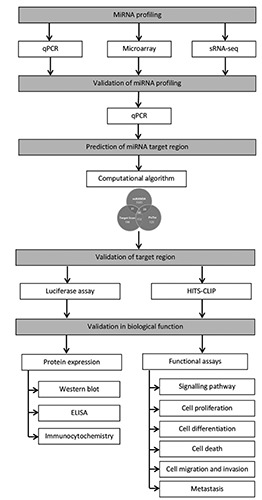
A proposed workflow as a guideline in predicting and validating the microRNA target site. miRNA, microRNA; qPCR, quantitative polymerase chain reaction; sRNA-seq, small RNA sequencing; HITS-CLIP, high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation.
MicroRNA profiling in cancers
To determine the target region and the biological effects of miRNA, a list of potential miRNAs is required to prepare prior to further characterization.30 Up till today, there are three methods in miRNA profiling which is quantitative polymerase chain reaction (qPCR), hybridisation-based methods such as miRNA profiling using microarray, and high-throughput sequencing such as small RNA sequencing (sRNA-Seq).
Quantitative PCR has been employed to profile miRNAs based on highly reported miRNAs related to cancers. Many companies have developed qPCR-based microarray for the miRNA expression detection. For example, miRStarTM Human Cancer Focus miRNA PCR Array (Arraystar) can test up to 184 miRNAs mostly that are related to cancer to be profiled in parallel. This array uses a panel that includes eight miRNA references to provide a better data quality and quantity, and also to normalize the PCR data. Therefore, qPCR is a powerful tool to analyze the expression level of cancer-related miRNAs quickly and conveniently where it requires less than six hours.31 The obstacle to perform this highly parallel qPCR is the optimal reaction conditions that may be different between miRNAs in primer annealing. Yet, the use of locked nucleic acid (LNAs) into primers is an effective strategy to optimize miRNA primer hybridization conditions. However, this predesigned miRNA cancer panel could not be used to identify novel miRNAs. This method is not suitable as a discovery tool for those who are looking for a new list of miRNAs related to a cancer but it is suitable to be used as a validation method.
MiRNA microarray is defined as a high-throughput technology used primarily in the analysis of gene expression. It was an established method used to analyze large numbers of miRNAs in the past.32,33 In this method, 3’ end of miRNAs are tagged with fluorophore- labeled nucleotides using T4 RNA ligase. There is a step whereby the labeled miRNA will hybridize to separately arrayed probes on slides or beads. After removing the non-specific binding, individual miRNAs can be detected. Till today, this method is still being chosen by many scientists due to its low price as compared to other methods, able to produce higher number of parallel measurement, and it could also compare relative abundance of specific miRNAs between two different states.27,28 However, the profiling results by this method produced lower specificity compared to small RNA-sequencing.34-37
Small RNA-sequencing (sRNAseq) is a new technique used to isolate and sequence many types of known and novel small RNA (sRNA) including miRNA. Usually the method starts with sRNA complementary DNA (cDNA) library preparation from the RNA sample of interest. Then, it is followed by parallel sequencing of millions of individual cDNA molecules from cDNA library on a single run. The sequence reads will be analyzed by bioinformatics to identify both known and novel microRNAs in the datasets, and specifies relative quantification by a digital approach. The major advantages of this method are it could produce precised miRNA sequence identification even for novel miRNAs, and also able to identify and distinguish between miRNA variants with high accuracy and sensitivity. Even though it is very helpful, however, it requires expertise in bioinformatics analysis and additional time to complete sequencing-based analysis.31
Without performing miRNA profiling, the list of whole human miRNAs could also be retrieved from miRBase 21 (www.mirbase. org), as described previously.38 The miRNA expression profiling dataset can also be downloaded from Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/geo) for focused miRNA selection, as all the data derived from experimental study performed on microarray, qPCR, and high-throughput sequencing.39
Validation of miRNA profiling and small RNA sequencing
Quantitative PCR is considered as a gold standard and established method in gene expression detection and quantitation to validate miRNA expression profiling and small RNA sequencing. There are two types of qPCR which is TaqMan qPCR and SYBR-greenbased qPCR. In TaqMan qPCR, a stem-loop primer is used in the reverse transcription reactions that are specific to the 3′ end of the miRNA. The forward and reverse primers will subsequently be used for amplification. While the DNA polymerase working on the template, the TaqMan probe will be hydrolyzed by Taq polymerase and freed the fluorescent dye from the quencher, resulting in light emission. In SYBR-green-based qPCR, to generate miRNA cDNA, enzymatic addition of a poly(A) tail is used. For PCR amplification, both specific forward and reverse primers are used to anneal to the 3′ miRNA. Then, SYBR green dye will bind to each double-stranded DNA product to give fluorescent intensity.
Prediction of miRNA target region
According to Bartel,21 computational approaches can accurately predict miRNAs with their targeted mRNAs based on the thermodynamic rule and the evolution of miRNA recognition element (MRE) sequence in silico. Besides 3’UTR, target sites of miRNA can also happen in 5’UTR and open reading frame (ORF).40-42 However, in this review, we are focusing more on 3’UTR since ORF and 5’UTR appear to be less effective and less frequent, and most target mRNAs studied to date are regulated through 3’UTR interaction.21
There are several computational algorithms that predict target MRE sequences with different measures that include basematched, target availability and evolutionary conservation of targeted sites. Basic characteristics of prediction methods as seen in Table 2 where it can compute all the predicted miRNA targeted sites of the gene by simply enter the Gene Symbol.28,43-46 All algorithms used can predict up to hundred miRNA’s targeted sites and the predicted binding sites usually distinct from one to another. Hence, the best possible way recommended in determining predicted miRNA binding site is by combining at least two or three algorithms that predict the same miRNA binding sites and identify the overlapping results among them.47
Table 2.
Computational methods for miRNA target prediction. 3’UTR, 3’untranslated region.
| Name | Targets on mRNA | Species | URL | Reference |
|---|---|---|---|---|
| DIANA Micro-T | 3’UTR | Human, mouse, rat and chicken | http://diana.imis.athena-innovation.gr/DianaTools/index.php | (43) |
| miRTar | Full length | Human | http://mirtar.mbc.nctu.edu.tw/human/ | (28) |
| TargetScan | 3’UTR | Human, mouse, rat, dog and chicken | http://www.targetscan.org/vert_71 | (44) |
| microrna.org | 3’UTR | Human, mouse, rat, roundworm and fruitfly | http://34.236.212.39/microrna/home.do | (45) |
| miRDB | 3’UTR | Human, mouse, rat, dog and chicken | http://www.mirdb.org | (46) |
3’UTR, 3’untranslated region.
Validation of the target regions
According to Martinez-Sanchez & Murphy (2013), once the predicted binding site of miRNA is determined, its functionality in biological model can be validated.48 As stated before, the computational algorithm could predict a high number of miRNA binding sites. To identify and verify these target interactions between miRNA and mRNA, luciferase reporter assay has become a favourable method among researchers. It is a recommended assay as it could produce rapid and reproducible results to determine the functional and non-functional interaction sites.
Generally, before performing this assay, 3’UTR of the transcript of interest is cloned downstream of the luciferase ORF contained in the reporter plasmid. Then, the recombinant plasmid together with their miRNAs of interest will be transiently transfected into host cells. After transfection is done, the miRNA will possibly bind to the mRNA specific targeted site, causing the reporter protein production to repress and hence, reducing the luciferase expression or activity. This is normally measured using fluorescent after 24 to 48 h post-transfection. To study the regulation of the selected miRNA, an antagonist to the miRNA could be used causing the action of miRNA to be inhibited, thus resulting the recovery of luciferase expression. This type of experiment could conclude that the luciferase repression is most likely due to the presence of the studied miRNA. Direct interaction of miRNAmRNA can be further verified by introducing mutated target site in the reporter assay. Hence, it could reduce or eliminate any miRNAmediated effects.48
Even though luciferase reporter assay is the best method to determine the functional target site biologically, however it can cause misleading assessment of targets.47 There is a possibility situation that leads to wrong interactions when there are two molecules with complementary sequence bind together. It can be caused by the wrong cofactor environment. The other disadvantages of this method are it needs a large workforce in terms of choosing the region to be cloned, cloning processes and multiple protocols to be optimized in cell transfection.42,49,50 Therefore, to validate interaction between miRNA-binding site determined by reporter assays, further experiment should be tested.
There is another experiment could be used to validate miRNAmRNA interactions, which is miRNA pull-down strategy, and this could be done even without the use of reporter assay.51,52 They demonstrated that miRNA-mRNA target pairs at the 3′UTR of the targeted mRNAcould be immunoprecipitate the RISC components with Argonaute (AGO) or TNRC6.45,53-55 To identify the complex of AGO-bound miRNA-mRNA, high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) was a powerful technique prior to immunoprecipitation. It could crosslink RNA to associated RNA-binding proteins by utilizing ultraviolet irradiation This technique was able to provide an extensive perception to the location of mRNA targeted by miRNA.55 Consequently, this technique works best in identifying an operative miRNA targets either based on their physical interaction in vitro or in vivo.
Validation in biological function
Once the target site of a given miRNA has been validated experimentally, it can be revealed that there is biologically gene regulation. 47 Theoretically, when the expression of miRNA is downregulated, the mRNA is predicted to be upregulated, and vice versa. Hence, the functionally inverse relationship between miRNAs and mRNAs target site can be validated by several method.
To look for the effects of validated targets towards miRNA and mRNA co-expression, it can be simply demonstrated by Northern blot analysis or qPCR by using total RNA from specific cell type, together with their specific primers of miRNA or targeted mRNA. Since the interaction of miRNA-mRNA is negatively regulated, the overexpression of miRNA would reduce the expression of the targeted mRNA, and vice versa. Another experimental technique to demonstrate miRNA and mRNA co-expression, is in situ hybridization whereby it can be utilized in tissue or cell of relevant samples.56
If the mRNA given was the real target of specific miRNA, the amount of protein expression could also change. Usually loss-offunction or gain-of-function (over-expression) experiments are the typical approach by transiently transfect miRNA mimics or inhibitors into a cell type which expressing the putative target protein. 39 Subsequently, western blot analysis using a specific antibody against protein can be utilized to look for the protein expression. 14 Another alternative experiment can be used to quantify difference in protein expression is ELISA or immunocytochemistry. So, with the presence of miRNA mimics or inhibitors, the target protein levels or activities should be easily detected. With the presence of miRNA mimics in cells, the endogenous expression of targeted mRNA would be reduced. However, with the presence of miRNA inhibitors, the endogenous expression of targeted miRNA would be reduced, hence, increased the expression of mRNA.
Once regulation of a target gene by a given miRNA has been confirmed experimentally, the regulation in biological function becomes necessarily to be demonstrated. Usually the assays includes signalling pathway, cell proliferation, cell differentiation, cell death, cell migration and invasion, metastasis, and others. For example, a study showed that the expression of miR-130a were up regulated in high grade serous ovarian cancer (HGSOC) tissues by using microarray and qPCR.57 However, with the presence of antimiR- 130a, it could reverse cell proliferation, migration and invasion. By using several algorithms which is TargetScan 5.1, miRDB and microrna.org to find potential miRNA-mRNA interaction, three potential target sites at 3’UTR has been confirmed by luciferase assay. Intraventricular injection of a miR-130a into nude mice enhanced tumor growth and metastasis in vivo.
Conclusions
The interactions of miRNA-mRNA contribute to the complexity and constitute the biological network of functionally-associated molecules in pathological events involved in cancer. A computational prediction algorithm has been developed recently. However only a small proportion of the interaction have been functionally validated. Even though there are limitations in the method discussed, following the workflow may lead to relevant target identification biologically, where understanding the functions may assist undoubted therapeutic potentials.
Acknowledgments
This work was supported by grants from University Kebangsaan, Malaysia (project code: FF-2017-192) and the Ministry of Higher Education, Malaysia (FRGS/1/2014/SKK01/UKM/02/2).
References
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009;10:126-39. [DOI] [PubMed] [Google Scholar]
- 2.Lee R, Feinbaum RL, Ambrost V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [DOI] [PubMed] [Google Scholar]
- 3.Lujambio A, Lowe S. The microcosmos of cancer Amaia. Nature 2012;482:347-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012;148:1172-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011;469:336-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci 2004;101:2999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene 2006;25:6220-7. [DOI] [PubMed] [Google Scholar]
- 8.Guo J, Miao Y, Xiao B, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol 2009;24:652-7. [DOI] [PubMed] [Google Scholar]
- 9.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A 2005;102:19075-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65:7065-70. [DOI] [PubMed] [Google Scholar]
- 11.Michael MZ, O’Connor SM, van Holst Pellekaan NG, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 2003;1:882-91. [PubMed] [Google Scholar]
- 12.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res 2008;14:2690-5. [DOI] [PubMed] [Google Scholar]
- 13.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 micrornas in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753-6. [DOI] [PubMed] [Google Scholar]
- 14.Saghir FSA, Rose IM, Dali AZHM, et al. Gene expression profiling and cancer-related pathways in Type I endometrial carcinoma. Int J Gynecol Cancer 2010;20:724-31. [DOI] [PubMed] [Google Scholar]
- 15.Mokhtar NM, Saghir FS, Dali AM, et al. Differentially expressed genes in endometroid endometrial carcinoma using microarray: a preliminary report. Reprod Fertil Dev 2008;20:89. [Google Scholar]
- 16.Gennarino VA, D’Angelo G, Dharmalingam G, et al. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res 2012;22:1163-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci 2007;104:15805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep 2009;10:400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majid S, Dar AA, Saini S, et al. MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS One 2013;8:e67686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaman MS, Thamminana S, Shahryari V, et al. Inhibition of PTEN gene expression by oncogenic miR-23b-3p in renal cancer. PLoS One 2012;7:e50203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalmay T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem 2013;54:29-38. [DOI] [PubMed] [Google Scholar]
- 23.Orang AV, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014;2014:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 2010;9:775-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015;12:697. [DOI] [PubMed] [Google Scholar]
- 26.Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016;44:D239-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao F, Zuo Z, Cai G, et al. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res 2009;37:105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu JB, Chiu C, Hsu S, et al. miRTar : an integrated system for identifying miRNA-target interactions in human. BMC Bioinformatics 2011;12:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosik KS, Krichevsky AM. The elegance of the microRNAs: a neuronal perspective. Neuron 2005;47:779-82. [DOI] [PubMed] [Google Scholar]
- 30.Berezikov E, Cuppen E, Plasterk RHA. Approaches to microrna discovery. Nat Genet 2006;38:S2. [DOI] [PubMed] [Google Scholar]
- 31.Chugh P, Dittmer DP. Potential pitfalls in microRNA profiling. Wiley Interdiscip Rev RNA 2012;3:601-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci 2004;101:9740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012;13:358-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manikandan M, Deva Magendhra Rao AK, Arunkumar G, et al. Oral squamous cell carcinoma: MicroRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol Cancer 2016;15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hur K, Toiyama Y, Schetter AJ, et al. Identification of a metastasis- specific microRNA signature in human colorectal cancer. JCNI 2015;107:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leidinger P, Hart M, Backes C, et al. Differential blood-based diagnosis between benign prostatic hyperplasia and prostate cancer: miRNA as source for biomarkers independent of PSA level, Gleason score, or TNM status. Tumor Biol 2016;37:10177-85. [DOI] [PubMed] [Google Scholar]
- 37.Hironaka-Mitsuhashi A, Matsuzaki J, Takahashi R, et al. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. PLoS One 2017;12:e0187638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh J, Tabunoki H. Comprehensive analysis of human microRNA target networks. BioData Min 2011;4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satoh J. Molecular network analysis of human microRNA targetome: from cancers to Alzheimer’s disease. Biodata Min 2012;5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Git A, Dvinge H, Salmon-Divon M, et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. Rna 2010;16:991-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RHA. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res 2004;32:6284-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lytle JR, Yario T, Steitz J. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci U S A 2007;104:9667-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Georgakilas MDPG, Kostoulas N, Vlachos IS, et al. DIANAmicroT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013;41:169-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betel D, Wilson M, Gabow A, et al. The microRNA. Org Resour Targets Express 2008;36:149-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucl Acid Res 2015;43:146-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuhn DE, Martin MM, Feldman DS, et al. Experimental validation of miRNA targets. Methods 2008;44:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Sanchez A, Murphy C. MicroRNA Target identification- experimental approaches. Biology (Basel) 2013;2:189-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendrickson DG, Hogan DJ, McCullough HL, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol 2009;7:25-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong YW, Cannell IG, de Moor CH, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci U S A 2008;105:8866-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beitzinger M, Peters L, Zhu JY, et al. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol 2007;4:76-84. [DOI] [PubMed] [Google Scholar]
- 52.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. Rna 2007;13:1198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010;141:129-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karginov FV, Conaco C, Xuan Z, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A 2007;104:19291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res 2011;39:6845-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wheeler GUY, Valoczi A, Havelda Z, Dalmay T. In situ detection of animal and plant microRNAs. DNA Cell Biol 2007;26:251-5. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Zhang X, Tang W, et al. miR-130a upregulates mTOR pathway by targeting TSC1 and is transactivated by NF-κB in high-grade serous ovarian carcinoma. Cell Death Differ 2017;1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


