Abstract
Dosage compensation has evolved in concert with Y-chromosome degeneration in many taxa that exhibit heterogametic sex chromosomes. Dosage compensation overcomes the biological challenge of a “half dose” of X chromosome gene transcripts in the heterogametic sex. The need to equalize gene expression of a hemizygous X with that of autosomes arises from the fact that the X chromosomes retain hundreds of functional genes that are actively transcribed in both sexes and interact with genes expressed on the autosomes. Sex determination and heterogametic sex chromosomes have evolved multiple times in Diptera, and in each case the genetic control of dosage compensation is tightly linked to sex determination. In the Anopheles gambiae species complex (Culicidae), maleness is conferred by the Y-chromosome gene Yob, which despite its conserved role between species is polymorphic in its copy number between them. Previous work demonstrated that male An. gambiae s.s. males exhibit complete dosage compensation in pupal and adult stages. In the present study, we have extended this analysis to three sister species in the An. gambiae complex: An. coluzzii, An. arabiensis, and An. quadriannulatus. In addition, we analyzed dosage compensation in bi-directional F1 hybrids between these species to determine if hybridization results in the mis-regulation and disruption of dosage compensation. Our results confirm that dosage compensation operates in the An. gambiae species complex through the hypertranscription of the male X chromosome. Additionally, dosage compensation in hybrid males does not differ from parental males, indicating that hybridization does not result in the mis-regulation of dosage compensation.
Keywords: Anopheles, dosage compensation, hybridization, RNAseq
Introduction
Sex chromosomes evolved independently from autosomes multiple times within Diptera (Bachtrog et al. 2014; Vicoso and Bachtrog 2015). Although X chromosomes largely retain their original gene content (Vicoso and Charlesworth 2006), sexually antagonistic mutations that occur on nascent Y-chromosomes suppress recombination with the X (Rice 1996). This leads to Y-chromosome degeneration as male-biased genes move from the X to the Y chromosome or the autosomes or are lost (Parisi et al. 2003), and eventually the formation of heterogametic sex chromosomes (Bachtrog 2013). Old Y-chromosomes harbor few protein coding genes and are repeat-rich, hampering, until recently, their sequencing and characterization (Mahajan and Bachtrog 2017).
As neo-Y chromosomes degenerate, X-linked genes that are not female-biased in their expression must be equalized or compensated for in the heterogametic sex. The evolution of dosage compensation (DC) has occurred in concert with Y-chromosome degeneration in many taxa that exhibit heterogametic sex determination (Disteche 2012; Graves 2015). DC overcomes the biological challenge of a “half dose” of X chromosome gene transcripts (or Z chromosome transcripts in ZZ/ZW systems) in the heterogametic sex. The need to equalize gene expression of a hemizygous X with that of autosomes arises from the fact that the X chromosomes retain hundreds of functional genes that are actively transcribed in both sexes.
In Drosophila and Anopheles, the regulation of DC is closely tied to the sex-determination pathway despite striking differences in sex determination between these genera. In Drosophila females, a double dose of X chromosome-linked signal elements (XSE) initiates female-specific pre-mRNA splicing of sex lethal (Sxl) transcripts. Sxl in turn regulates sex-specific splicing of transformer (tra), which, along with its nonsex-specific cofactor transformer2 (tra2), promotes female-specific splicing of doublesex (dsx), and fruitless (fru) pre-mRNAs. Male and female isoforms of dsx and fru modulate the expression of genes involved in sexually dimorphic morphologies and physiologies. A single dose of XSE fails to initiate the cascade of female-specific splicing and male transcripts are produced (Penalva and Sanchez 2003). Although male and female isoforms of DSX share the same DNA-binding domain, they differ in their C-terminal domains that confer sex-specific gene regulation (Clough et al. 2014).
Dsx and fru are highly conserved in insects despite their variety of sex-determination mechanisms (Wilkins 1995; Salz and Erickson 2010; Herpin and Schartl 2015). In mosquitoes (Diptera: Culicidae), sex determination is controlled by a M-factor located on the Y chromosome in the subfamily Anophelinae, which has heterogametic sex chromosomes (Krzywinska et al. 2016), or on the homomorphic sex-determining chromosome of the subfamily Culicinae (Hall et al. 2016). Transcription of the M-factor during early embryo development activates a cascade of sex-specific gene splicing that controls male development through sex-specific splicing of dsx and fru.
In the Anopheles gambiae species complex, the M-factor Yob is located within the male-determining locus of the Y chromosome. Yob expression is male-specific, begins within 2.5 h of embryo oviposition, and occurs prior to dsx splicing by up to 6 h (Krzywinska et al. 2016). This indicates that Yob is either a direct or indirect upstream regulator of dsx splicing. Yob is the only protein-coding gene on the Y chromosome that is shared by all members of the An. gambiae species complex (Hall et al. 2016). Yob is polymorphic in copy number between An. gambiae complex species and within An. gambiae s.s. (Hall et al. 2016). An. stephensi, which diverged from the An. gambiae species complex ∼30 Ma, lacks a Yob homolog, indicating a recent origin of Yob in the An. gambiae complex lineage (Hall et al. 2016).
Bernardini et al. (2017) were able introgress the An. gambiae Y chromosome into an An. arabiensis genetic background using an F1 × F0 crossing strategy, which allowed them to overcome a severe bottleneck associated with this cross. They found that when isolated, An. gambiae and An. arabiensis Y-chromosomes are interchangeable; the An. gambiae Y chromosome has no effect on An. arabiensis gene expression and its introgression does not cause male sterility. Thus, by itself a heterospecific Y-chromosome does not appear to disrupt dosage compensation in this cross. However, we do not know how dosage compensation is affected in F1 hybrids when the genetic background of the autosomes is not homozygous.
DC of the hemizygous X in male Drosophila occurs through the binding of the male-specific lethal (MSL) complex to the X. The MSL complex is comprised of four proteins encoded by the male-specific lethal (msl) genes 1–3 and maleless (mle), all of which cause male inviability when mutated (Penalva and Sanchez 2003). Sxl mutations, or genes impacting its regulation, result in overexpression of genes on the X chromosome. This results in female lethality during embryogenesis (Lucchesi and Skripsky 1981; Gergen 1987; Hilfiker et al. 1995; Erickson and Quintero 2007) likely due to the mis-regulation of DC (Cline 1993; Biedler and Tu 2016).
Like Drosophila, An. stephensi and An. gambiae s.s exhibit complete DC through the hypertranscription of the X chromosome (Jiang et al. 2015; Rose et al. 2016). Ectopic injections of Yob mRNA into An. gambiae embryos causes female lethality due to abnormal X chromosome transcription, but has no effect on males. Additionally, knockdown of Yob in male An. gambiae embryos results in 100% mortality (Krzywinska et al. 2016). Krzywinska et al (2016) suggested that these results implicate Yob as an upstream regulator of DC in Anopheles gambiae due to the interconnected roles of sex-determination genes and DC regulatory pathways in other insects. Orthologs of the five Drosophila genes that encode the MSL complex are present in the An. gambiae transcriptome. However, Anopheles msl-1 and msl-2 are highly diverged from their Drosophila orthologs (Rose et al. 2016), which may indicate that they may have different functions in Anopheles.
Studies in Drosophila initially implicated defects in DC regulation in hybrid male lethality (Chatterjee et al. 2007; Rodriguez et al. 2007; Bachtrog 2008) under a model where MSL proteins and their binding sites evolved on the D. melanogaster lineage in a way that a heterospecific mix of MSL complex proteins in F1 D. melanogaster–D. simulans hybrid males did not effectively regulate DC. Barbash (2010) subsequently refuted this hypothesis through a genetic screen of mle and msl mutant lines.
The An. gambiae species complex harbors the most important vectors of human malaria and has recently emerged as a model system for the study of speciation genetics (Neafsey et al. 2015). However, little is known about dosage compensation regulation in Anopheles: sex determination differs drastically between Drosophila and Anopheles, our knowledge of the link between Yob expression and DC regulation is incomplete, and msl-1 and msl-2 are highly diverged in sequence, and perhaps function, between Drosophila and Anopheles.
In the present study, we explored whether DC regulation has diverged between members of An. gambiae species complex, and is mis-regulated in their hybrids, through an analysis of genome-wide gene expression in three sister species: An. coluzzii, An. arabiensis, and An. quadriannulatus, as well as reciprocal F1 hybrids derived from An. coluzzii × An. arabiensis and An. coluzzi × An. quadriannulatus crosses.
Materials and Methods
Mosquito Rearing
Mosquitoes were derived from the SUA2La (An. coluzzii), SANGQUA (aka SANGWE, An. quadriannulatus), and DONGOLA (An. arabiensis) strains, or from bidirectional crosses between An. coluzzii and An. arabiensis, as well as An. coluzzii and An. quadriannulatus. Cross directions are indicated by female × male. Mosquitoes were reared in 12 h light/12 h dark cycles at 25 ˚C and 70–80% relative humidity with access to 5% sucrose solution. Colonies were blood fed via an artificial membrane with warmed, defibrinated sheep blood (Hemostat Laboratories, Dixon, CA). Adults were provided wetted filter paper during the period of 48–72 h postfeeding for oviposition. Larvae were fed Tetra brand TetraColor Tropical Crisps (fish food) ad libitum until pupation. Mosquitoes were collected 12 h postpupation and sexed prior to RNA extraction. Bidirectional crosses were performed by sexing 50–100 male and female pupae from the respective parental colonies prior to mixing them in adult cages. Rearing conditions of F1 larvae and collection of F1 pupae were performed as described above.
RNA Isolation and Sequencing
Mosquitoes were sexed 12 h postpupation. Total RNA extractions were performed on five pupae per replicate using a Qiagen RNeasy Mini kit (Qiagen, Redwood City, CA). RNA samples were quantified using a NanoDrop spectrophotometer and their quality was assessed using an Agilent Bioanalyzer. Equimolar amounts of two to four extraction products (10 or 20 total mosquito pupae) were combined to form a single biological replicate. Pooled RNA samples were again run on the Agilent Bioanalyzer and NanoDrop spectrophotometer prior to library prep and sequencing. Library prep was performed using an Illumina TruSeq RNA library prep kit (Illumina Inc., San Diego, CA). Two biological replicates for each strain, cross, and sex (28 libraries total) were sequenced on seven lanes of Illumina HiSeq 2500 machine using 125 bp single-end chemistry. Raw RNA sequencing reads are available on the NCBI Sequence Read Archive at project accession number PRJNA475281.
Pseudo-Genome Construction
The first step of our analysis was to construct an An. gambiae-based pseudo-genome for each parental species. By incorporating SNPs from each species into the AgamP4 genome, we were able to use the AgamP4 coordinates and gene set as a common framework to quantify transcript abundance. FastQC package version 0.11.4 (Babraham Bioinformatics) was used to examine read quality. Illumina TruSeq adapters were removed using Trimmomatic version 0.30 (Bolger et al. 2014), while simultaneously soft-clipping the reads from both 5′ and 3′ ends to an average phred quality score of 20, with no single bp in a four bp window below a phred quality score of 10. Only reads ≥ 50 bp were retained for alignment and subsequent analyses.
Trimmed RNAseq data from the three species were initially aligned to their respective, species-specific genomes, which were obtained from VectorBase.org (AcolM1 for An. coluzzii, AaraD1 for An. arabiensis, and AquaS1 for An. quadriannulatus, Giraldo-Calderon et al. 2015). Initial alignments were performed using the splice-aware alignment software STAR version 2.4.2a using default parameters with the exception of the option “–outFilterMismatchNoverLmax 0.1” (Dobin et al. 2013). Next, an index was created for each alignment, which annotated all splice junctions found during the first alignment round. This index informed the second alignment with STAR (as above), which realigned the reads around splice junctions. Picard tools version 2.8.1 (http://broadinstitute.github.io/picard/; last accessed November 3, 2016) was used to mark and remove PCR duplicates.
BAM alignments from each of the three species (two biological replicates of males and females) were then merged using Picard MergeBamAlignment, and SAMtools/BCFtools version 1.3 (Li et al. 2009) was used to call variants within each merged alignment using the BCFtools “call” tool. This process was informed by a prior mutation rate of 0.02. Next, GATK was used to filter variant call files for only biallelic SNPs. The vcfutils.pl varFilter function, a component of the BCFtools package, was used to filter SNPs using default parameters with the exception of not filtering for strand bias, as this can be expected in RNAseq data.
Filtered SNPs were converted to the genomic coordinates of the An. gambiae s.s. PEST strain AgamP4 genome (Holt et al. 2002) using the Picard Tools (github.com/broadinstitute/picard) LiftoverVCF function and a chain file to match base-pair coordinates between genomes. Chain files were created from pair-wise multiple-alignment files between the An. coluzzii, An. arabiensis, and An. quadriannulatus genomes and the An. gambiae s.s. PEST reference originally published by Fontaine et al. (2015), which were downloaded from VectorBase.org. Once SNPs were converted to AgamP4 coordinates, they were incorporated into the AgamP4 genome using the GATK FastaAlternateReferenceMaker to create a pseudo-genome for each species using the AgamP4 backbone/coordinates.
Next, we again performed a STAR 2-pass alignment (as above) for each parental library using the species-specific, AgamP4-baesed pseudo-genome as a reference. SNPs were called using the methods described above, and were then incorporated into the previous pseudo-genome reference to further populate the AgamP4-based pseudo-genome of each species with variation from that strain. Subsequently, the AgamP4-based pseudo-genomes were used as a reference to create a cDNA pseudo-genome for each species using the generate_transcripts function of rSeq version 0.2.2 (Jiang and Wong 2009; Salzman et al. 2011). Gene transcript coordinates were derived from the AgamP4.4 gene set (VectroBase.org).
Quantification of Gene Expression
Parental species libraries (An. coluzzii, An. arabiensis, and An. quadriannulatus) were aligned to their respective cDNA pseudo-genomes using the –very-sensitive mode of Bowtie 2 version 2.2.9 with default parameters (Langmead and Salzberg 2012). Only uniquely mapped reads were retained to calculate transcript abundances.
RNAseq reads from F1 hybrids were aligned to diploid pseudo-genomes comprised of the AgamP4.4 based cDNA pseudo-genomes of each parental species. rSeq version 0.2.2 (Jiang and Wong 2009; Salzman et al. 2011) was used to create cDNA genomes from the AgamP4 based pseudo-genomes of each parental species (as above). The maternal and paternal cDNA genomes for each hybrid were then merged into a diploid genome using the ASE-TIGAR program (Nariai et al. 2016, http://nagasakilab.csml.org/ase-tigar/; last accessed August 29, 2016), which concatenates the cDNA genomes of the maternal and paternal strains into one genome file. RNAseq reads from F1 hybrids were assessed for quality and trimmed using FastQC and Trimmomatic as described above. Reads were mapped to the diploid, biparental cDNA pseudo-genomes for the respective cross using the –very-sensitive mode Bowtie 2 version 2.2.9 with default parameters (Langmead and Salzberg 2012). F1 hybrid reads were allowed to align up to 100 times across the diploid reference genome. ASE-TIGAR was used to identify the most likely parental genome of origin and location of each transcript. Thus, only the highest probability map position of each transcript was quantified. Total transcript abundance (maternal + paternal expression in F1 hybrids) was used for the subsequent analysis of dosage compensation.
Analysis of Dosage Compensation
Transcript abundances were standardized by converting them to RPKMs (reads per kilo-base of transcript per million reads) in R (www.cran.r-project.org, R Core Team 2013). Two methods were used to assess dosage compensation in species and F1 hybrid samples. First, the ratio of median expression of X-linked and autosomal genes in each sample was determined. This was performed by dividing the RPKM value of each X-linked gene by the median RPKM expression level of all autosomal loci. The “boot” package in R (Davison and Hinkley 1997; Canty and Ripley 2015) was used to calculate 95% confidence intervals of the median of this distribution for each sample using 10,000 replicates. X to autosomal (X:A) median gene expression was analyzed at increasing minimum RPKM values between zero and 10. In order to be included in the analysis at a specific RPKM threshold, a gene had to exceed the specified RPKM level in all samples in a species comparison (ex. An. coluzzii, An. arabiensis, and their hybrids). Because of this, the number of genes included in this analysis differs for An. coluzzi between the An. coluzzi–An. arabiensis and the An. coluzzi–An. quadriannulatus species comparisons. An analysis of X:A median gene expression without RPKM cut-off (RPKM ≥ 0.0) would also include transcriptionally inactive genes which would bias the analyses (Kharchenko et al. 2011). Additionally, work in Drosophila has shown that genes with low expression have a lower degree of dosage compensation (McAnally and Yampolsky 2010). Thus, by analyzing X:A expression ratios at increasing minimum RPKM thresholds (RPKM > 0.0, 0.2, 0.5, 1.0, 2.0, 5.0, 10.0), we were able to identify the minimum RPKM level that removes bias during subsequent gene expression analyses.
A comparison of median X:A gene expression ratios between males and females shows if genes on the male hemizygous X chromosome is hyperexpressed to match the level of X chromosome expression in females. In females, we expect X:A expression ratios to be at or near one. If the X chromosome is hyperexpressed in males, the X:A expression ratio should be similar to that of females. If dosage compensation is absent, the X:A expression ratio of males should be half that of females. X:A expression ratios in males and females were also compared with the expression ratio of the second and third chromosomes (2:3). We expect the 2:3 expression ratio to be equal in both sexes. Thus, 2:3 median expression serves as an internal standard with which to compare X:A median expression. The 2:3 expression ratios were calculated by correcting the RPKM value of each chromosome 2 gene by the median RPKM of all chromosome 3 genes. The 2:3 expression ratios were also analyzed at increasing minimum RPKM thresholds (RPKM > 0.0, 0.2, 0.5, 1.0, 2.0, 5.0, 10.0).
The 95% confidence intervals of the median of the expression ratio distribution was calculated using the “boot” package in R using 10,000 replicates (Davison and Hinkley 1997, Canty and Ripley 2015). A Kruskal–Wallis test was used to test for significant differences between the median of the X:A expression ratio distribution between each sample (Kruskal and Wallis 1952). We used an ANOVA to test for significant differences in the mean of the X:A expression ratio distribution within and between each sample in a species comparison. The same tests were performed to analyze significant differences between the distributions of 2:3 expression ratios between samples. Where appropriate, a Tukey’s post-hoc test (Tukey 1949) was performed to test for significant differences in the means of pair-wise comparisons, and a Dunn’s test (Dunn 1964) was used to test for significant differences in the medians of pair-wise comparisons.
Additionally, we calculated male to female (M:F) expression ratios of X-linked and autosomal genes. Genes were only included in this analysis if they exceeded a RPKM of 10.0 across all samples in a comparison (see Results). The expectation of this approach is that the M:F ratio of X-linked and the M:F ratio of autosomal genes will not differ if dosage compensation is operating effectively. M:F expression ratios were calculated by first calculating the mean RPKM expression of each gene between the two biological replicates for each sample. Then, the male to female expression ratio was calculated for each gene within a species or F1 hybrid. Medians and 95% confidence intervals were calculated for X-linked and autosomal distributions separately. The 95% confidence intervals of the median of each M:F expression ratio distribution were calculated as described above. An ANOVA was used to test for significant differences between the means of the X-linked, M:F expression ratio distribution and the autosomal M:F expression ratio distribution within each species and F1 hybrid. Similarly, a Kruskal–Wallis test was used to test for significant differences between the medians of the X-linked, M:F expression ratio distribution and the autosomal M:F expression ratio distribution within each species and F1 hybrid.
Results
RNA Sequencing and Alignment
The sequencing effort yielded an average of 67.23 million reads per sample, though this number is skewed by one sample (COLZ × QUAD Female 2), which had 248.18 million reads (supplementary tables S1 and S2, Supplementary Material online). If this sample is excluded, the mean number of reads per sample is 60.53 million, ranging from 51.73 to 75.01 million. The mean mapping efficiency of parental libraries to their respective, species-specific reference genomes was 85.5% (min. 80.9%, max 88.7%, supplementary table S1, Supplementary Material online). The mapping efficiency of parental species libraries increased when aligned to their respective AgamP4-based pseudo-genomes (mean 97.3%, min. 95.2%, max 99.1%, supplementary table S1, Supplementary Material online).
The mapping efficiency of all libraries to their respective cDNA pseudo-genomes was reduced in comparison to either DNA genome alignment (mean 63.8%, min. 62.3%, max 65.2%, supplementary table S1, Supplementary Material online). However, this reduction was uniform across species and may result from an incomplete AgamP4 gene annotation. F1 hybrid RNAseq reads were aligned to the respective biparental cDNA pseudo-genome for that cross. After filtering BAM alignments with ASE-Tigar, the average mapping efficiency for F1 hybrid libraries was 64.3% (min. 62.3%, max. 66.7%, supplementary table S2, Supplementary Material online). Thus, the mapping efficiency of F1 hybrid and parental libraries to the cDNA pseudo-genomes was equivalent.
Dosage Compensation in An. coluzzii, An. arabiensis, and An. quadriannulatus
The analysis of dosage compensation can be biased when genes with very low expression are included. To overcome this challenge, we calculated X: A and chromosome 2:3 gene expression ratios at increasing minimum RPKM thresholds from zero to 10. The number of genes included in the An. coluzzii–An. arabiensis and An. coluzzii–An. quadriannulatus comparison data sets do not differ substantially at any RPKM threshold (supplementary table S3, Supplementary Material online). The medians of the X:A and 2:3 distributions were tested for significant differences at each RPKM threshold.
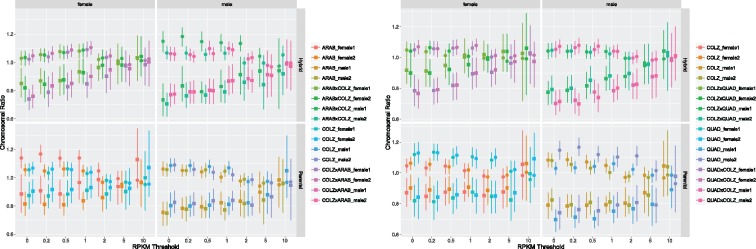
X:A median expression ratios approach 1.0 for male and female An. coluzzii, An. arabiensis, and An. quadriannulatus as the RPKM threshold increases (supplementary tables S4 and S5, Supplementary Material online), and at RPKM > 10, all 95% confidence intervals of the median exression ratios include 1.0 (fig. 1 and table 1). The same is true for 2:3 median expression ratios for males and females of all parental species (fig. 1;supplementary tables S8 and S9, Supplementary Material online). This result demonstrates that at RPKM > 10, 2:3 median expression ratios serve as an appropriate control with which to compare X:A expression in the effort to determine if dosage compensation is active in males.
Fig. 1.
—Scatter plot of X:A (squares) and 2:3 (circles) expression ratios at increasing minimum RPKM thresholds for the An. coluzzii–An. arabiensis species comparisons (left) and the An. coluzzii–An. quadriannulatus species comparisons. Panels separate male and female (x axis), and parental and hybrid samples (y axis). Lines represent the 95% confidence interval of the median for each distribution.
Table 1.
Median X:A Expression Ratios, and Their 95% Confidence Intervals (CI), for Each Sample of the An. coluzzii–An. arabiensis Species Comparison and the An. coluzzii–An. quadriannulatus Species Comparison at RPKM > 10
| Species / F1 Hybrid | Sex | Biological Replicate | Median | 95% CI | Species/F1 Hybrid | Sex | Biological Replicate | Median | 95% CI |
|---|---|---|---|---|---|---|---|---|---|
| QUAD | Female | 1 | 0.96 | (0.89, 1.01) | ARAB | Female | 1 | 1.13 | (0.93, 1.35) |
| QUAD | Female | 2 | 0.98 | (0.92, 1.04) | ARAB | Female | 2 | 0.97 | (0.79, 1.13) |
| QUAD | Male | 1 | 0.99 | (0.92, 1.04) | ARAB | Male | 1 | 0.99 | (0.79, 1.14) |
| QUAD | Male | 2 | 0.93 | (0.86, 1) | ARAB | Male | 2 | 0.96 | (0.75, 1.13) |
| COLZ | Female | 1 | 0.98 | (0.93, 1.03) | COLZ | Female | 1 | 1.00 | (0.77, 1.19) |
| COLZ | Female | 2 | 1.00 | (0.94, 1.07) | COLZ | Female | 2 | 1.07 | (0.87, 1.33) |
| COLZ | Male | 1 | 0.97 | (0.9, 1.03) | COLZ | Male | 1 | 1.05 | (0.86, 1.3) |
| COLZ | Male | 2 | 0.99 | (0.94, 1.03) | COLZ | Male | 2 | 0.95 | (0.7, 1.13) |
| QUAD × COLZ | Female | 1 | 1.02 | (0.97, 1.09) | ARAB × COLZ | Female | 1 | 1.03 | (0.87, 1.19) |
| QUAD × COLZ | Female | 2 | 1.02 | (0.96, 1.06) | ARAB × COLZ | Female | 2 | 1.04 | (0.9, 1.18) |
| QUAD × COLZ | Male | 1 | 0.99 | (0.94, 1.04) | ARAB × COLZ | Male | 1 | 0.94 | (0.67, 1.17) |
| QUAD × COLZ | Male | 2 | 1.01 | (0.96, 1.05) | ARAB × COLZ | Male | 2 | 0.92 | (0.77, 1.06) |
| COLZ × QUAD | Female | 1 | 1.00 | (0.95, 1.05) | COLZ × ARAB | Female | 1 | 0.98 | (0.84, 1.1) |
| COLZ × QUAD | Female | 2 | 0.99 | (0.94, 1.06) | COLZ × ARAB | Female | 2 | 1.00 | (0.83, 1.15) |
| COLZ × QUAD | Male | 1 | 0.99 | (0.93, 1.04) | COLZ × ARAB | Male | 1 | 0.99 | (0.82, 1.16) |
| COLZ × QUAD | Male | 2 | 1.01 | (0.96, 1.07) | COLZ × ARAB | Male | 2 | 0.97 | (0.8, 1.15) |
| ANOVA F-value | 1.83 | ANOVA F-value | 0.38 | ||||||
| ANOVA Pr(>F) | 0.03 | ANOVA Pr(>F) | 0.984 | ||||||
| Kruskal–Wallis X2 | 31.55 | Kruskal–Wallis X2 | 5.34 | ||||||
| Kruskal–Wallis P value | 0.01 | Kruskal–Wallis P value | 0.989 | ||||||
Note.—The results of the between sample ANOVA (F-value and probability > F) and Kruskal–Wallis test (X2 and P value) are reported.
We tested for significant differences between the medians of the X:A and 2:3 expression ratio distributions in males and females of each parental species and found no significant differences at a minimum RPKM > 10.0 (X2 P value > 0.05, supplementary tables S8 and S9, Supplementary Material online). This result was confirmed through pair-wise tests for differences between of X:A and 2:3 median expression ratios between males and females, with in each parental species. X:A an 2:3 median expression ratios did not differ between any pair-wise comparisons of males and females at RPKM > 2.0 (supplementary tables S10 and S11, Supplementary Material online).
Another way to explore whether dosage compensation is operating is by comparing directly the median male to female expression ratios of X-linked and autosomal genes. Due to the underrepresentation of male-biased on the X-chromosome (Magnusson et al. 2011; Magnusson et al. 2012), we expect the male to female expression ratios of X-linked genes to be female-biased, but nonetheless close to one. Based on the findings of the chromosome ratio analyses (above), only genes with a minimum RPKM > 10.0 across all samples in a species comparison were included in this analysis.
The median M:F expression ratios ranged between 0.89 and 0.99 for X-linked genes in parental species, and between 0.91 and 1.05 for autosomal loci (table 2 and fig. 2). An ANOVA found no significant difference between the means of the X-linked and autosomal distributions, however, a Dunn’s test found significant differences between the medians of the distributions (table 2). Despite these differences, all values are close to 1.0. In each species, the median of the M:F expression ratio distribution was lower for X-linked genes versus autosomal genes (fig. 2), indicating, as expected, a higher proportion of male-biased genes expressed on the autosomes compared with the X chromosome. An additional explanation for this pattern (though not mutually exclusive) is a higher proportion of female-biased genes on the X chromosome relative to the autosomes. However, previous studies found that genes with female-biased expression in An. gambiae are randomly distributed amongst the X chromosome and autosomes (Magnusson et al. 2012).
Table 2.
Comparison of M:F Expression Ratio Distributions, Separated by X-Linked and Autosomal Genes
| X-Linked |
Autosomal |
ANOVA |
Dunn’s Test |
|||||
|---|---|---|---|---|---|---|---|---|
| Species/F1 Hybrid | Median | 95% CI | Median | 95% CI | F-Value | Pr(>F) | X2 | P Value |
| COLZ | 0.99 | (0.97, 1) | 1.05 | (1.05, 1.06) | 0.09 | 0.904 | 47.29 | 0.000 |
| ARAB | 0.91 | (0.87, 0.95) | 0.91 | (0.9, 0.92) | 0.05 | 0.997 | 6.79 | 0.005 |
| COLZ × ARAB | 0.95 | (0.93, 0.97) | 0.98 | (0.97, 0.98) | 0.04 | 0.999 | 7.45 | 0.003 |
| ARAB × COLZ | 0.90 | (0.84, 0.95) | 1.00 | (0.99, 1.02) | 0.07 | 0.968 | 6.59 | 0.005 |
| COLZ | 0.99 | (0.97, 1) | 1.04 | (1.04, 1.05) | 0.08 | 0.993 | 35.16 | 0.000 |
| QUAD | 0.89 | (0.83, 0.94) | 0.96 | (0.95, 0.97) | 0.05 | 0.999 | 25.54 | 0.000 |
| COLZ × QUAD | 1.07 | (1.03, 1.11) | 1.11 | (1.1, 1.12) | 0.10 | 0.976 | 10.09 | 0.001 |
| QUAD × COLZ | 0.87 | (0.85, 0.89) | 0.90 | (0.89, 0.9) | 0.05 | 0.999 | 13.00 | 0.000 |
Note.—Genes are only included if they have an expression level >10.0 RPKM in all samples within a comparison (An. coluzzii–An. arabiensis or An. coluzzii–An. quadriannulatus). F values and P values (Pr(>F)) are reported for an ANOVA, which was used to test for significant differences between the means of the X-linked and autosomal M:F expression ratio distributions. X2 and P values are reported for the Dunn’s test, which was used to test for significant differences between the medians of the X-linked and autosomal M:F expression ratio distributions.
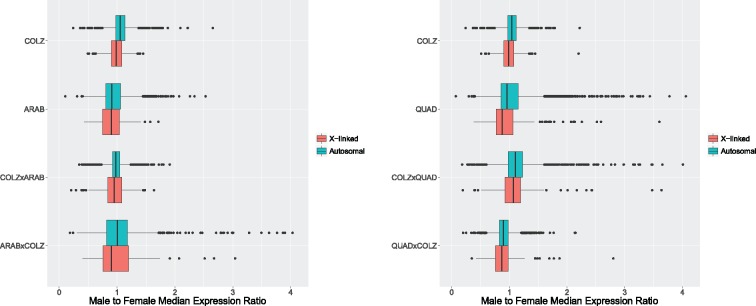
Fig. 2.
—Box plot of M:F expression ratios distributions for the An. coluzzii–An. arabiensis species comparisons (left) and the the An. coluzzii–An. quadriannulatus species comparisons (right), separated by X-linked (pink) and autosomal genes (blue).
These results confirm that dosage compensation is acting to balance X chromosome and autosomal gene expression in hemizygous males of An. coluzzi, An. arabiensis, and An. quadriannulatus, and that X chromosome dosage is equivalent between males of these species.
Dosage Compensation in Anopheles gambiae Complex Hybrids
Like parental species, X:A and 2:3 median expression ratios approach 1.0 for male and female F1 hybrids as the RPKM threshold increases regardless of the parental species or direction of the cross (supplementary tables S4 and S5, Supplementary Material online). At RPKM > 10 the 95% confidence intervals of the X:A median expression ratios include 1.0 for all hybrids (fig. 1 and table 1). The same is true for 2:3 median expression ratios (supplementary tables S6 and S7, Supplementary Material online).
The X:A median expression ratio of bidirectional hybrids between An. coluzzii and An. arabiensis do not differ significantly from parental species at RPKM > 0.2 (supplementary table S4, Supplementary Material online) or at RPKM > 0.5 for 2:3 expression ratios (supplementary table S6, Supplementary Material online). A Kruskal–Wallis test found no significant differences between median X:A and 2:3 expression ratios within both males and female hybrids at RPKM > 10 (supplementary table S8, Supplementary Material online). Additionally, between sex comparisons of median X:A and 2:3 expression ratios in F1 hybrids found no significant differences at RPKM > 10 (supplementary table S10, Supplementary Material online).
Similar patterns were found in bidirectional hybrids between An. coluzzi and An. quadriannulatus. The median X:A expression ratios of F1 males and females did not differ from parental species at RPKM > 10 (supplementary table S5, Supplementary Material online), and median 2:3 expression ratios did not differ above RPKM > 0.5 (supplementary table S7, Supplementary Material online). No significant differences were found between median X:A and 2:3 expression ratios within both males and female hybrids at RPKM > 10 (supplementary table S9, Supplementary Material online), with on exception (COLZ × QUAD Female 1, Kruskal–Wallis P value = 0.036). However, this difference is not significant after Bonferroni correction, and this result has no relevant bearing on our dosage compensation analyses. Finally, between sex comparisons of median X:A and 2:3 expression ratios in F1 hybrids found no significant differences above RPKM > 0.5 (supplementary table S11, Supplementary Material online).
The median M:F expression ratios of ranged between 0.87 and 1.07 for X-linked genes in hybrids, and between 0.90 and 1.11 for autosomal loci (table 2 and fig. 2). Corresponding to the analysis of parental species, an ANOVA found no significant difference between the means of the X-linked and autosomal distributions, but a Dunn’s test found significant differences between the medians (table 2).
These results demonstrate that hybridization between An. coluzzii and An. arabiensis, and An. coluzzii and An. quadriannulatus, does not result in the mis-regulation and disruption of dosage compensation in F1 hybrid males.
Discussion
We analyzed dosage compensation in the closely related species within the Anopheles gambiae complex; An. coluzzii, An. arabiensis, and An. quadriannulatus, as well as in their F1 hybrids. Dosage compensation was measured by comparing the ratio of male to female gene expression on the X-chromosome and autosomes, and by comparing X-chromosome versus autosome median gene expression levels in males and females. By analyzing bidirectional crosses between An. coluzzii–An. arabiensis and An. coluzzii–An. quadriannulatu, we tested if the species from which the Y or X chromosome is inherited affects dosage compensation in male hybrids. Specifically, these comparisons tested if the observed variation in the M locus between these closely related species may impact dosage compensation and if trans-acting factors from the heterozygous autosomes of hybrids affected X chromosome transcription differently depending on which parental species the X chromosome was inherited from.
Our results correspond with previous studies that compared X:A and M:F expression ratios in An. stephensi and An. gambiae s.s. to demonstrate that dosage compensation acts in males through the hypertranscription of the hemizygous X (Jiang et al. 2015; Rose et al. 2016). Jiang et al. (2015) reported median X:A expression ratios in An. stephensi males and females between 0.92 and 0.98 at minimum RPKM thresholds between 0.0 and 4.0, with no significant differences between median X and autosomal expression in males or females above a minimum RPKM > 2.0. In addition, these authors compared expression ratios between An. stephensi X-linked genes and their one-to-one orthologs in Ae. aegypti, a species with homomorphic sex chromosomes. The median RPKM ratio of X-linked An. stephensi genes to their Ae. aegypti orthologs was close to one in both sexes after normalization, indicating full dosage compensation of these genes on the hemizygous X chromosome of An. stephensi males.
The evolution of sex determination and heteromorphic sex chromosomes evolved independently between Drosophila and Anopheles mosquitoes. Drosophila and Anopheles differ in their mechanisms of sex-determination (dosage vs. the Yob M-locus), and the mechanisms controlling dosage compensation in Anopheles differ from those in Drosophila. Sxl controls sex-determination in Drosophila, however, the Anopheles gambiae homolog does not have sex-specific transcripts (Saccone et al. 2002), and is not involved in sex determination (Hall et al. 2016). Additionally, while the orthologs of Drosophila MSL complex proteins are transcribed in An. gambiae, they are highly divergent, and their role, or lack thereof, in dosage compensation is unknown (Rose et al. 2016).
The independent evolution of sex chromosomes and dosage compensation within the Culicidae (Biedler and Tu 2016), and the rapid evolution of the M-locus in the An. gambiae species complex (Hall et al. 2016), left open the question whether dosage compensation is disrupted in An. gambiae complex hybrids. Our analysis of median X:A and 2:3 gene expression ratios showed that dosage compensation is not disrupted in An. coluzzi–An. arabiensis or An. coluzzii–An. quadriannulatus hybrids, irrespective of the direction of the cross. Median X:A gene expression ratios did not differ between males and females of these hybrids; all were near 1.0, indicating complete compensation.
These results demonstrate that genes on the male X chromosome are hyperexpressed to match (on average) the expression of autosomal genes. Median male to female expression ratios for X-linked and autosomal loci differ significantly within all samples in this study (parental species and hybrids) but both of these ratios are near one. This finding is in agreement with prior work on dosage compensation in An. gambiae s.s. and An. stephensi (Jiang et al. 2015; Rose et al. 2016) which showed female-biased expression of X linked loci. In all species and hybrids in the present study, median M:F expression ratios are lower (female-biased) for X-linked genes compared with autosomal loci. This pattern could result from a number of evolutionary pressures that we discuss below.
Rose et al.’s (2016) analysis of dosage compensation in An. gambiae s.s. (M form) larvae and pupae found evidence for complete dosage compensation in both developmental stages. Median X:A expression ratios were found to be close to one in male and female larvae and pupae. Although they note differential abundances of genes between the X chromosome and autosomes that are sex-biased in their expression, autosomal genes in this study were approximately equally expressed in both sexes, and X-linked genes were slightly female biased. Female-biased expression of X-linked genes was strongest in larvae compared with the pupae.
Female-biased expression of X-linked genes could result from an absence of dosage compensation in male testes (Baker and Russell 2011; Rose et al. 2016), an underrepresentation of male-biased genes on the X chromosome (Magnusson et al. 2012), X chromosome inactivation in the male germline (Magnusson et al. 2012), tissue-specific dosage effects in males (Baker and Russell 2011), or a combination of these factors. In An. gambiae s.s. genes with testes-specific expression are underrepresented on the X chromosome, whereas genes with ovary-specific expression are not (Baker and Russell 2011). Furthermore, male-biased, somatically expressed genes are underrepresented on the X chromosome (Baker and Russell 2011; Magnusson et al. 2012).
Spermatogenesis occurs primarily during the 4th instar larvae and pupal stages in Anopheles mosquitoes (Clements 1992; Krzywinska and Krzywinski 2009) and no genes that show male-biased expression during these developmental stages are X-linked (Magnusson et al. 2011). An analysis of gene expression in the An. gambiae male pupae germline performed by Rose et al (2016) suggests that, like Drosophila and other Diptera (Meiklejohn and Presgraves 2012; Vicoso and Bachtrog 2015), the X chromosome is not hyperexpressed in the testes, and thus does not show dosage compensation in this tissue (Baker and Russell 2011; Rose et al. 2016). Sterile F1 male hybrids in the An. gambiae complex exhibit atrophied testes, malformed sperm, or a total lack of sperm due to the disruption of meiosis (Slotman et al. 2004). However, their somatic tissues appear to develop normally.
The goal of our analysis was to confirm that dosage compensation takes place in the closely related members of the An. gambiae species complex, and to discern whether or not dosage compensation is mis-regulated or otherwise disrupted in F1 hybrid males. Our results show female-biased expression of X-linked loci in both hybrids and pure species males, and no significant differences in X:A and 2:3 median expression levels between hybrid and pure species males. These results indicate that dosage compensation behaves normally in hybrid males:nonsex-biased X-linked genes are hyper expressed in somatic tissues and testes have 1X expression of nonsex biased, X-linked genes.
The rapid radiation of species in the An. gambiae species complex provides a unique study system for investigating fundamental questions in evolutionary biology and genetics. Despite ongoing introgression and low levels of genetic differentiation between members of the An. gambiae complex (Fontaine et al. 2015), reproductive and ecological isolation has allowed for local adaptation, behavioral divergence, and structural reorganization of their genomes (Neafsey et al. 2015). This study demonstrates that despite the genetic and ecological divergence between An. gambiae complex member species, dosage compensation operates in pure species and hybrid males to balance X-linked and autosomal gene expression.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was funded by a National Science Foundation Doctoral Dissertation Improvement Grant (award # 1601675) to K.C. Deitz and M.A. Slotman and a Texas A&M University Genomics Seed Grant to K.C. Deitz and M.A. Slotman.
Literature Cited
- Bachtrog D. 2008. Positive selection at the binding sites of the male-specific lethal complex involved in dosage compensation in Drosophila. Genetics 180(2):1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 14(2):113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, et al. 2014. Sex Determination: why so many ways of doing it? PLoS Biol. 12(7):e1001899.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Russell S.. 2011. Role of testis-specific gene expression in sex-chromosome evolution of Anopheles gambiae. Genetics 189(3):1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash DA. 2010. Genetic testing of the hypothesis that hybrid male lethality results from a failure in dosage compensation. Genetics 184(1):313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini F, et al. 2017. Cross-species Y-chromosome function between malaria vectors of the Anopheles gambiae species complex. Genetics 207(2):729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedler JK, Tu Z.. 2016. Chapter two: sex determination in mosquitoes. Adv Insect Physiol. 51:37–66. [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canty A, Ripley B.. 2015. boot: Bootstrap R. S-plus. functions. R package version 1.3-1.7. CRAN Repository.
- Chatterjee RN, et al. 2007. Drosophila simulans Lethal hybrid rescue mutation (Lhr) rescues inviable hybrids by restoring X chromosomal dosage compensation and causes fluctuating asymmetry of development. J Genet. 86(3):203–215. [DOI] [PubMed] [Google Scholar]
- Clements AN. 1992. Spermatogenesis and the structure of spermatozoa In: Clements AN, editor. The biology of mosquitoes: development, nutrition and reproduction. London: Chapmann and Hall; p. 333–335. [Google Scholar]
- Cline TW. 1993. The Drosophila sex determination signal: how do flies count to two? Trends Genet. 9(11):385–390. [DOI] [PubMed] [Google Scholar]
- Clough E, et al. 2014. Sex- and tissue-specific functions of Drosophila doublesex transcription factor targets. Dev Cell 31(6):761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AC, Hinkley DV.. 1997. Bootstrap methods and their applications. Cambridge: Cambridge University Press. [Google Scholar]
- Disteche CM. 2012. Dosage compensation of the sex chromosomes. Annu Rev Genet. 46:537–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn OJ. 1964. Multiple comparison using rank sums. Technometrics 6(3):241–252. [Google Scholar]
- Erickson JW, Quintero JJ.. 2007. Indirect effects of ploidy suggest X chromosome dose, not the X: a ratio, signals sex in Drosophila. PLoS Biol. 5(12):e332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, et al. 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347(6217):1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen JP. 1987. Dosage compensation in Drosophila: evidence that daughterless and Sex-lethal control X chromosome activity at the blastoderm stage of embryogenesis. Genetics 117(3):477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderon GI, et al. 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43(D1):D707–D713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. 2015. In retrospect: twenty five years of the sex determining gene. Nature 528(7582):343–344. [DOI] [PubMed] [Google Scholar]
- Hall AB, et al. 2016. Radical remodeling of the Y chromosome in a recent radiation of malaria mosquitoes. Proc Natl Acad Sci U S A. 113(15):E2114–E2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Schartl M.. 2015. Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 16(10):1260–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker A, et al. 1995. The gene virilizer is required for female-specific splicing controlled by Sxl, the master gene for sexual development in Drosophila. Development 121(12):4017–4026. [DOI] [PubMed] [Google Scholar]
- Holt R, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298(5591):129–149. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wong WH.. 2009. Statistical inferences for isoform expression in RNA-seq. Bioinformatics 25(8):1026–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, et al. 2015. Complete dosage compensation in Anopheles stephensi and evolution of sex-biased genes in mosquitoes. Genome Biol Evol. 7(7):1914–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Xi R, Park PJ.. 2011. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet. 43(12):1167–1169. [DOI] [PubMed] [Google Scholar]
- Kruskal WH, Wallis WA.. 1952. Use of ranks in on-criterion variance analysis. J Am Stat Assoc. 47(260):583–621. [Google Scholar]
- Krzywinska E, Krzywinski J.. 2009. Analysis of expression in the Anopheles gambiae developing testes reveals rapidly evolving lineage-specific genes in mosquitoes. BMC Genomics 10:300.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinska E, et al. 2016. A maleness gene in the malaria mosquito Anopheles gambiae. Science 353(6294):67–69. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC, Skripsky T.. 1981. The link between dosage compensation and sex differentiation in Drosophila melanogaster. Chromosoma 82(2):217–227. [DOI] [PubMed] [Google Scholar]
- Magnusson K, et al. 2011. Transcription regulation of sex-biased genes during ontogeny in the malaria vector Anophles gambiae. PLoS One 6(6):e21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson K, et al. 2012. Demasculinization of the Anopheles gambiae X chromosome. BMC Evol Biol. 12:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Bachtrog D.. 2017. Convergent evolution of Y chromosome gene content in flies. Nat Commun. 8(1):785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAnally AA, Yampolsky LY.. 2010. Widespread transcriptional autosomal dosage compensation in Drosophila correlates with gene expression level. Genome Biol Evol. 2(0):44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Presgraves DC.. 2012. Little evidence for demasculinization of the Drosophila X chromosome among genes expressed in the male germline. Genome Biol Evol. 4(10):1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nariai N, Kojima K, Mimori T, Kawai Y, Nagasaki M.. 2016. A Bayesian approach for estimating allele-specific expression from RNA-seq data with diploid genomes. BMC Genomics 17(S1):2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, et al. 2015. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347(6217):1258522.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. 1989. Does postzygotic isolation result from improper dosage compensation?. Genetics 122(4):891–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, et al. 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299(5607):697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva LOF, Sanchez L.. 2003. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol Mol Biol Rev. 67(3):343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. 1996. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381(6579):232–234. [DOI] [PubMed] [Google Scholar]
- Rodriguez MA, et al. 2007. Species-specific positive selection of the male-specific lethal complex that participates in dosage compensation in Drosophila. PNAS 104(39):15412–15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, et al. 2016. Dosage compensation in the African malaria mosquito Anopheles gambiae. Genome Biol Evol. 8(2):411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone G, Pane A, Polito LC.. 2002. Sex determination in flies, fruitflies, and butterflies. Genetica 116(1):15–23. [DOI] [PubMed] [Google Scholar]
- Salz H, Erickson JW.. 2010. Sex determination in Drosophila: the view from the top. Fly 4(1):60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Jiang H, Wong WH.. 2011. Statistical modeling of RNA-seq data. Stat Sci. 26(1):62–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman MA, della TA, Powell JR.. 2004. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and Anopheles arabiensis. Genetics 167(1):275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey J. 1949. Comparing individual means in the analysis of variance. Biometrics 5(2):99–114. [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D.. 2015. Numerous transitions of sex chromosome in Diptera. PLoS Biol. 13(4):e1002078.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B.. 2006. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 7(8):645–653. [DOI] [PubMed] [Google Scholar]
- Wilkins AS. 1995. Moving up the hierarchy. A hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17(1):71–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.