Abstract
The interaction and organization of proteins in the sperm membrane are important for all aspects of sperm function. We have determined the interactions between 12 known mutationally defined and cloned sperm membrane proteins in a model system for reproduction, the nematode Caenorhabditis elegans. Identification of the interactions between sperm membrane proteins will improve our understanding of and ability to characterize defects in sperm function. To identify interacting proteins, we conducted a split-ubiquitin membrane yeast two-hybrid analysis of gene products identified through genetic screens that are necessary for sperm function and predicted to encode transmembrane proteins. Our analysis revealed novel interactions between sperm membrane proteins known to have roles in spermatogenesis, spermiogenesis, and fertilization. For example, we found that a protein known to play a role in sperm function during fertilization, SPE-38 (a predicted four pass transmembrane protein), interacts with proteins necessary for spermiogenesis and spermatogenesis and could serve as a central organizing protein in the plasma membrane. These novel interaction pairings will provide the foundation for investigating previously unrealized membrane protein interactions during spermatogenesis, spermiogenesis, and sperm function during fertilization.
Keywords: gamete biology, gametogenesis, sperm, sperm maturation, spermatid, spermatocyte
Summary Sentence
Novel interactions between 12 Caenorhabditis elegans sperm membrane proteins were identified using split-ubiquitin membrane yeast two-hybrid analyses to provide insight into membrane protein order during sperm development and function.
Introduction
Proper organization and localization of membrane proteins is necessary for sperm function. Despite the importance of membrane organization, our knowledge of how proteins are trafficked to the membrane, organized in the membrane, and reorganized during navigation of the reproductive tract is not well characterized. Both sperm and eggs contain specialized membrane regions that are necessary for function [1–7]. Multiprotein complexes have been proposed to mediate sperm function, particularly fusion [8–11]. To understand the protein complexes that form in the sperm membrane, we tested all possible interactions between 12 known sperm membrane proteins in the nematode Caenorhabditis elegans.
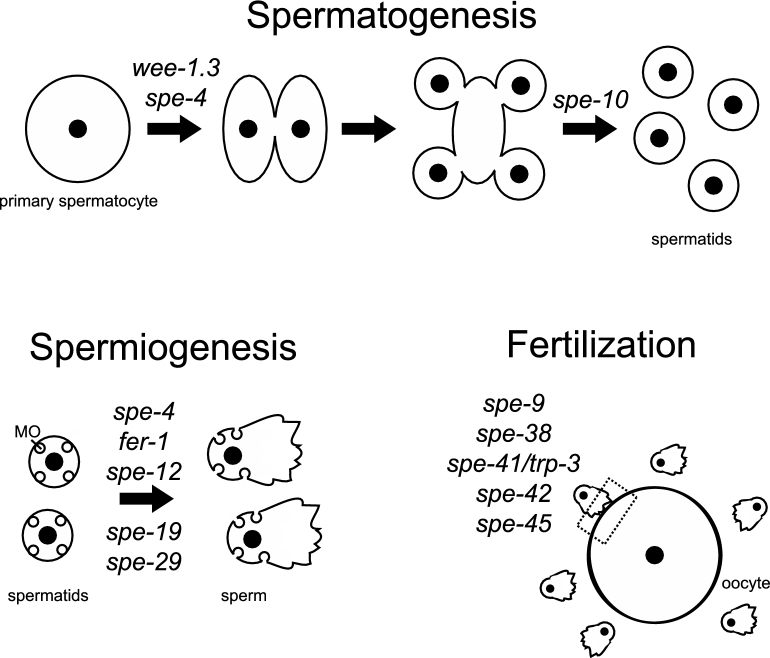
Sperm formation requires proper temporal progression of spermatogenesis and coordination of multiple individual and overlapping processes [12]. In meiosis I of C. elegans spermatogenesis, a primary spermatocyte divides into two secondary spermatocytes [13]. Then during meiosis II those secondary spermatocytes divide into two haploid spermatids each that are connected by a residual body [14]. The proper distribution of cellular components to the spermatids is aided by a fibrous body (FB)-membranous organelle (MO) complex [13, 14]. The MOs are Golgi-derived vesicles, as is the acrosome in mammalian sperm, that are localized near the plasma membrane of spermatids [13]. During spermiogenesis or postmeiotic sperm development, C. elegans sperm undergo a morphological change from a quiescent, round spermatid to an amoeboid sperm and gain motility, and the MOs fuse with the plasma membrane [13]. Membranous organelle fusion is viewed as being similar to the acrosome reaction [15]. The sperm development events that occur between anaphase II and residual body formation are analogous to spermiogenesis in other organisms, including vertebrates [12]. After spermiogenesis, the sperm are functional and can travel to the site of fertilization (the spermatheca) and fertilize the oocyte [13].
There are multiple mutationally defined, cloned, and well-characterized genes that are necessary for each developmental step of sperm development in C. elegans. Twelve of these genes encode proteins with predicted transmembrane domains (wee-1.3, spe-4, spe-10, fer-1, spe-12, spe-19, spe-29, spe-9, spe-38, spe-41/trp-3, spe-42, and spe-45) (Figure 1). Amongst these 12 genes, previous studies have only documented genetic interactions between three pairs: (1)spe-38 and spe-41/trp-3 [16], (2) fer-1 and spe-38 [17], and (3) fer-1 and spe-41/trp-3 [18]. Furthermore, only one pair of proteins, SPE-38 and SPE-41/TRP-3, has been shown to interact physically through split-ubiquitin yeast two hybrid assays [16]. Physical interactions between the remainder of the proteins encoded by these 12 genes have not been investigated. The lack of physical interaction data limits the ability to develop models of sperm membrane organization that can then be tested.
Figure 1.
Diagram of sperm development and function in C. elegans. During sperm development in C. elegans, primary spermatocytes divide into two secondary spermatocytes, which divide into two haploid spermatids each that are connected by a residual body [13, 14]. The membranous organelles (MOs) are Golgi-derived vesicles that are located near the plasma membrane of spermatids [13]. During spermiogenesis or post-meiotic sperm development, C. elegans sperm undergo a morphological change from a quiescent, round spermatid to an amoeboid sperm and gain motility, and the MOs fuse with the plasma membrane [13]. After spermiogenesis, the sperm are functional and can travel to the site of fertilization and fertilize the oocyte [13]. The approximate functional timing of the sperm genes that encode transmembrane proteins are indicated above the appropriate stage.
Previous high-throughput C. elegans interactome analyses have been conducted using yeast two hybrid assays [19, 20]. Unfortunately, transmembrane proteins are often not functional in traditional yeast two hybrid because they require insertion into a membrane; therefore, these datasets have not been helpful in developing models of the sperm membrane organization. For example, querying the 3864 known binary protein–protein interactions in the Worm Interactome database version 8 with the 12 sperm transmembrane proteins tested results in only a single positive interaction: SPE-10 and ABU-1 [20].
To develop a model for potential sperm membrane protein interactions that would increase our understanding of sperm membrane organization during development and function, we assayed the pairwise interactions between 12 full-length sperm transmembrane proteins using split-ubiquitin membrane yeast two hybrid (MYTH) assays. Our data reveal the potential of many of the sperm membrane proteins to interact with one another and provide evidence for the complexity of membrane interactions during sperm development. Network analysis reveals a sperm membrane cluster that could play a central role in organizing proteins at different stages of sperm development. Finally, our data provide the first comprehensive dataset about physical interactions between sperm membrane proteins and generate hypotheses about sperm membrane protein interactions that can be tested further.
Materials and methods
Yeast-two hybrid
The Dualsystems Biotech (Zurich, Switzerland) DUALmembrane Yeast Two-Hybrid System was used to perform the split-ubiquitin MYTH analyses. MYTH analysis is based on a split-ubiquitin approach where two distinct fragments of ubiquitin are fused to two proteins of interest. When an interaction occurs, ubiquitin is reconstituted, recognized by a deubiquitinating enzyme, and cleaved to release a transcription factor. The transcription factor then enters the nucleus and activates a reporter gene. An advantage MYTH analysis is that allows for the detection of full-length proteins in a membrane environment; however, it does have some of the disadvantages of classical yeast two hybrid including expression, autoactivation, and false positives [21, 22]. All assays were performed as per the manufacturer's instructions.
Twelve full-length genes necessary for spermatogenesis, spermiogenesis, or fertilization (fer and spe genes) that encode proteins with at least one predicted transmembrane domain (wee-1.3, spe-4, spe-10, fer-1, spe-12, spe-19, spe-29, spe-9, spe-38, spe-41/trp-3, spe-42, and spe-45) were cloned into the appropriate expression vectors based on their predicted or reported transmembrane domain structure (Figure 2). Please see the supplementary materials for primers and plasmids used for constructing the bait and prey plasmids (Supplementary Table S1). Cloning was performed using the SfiI restriction sites for both plasmid and PCR products. All genes were assayed as both bait and prey and for homotypic interactions.
Figure 2.
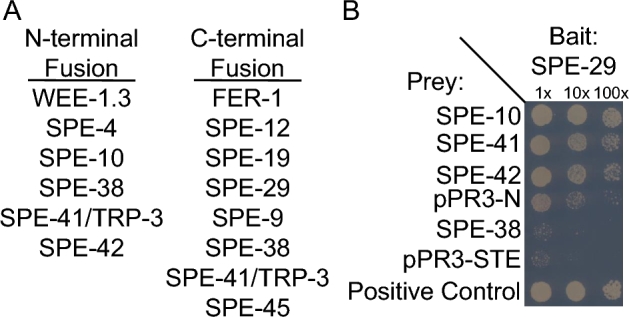
Sample results from a MYTH system to assay membrane interactions between sperm proteins. A split-ubiquitin based membrane yeast two-hybrid (MYTH) system was employed to detect interactions between SPE proteins containing a transmembrane domain. (A) Genes necessary for sperm development and function were cloned into expression vectors based on their predicted membrane topology in order to generate a protein with split-ubiquitin fused to the cytosolic terminus of the protein. (B) Yeast expressing the indicated bait and prey vectors were plated at three different dilutions for each experiment (1×, 10×, 100×). Empty prey vectors (pR3-N and pPR3-STE, respectively) were used to determine the background interactions. The positive control expresses a fusion of the yeast ER protein Alg5 to the N-terminus of ubiquitin (Nub).
Bait expression was confirmed by the DUALmembrane expression control assay. To confirm proper expression, each bait construct is co-transformed into the yeast strain NMY51 with either pAI-Alg5 or pDL2-Alg5 prey vectors. The pAI-Alg5 vector expresses a wild-type version of the N-terminal domain of ubiquitin (NubI) and pDL2-Alg5 expresses a mutant version of the N-terminal domain of ubiquitin with an Isoleucine to Glycine substitution at position 13 (NubG). If the bait protein is properly expressed along with NubI, it will reconstitute ubiquitin and lead to activation of reporter genes and colony growth on strong selection synthetic medium lacking leucine, tryptophan, histidine and adenine and will turn blue in the X-gal assay. Colony growth after co-expression of bait and NubI indicates the bait is functional and can be used in the analysis. Co-expression of bait with pDL-2 Alg5 should not activate reporter genes and result in little or no growth on strong selection medium. Any growth that is seen after co-expression with bait and pDL-Alg5 is considered background growth. 3-Amino-1,2,4-triazole was added to the medium to reduce the number of background colonies when necessary (Supplementary Table S2). All bait constructs were properly expressed, except SPE-41/TRP-3 bait that was cloned into the pBT-3-N vector (pBT3-N-SPE-41-N). As a result, pBT3-N-SPE-41-N was not included in the analysis.
After confirming bait expression, yeast containing the confirmed bait construct were transformed with prey plasmid and selected on synthetic medium lacking leucine and tryptophan. The resulting strain harboring both bait and prey plasmids was examined for their ability or inability to turn on the reporter genes and by their growth on synthetic medium lacking tryptophan, leucine, histidine, and adenine, and the number of colonies was recorded (Supplementary Table S2). Yeast expressing the indicating bait and prey vectors were plated at three different dilutions for each experiment (1×, 10×, 100×). Yeast cells transformed with bait construct and wild-type NubI from the pAI-Alg5 prey vector serve as a positive control for each replicate. Yeast cells transformed with bait construct and mutant NubG expressed from pDL2-Alg5 vector serve as negative control. The appropriate empty vector was also used as a negative control for each prey plasmid.
Colony quantification and interaction score determination
Colonies were counted using a dissecting microscope. Experimental samples were compared to the appropriate empty vector control to determine the number of colonies counted above background. The ratio of experimental colonies to empty vector colonies was computed, and this ratio is referred to as interaction score. Experimental groups with an interaction score greater that two were considered to have evidence indicating a positive interaction. If the empty vector control exhibited no colony growth, then the absolute number of colonies for the experimental group was equated to interaction score for those samples and only groups with greater than two colonies were considered positive. To be scored as a positive interaction, pairs had to show evidence of interactions in at least two dilutions in at least two independent experiments on two dates. All colony count data were entered in to Microsoft Excel. See supplementary materials for all raw data (Supplementary Table S2)
Network analysis
The sperm membrane protein interactome was imported into Cytoscape v3.4.0 (Supplementary Table S3) [23]. Duplicate edges (ignoring edge direction) and self-loops (homotypic interactions) were removed for network analysis. The network was treated as undirected, and NetworkAnalyzer was used to generate all network attributes under default settings (Supplementary Table S3). Interaction modules were detected using the MCODE (Molecular Complex Detection) v1.4.2 Cytoscape Application (Supplementary Table S4) [24].
Results
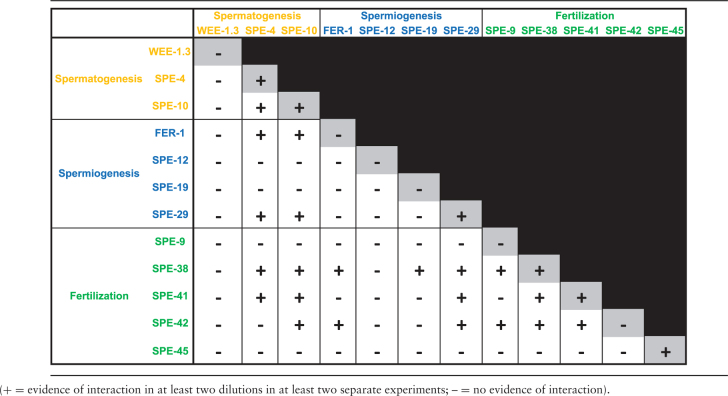
Twelve of the cloned fer and spe genes that are necessary for spermatogenesis, spermiogenesis, and fertilization and encode proteins with transmembrane domains were assayed using the split-ubiquitin MYTH system. Positive interactions were scored and quantified based on the number of colonies that grew on restricted medium plates (Figure 2). Of the possible 93 total heterotypic interactions between the 12 proteins, there were 21 interactions (22.6%) (Table 1). Furthermore, 6 of the 12 proteins (SPE-4, SPE-10, SPE-29, SPE-38, SPE-41/TRP-3, and SPE-45) displayed the ability to interact homotypically.
Table 1.
Summary of sperm membrane protein MYTH interactions.
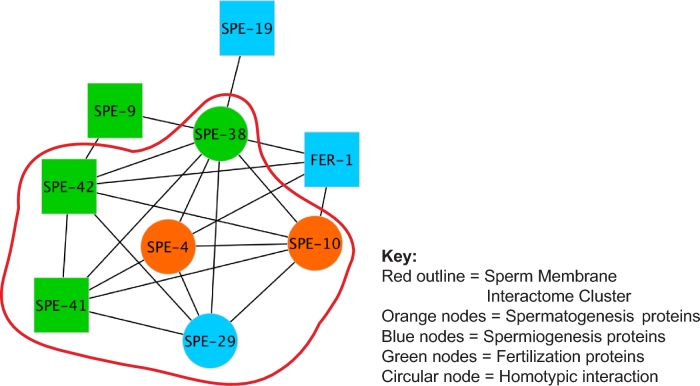
The nine proteins that did interact with other proteins heterotypically (SPE-4, SPE-10, FER-1, SPE-19, SPE-29, SPE-9, SPE-38, SPE-41/TRP-3, and SPE-42) define the sperm membrane protein interaction network or interactome (Figure 3). WEE-1.3, SPE-12, and SPE-45 did not interact with any of the other proteins tested, though SPE-45 did show the ability to interact homotypically. The resulting interactome has a density of 0.583, meaning that 58.3% of all possible interactions are realized in the network (Supplementary Table S4). The MCODE algorithm was used to identify interaction modules in the interactome. This analysis revealed that six of the interacting proteins (SPE-4, SPE-10, SPE-29, SPE-38, SPE-41/TRP-3, and SPE-42) form a cluster (Figure 3). These six proteins have 14 interactions between them and this cluster has both a density and correlation coefficient of 0.933 (Supplementary Table S4).
Figure 3.
Caenorhabditis elegans sperm membrane protein interactome. Network diagram generated from MYTH interaction data. The C. elegans sperm membrane interactome cluster is demarcated by a red outline. [blue nodes = genes necessary for spermiogenesis; orange nodes = genes necessary for spermatogenesis; green nodes = genes necessary for fertilization; circular nodes = proteins capable of homotypic interaction (self-looping node); square node = proteins not capable of homotypic interaction].
SPE-38 interacted with all nine proteins in the interactome, but not with WEE-1.3, SPE-12, and SPE-45 (Table 1). The high level of connectivity of SPE-38 is reflected in a betweenness centrality of 0.38, which is the highest for all interactome members (Table 2). If a member of a community has high betweenness centrality, they have a high probability of being necessary for to two neighbors to interact [25, 26]. SPE-38 is the only common interaction for all three proteins outside of the sperm membrane interactome cluster (FER-1, SPE-9, and SPE-19). The interaction of SPE-38 with these three proteins outside of the cluster results in a high neighborhood connectivity for FER-1, SPE-9, and SPE-19, despite their low number of direct interactions (Table 2). Neighborhood connectivity is a measure of how connected the neighbors are, not the individual protein itself [27].
Table 2.
Summary of C. elegans sperm membrane protein interactome network measures.
| Protein | Degree | Betweeness centrality | Closeness centrality | Neighborhood connectivity | Clustering coefficient | Topological coefficient |
|---|---|---|---|---|---|---|
| SPE-38 | 8 | 0.38 | 1 | 4.25 | 0.46 | 0.59 |
| SPE-10 | 6 | 0.025 | 0.8 | 5.5 | 0.8 | 0.69 |
| SPE-42 | 6 | 0.089 | 0.8 | 5 | 0.6 | 0.63 |
| SPE-4 | 5 | 0.018 | 0.73 | 5.6 | 0.8 | 0.7 |
| SPE-29 | 5 | 0.0071 | 0.73 | 6 | 0.9 | 0.75 |
| SPE-41/TRP-3 | 5 | 0.0071 | 0.73 | 6 | 0.9 | 0.75 |
| FER-1 | 4 | 0.0071 | 0.67 | 6.25 | 0.83 | 0.78 |
| SPE-9 | 2 | 0 | 0.57 | 7 | 1 | 0.88 |
| SPE-19 | 1 | 0 | 0.53 | 8 | 0 | 0 |
Discussion
The sperm transmembrane protein interaction analysis revealed that the proteins examined have the potential to form multi-protein complexes in the membrane. The cluster of proteins including SPE-4, SPE-10, SPE-29, SPE-38, SPE-41/TRP-3, and SPE-42 could represent a key group of proteins that interact in the membrane at various stages of sperm development. Of these proteins in the cluster, spe-4 and spe-10 are necessary for spermatogenesis, spe-29 is necessary for spermiogenesis, and spe-38, spe-41/trp-3, and spe-42 are necessary for fertilization (Figure 1). Despite the genetic data indicating distinct roles for these genes during sperm development, these data indicate that these proteins can interact physically.
Prior to this study, there were no published data indicating the physical interaction between any full-length sperm membrane proteins. However, previous studies by Singaravelu et al. identified an interaction between a truncated version of SPE-38 and SPE-41/TRP-3 [16]. In those studies, Singaravelu et al. used full-length SPE-38 with a ubiquitin fusion on the C-terminus and found no interaction with full-length SPE-41/TRP-3 with a ubiquitin fusion on the C-terminus [16], which we confirmed in this study. However, we did detect interactions when ubiquitin was fused at the C-terminus of SPE-38 and the N-terminus of SPE-41/TRP-3 and vice versa. This indicates that the terminal that the ubiquitin fusion is placed on can impact the detection of interaction in the membrane.
The protein–protein interaction data verify the previously reported genetic data for both spe-38 interacting with spe-41/trp-3 and fer-1 interacting with spe-38 interactions. Epistasis experiments indicated that spe-38 and spe-41/trp-3 interact and in spe-38(eb44) mutants, SPE-41/TRP-3 are mislocalized [16]. As discussed above, our data indicate that SPE-38 and SPE-41/TRP-3 interact physically. Therefore, the mislocalization of SPE-41/TRP-3 in spe-38(eb44) mutants could be because SPE-38 and SPE-41/TRP-3 can no longer interact normally in sperm. Additionally, fer-1 has been shown to genetically interact with spe-38 [17]. Mutations in fer-1 prevent MO fusion in developing sperm [28], and both SPE-38 and SPE-41/TRP-3 ar e mislocalized in the sperm of fer-1(hc1) animals [17, 18]. Based on the results of our study, we hypothesize that SPE-38 is interacting directly with FER-1 in the MO; and that, in fer-1(hc1) mutants, disruption of this interaction is maintained thus preventing proper SPE-38 localization.
Unlike the interaction between FER-1 and SPE-38, our data do not indicate that FER-1 and SPE-41/TRP-3 interact physically, despite genetic data indicating an interaction [18]. We hypothesize that the failure of SPE-41/TRP-3 to relocalize from the MO to the plasma membrane and pseudopod is indirectly caused by the lack of MO fusion in fer-1(hc1) mutants, not the loss of physical interaction between FER-1 and SPE-41/TRP-3. In fer-1(hc1) mutants, SPE-38 and SPE-41/TRP-3 do not have similar localization patterns despite interacting physically. We hypothesize that the interaction between SPE-38 and SPE-41/TRP-3 is transient and regulated by additional proteins.
Of the 12 proteins tested, SPE-38 had the highest number of interactions. This is reflected in SPE-38 having the highest degree of betweenness centrality and could indicate a central role organizing proteins in the sperm membrane. SPE-38 is a novel membrane protein with unknown biochemical function [17]. The protein spans the membrane four times but lacks the CD domain of tetraspanin proteins that are known to organize proteins in the membrane [17]. spe-38(eb44) mutants are completely sterile, while spe-41/trp-3 mutants [18], spe-42 [29], and some spe-9 mutants are weakly fertile [30]. Our data show that SPE-38 can interact with all of these proteins. spe-38(eb44) mutants have altered SPE-41/TRP-3 localization [16] and it is possible the SPE-9 and SPE-42 function may be defective in these mutants. The centrality of SPE-38 to the interaction network is what could result in the complete sterility of spe-38 mutants by indirectly disrupting other proteins in the sperm membrane.
SPE-4, SPE-10, and SPE-42 encode proteins with domains that could mediate protein–protein interactions and form a core membrane organizing cluster. spe-4 encodes for a protein containing a presenilin domain similar to gamma-secretase [31]. Gamma-secretase cleaves intramembrane proteins with a broad range of substrates [32]. spe-10 encodes for a protein containing a DHHC-CRD Zinc finger motif that is hypothesized to act as a palmitoyl transferase [33]. Many proteins tested in the interactome are predicted to have palymitoylation sites using CSS-Palm 2.0 predication software [34]. spe-42 encodes a C4C4-type ring finger domain that generally mediates protein–protein interaction and could also act as a ubiquitin ligase [29]. spe-42 also encodes for a dendritic cell specific transmembrane protein (DC-STAMP) domain [29]. DC-STAMP is a seven transmembrane protein that is necessary for osteoclast cell–cell fusion [35, 36].
MYTH studies suffer from a number of experimental challenges including issues with expression, autoactivation, and false positives [21, 22]; however, several points of data support to our results. First, we do not see broad nonspecific interactions between constructions as illustrated by the fact that WEE-1.3, SPE-12, and SPE-45 do not interact with other proteins regardless of its expression as a bait or prey. Second, there should be no false positive interactions between proteins that are not localized in the same cell type or proteins that do not share subcellular location patterns since all of the proteins tested here are expressed in sperm [37, 38] and every protein other than WEE-1.3 has nonnuclear subcellular localization [39]. Finally, we tested all interactions and both bait–prey combinations and provided all of the raw data to allow for reinterpretation based on different thresholds for interaction. Bait–prey swapping is a potential way to confirm the binary interaction between two proteins; however, many swapping bait–prey combinations have been shown to alter the interactions detected [40, 41].
Due to the highly interconnected nature of the interactome it would be reasonable to predict that more epistasis experiments would show genetic interactions between the genes of interest. However, the lack of epistatic interaction could be due to the nature of the sperm sterile (spe) phenotypes examined. Analyzing an allelic series for epistatic interactions would provide more insight. In addition, the developmental stage and membrane dynamics of each stage much be taken into account. Immunofluorescence data show that FER-1, SPE-9, SPE-38, and SPE-41/TRP-3 undergo dynamic relocalization during spermiogenesis [16, 17, 42, 43]. Thus, the protein–protein interactions are not likely to be static. Our data are essential for developing a model for protein interactions that can be tested to determine how sperm membrane proteins interact during the morphological shifts associated with spermatogenesis and spermiogenesis and during sperm migration in the hermaphrodite reproductive tract.
The sperm membrane in other species is known to contain specialized domains that are modified during sperm development and transit through the reproductive tract [44]. These domains are generated during development through protein interactions with molecular chaperones, protein–protein interactions in the sperm membrane, and regulation of cholesterol in the sperm membrane. Molecular chaperones are necessary to form and maintain multiprotein complexes [45]. The disruption of the testis specific chaperones calmegin and calsperin in mice leads to the disruption of ADAM complexes in the sperm membrane and fertility defects [46–48]. The genetic deletion of single protein of multiprotein complexes can lead to the loss of other complex members on the sperm surface [49]. For example, the targeted removal of ADAM2 from sperm leads to the loss of ADAM3 from the sperm membrane [50]. In addition, the targeted elimination of sperm membrane proteins can lead to spermatogenesis arrest resulting from improper processing of proteins during spermatogenesis, as is the case with HSPA2 [44]. Based on our interactome data, a disruption of protein function or localization could influence the function or localization of multiple other proteins. This is even true for proteins such as FER-1, SPE-9, and SPE-19 that have a low number of interactions, but high neighborhood connectivity, meaning that disrupting them could influence their neighbors and additional other proteins connected to those neighbors.
The sperm membrane during spermatogenesis, spermiogenesis, and fertilization is dynamic. The C. elegans sperm membrane interactome data presented here provide information about the types of interactions between membrane proteins that could be occurring during spermatogenesis, spermiogenesis, and sperm function during fertilization (Figure 4). Genes that are defined as necessary in earlier steps of sperm development may also be functional at later stages. Follow-up experiments will be necessary to validate these interactions at different developmental time points. There are additional transmembrane proteins that are expressed in sperm [37] and it is likely that some are necessary sperm function. Future genetic analysis will aid in defining the functions of those genes. This work provides a means to develop hypotheses about how the sperm membrane is organized and functions and is rationale for future experimentation. The molecular genetic tools available in C. elegans provide an excellent opportunity to analyze allelic series of each gene and determine how the variants influence sperm function.
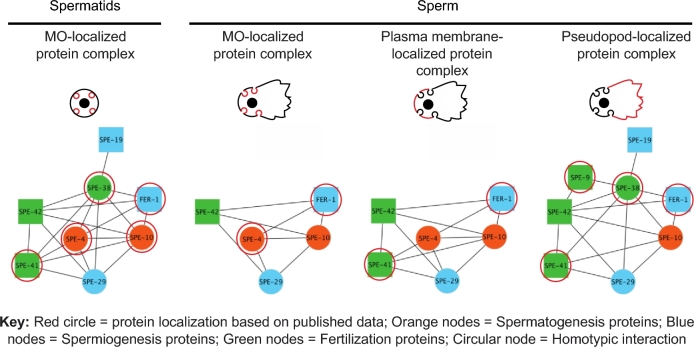
Figure 4.
Caenorhabditis elegans sperm membrane interactions during development. Sperm protein complex composition changes depending on the stage of sperm development. In spermatids, FER-1, SPE-4, SPE-10, SPE-38, and SPE-41/TRP-3 are localized to the MO (circled in red). The data presented here indicated that these proteins interact with SPE-19, SPE-29, and SPE-42 and form a sperm membrane complex in the MO of spermatids. Similar sperm membrane complex models are presented for MO, plasma membrane, and pseudopod localization by adding the interaction data presented here with the known protein localization. [red circle = protein localization based on published data; blue nodes = genes necessary for spermiogenesis; orange nodes = genes necessary for spermatogenesis; green nodes = genes necessary for fertilization; circular nodes = proteins capable of homotypic interaction (self-looping node); square node = proteins not capable of homotypic interaction].
Supplementary data
Supplementary data are available at BIOLRE online.
Supplementary Table S1. Summary of the primers, primer sequences, and vectors utilized for constructing bait (a) and (b) prey plasmids used in split-ubiquitin membrane yeast two hybrid (MYTH) system.
Supplementary Table S2. Full summary of interactome and raw data from split-ubiquitin (MYTH) assays. (a) Full table of interactions bait and prey orientations. (+ = evidence of interaction in at least two dilutions in at least two separate experiments; – = no evidence of interaction). (b) Full table of colony counts and interaction scores for all experiments. [bait = bait vector plus insert; date = date of experiment (MM-DD-YY format); 3AT_conc = concentration of 3-Amino-1,2,4-triazole in synthetic medium; prey = prey vector plus insert; group = experimental group (positive = positive control, negative = negative control, empty = empty vector negative control, exp = experimental group); dilution = dilution used in experiment; colonies = colonies counted under microscope (200 colonies is the maximum value); interaction score = ratio of experimental colonies to empty vector colonies].
Supplementary Table S3. Caenorhabditis elegans sperm membrane interactome network file for Cytoscape import [BAIT = source node; PREY = target node; interaction = interaction type; SELF = “y” means that protein can interaction homotypically (self-looping node) and “n” means that the protein does not interact homotypically].
Supplementary Table S4. Network measures for C. elegans sperm membrane interactome network [(a) Cytoscape node attributes, (b) Cytoscape network analysis, (c) MCODE analysis, and (d) Cytoscape network analysis (NA) of sperm membrane interactome cluster (cluster)].
Acknowledgments
We thank Dr Thomas Lombardi (Department of Computing and Information Studies, Washington and Jefferson College) for consultation and advice on network analyses, Dr Gunasekaren Singaravelu for advice analyzing MYTH data, Dr Xue Mei for critical reading of the manuscript, and Dr Amber R. Krauchunas for advice analyzing and summarizing interactome data and discussion and critical reading of the manuscript. Work in the Singson Lab is supported by an NIH grant (R01 HD054681).
Notes
Conference Presentations:
Marcello MR, Druzhinina M, and Singson A. “Mapping the Sperm Membrane Protein Interactome in C. elegans.” Fertilization and Activation of Development Gordon Research Conference. 2015; Holderness, New Hampshire (Invited Oral Presentation).
Marcello MR, Druzhinina M, and Singson A. “Mapping the Sperm Membrane Protein Interactome.” American Society of Andrology 39th Annual Conference. 2014; Atlanta, Georgia (Poster Presentation).
Marcello MR, Druzhinina M, and Singson A. “Construction of a C. elegans Spermiogenesis and Sperm Function Protein-Protein Interaction Network.” Fertilization and Activation of Development Gordon Research Conference. 2011; Plymouth, New Hampshire (Poster Presentation).
Marcello MR, Druzhinina M, Singaravelu G, and Singson A, “Development of a C. elegans Spermiogenesis and Sperm Function Protein Interaction Network.” 18th International C. elegans Meeting. 2011; Los Angeles, California (Poster Presentation).
Edited by Dr. Monika A. Ward, PhD, University of Hawaii John A. Burns School of Medicine
References
- 1. Sato K, Iwasaki T, Ogawa K, Konishi M, Tokmakov AA, Fukami Y. Low density detergent-insoluble membrane of Xenopus eggs: subcellular microdomain for tyrosine kinase signaling in fertilization. Dev Camb Engl 2002; 129:885–896. [DOI] [PubMed] [Google Scholar]
- 2. Travis AJ, Merdiushev T, Vargas LA, Jones BH, Purdon MA, Nipper RW, Galatioto J, Moss SB, Hunnicutt GR, Kopf GS. Expression and localization of caveolin-1, and the presence of membrane rafts, in mouse and Guinea pig spermatozoa. Dev Biol 2001; 240(2):599–610. [DOI] [PubMed] [Google Scholar]
- 3. Treviño CL, Serrano CJ, Beltrán C, Felix R, Darszon A. Identification of mouse trp homologs and lipid rafts from spermatogenic cells and sperm. FEBS Lett 2001; 509(1):119–125. [DOI] [PubMed] [Google Scholar]
- 4. Belton RJ, Adams NL, Foltz KR. Isolation and characterization of sea urchin egg lipid rafts and their possible function during fertilization. Mol Reprod Dev 2001; 59(3):294–305. [DOI] [PubMed] [Google Scholar]
- 5. Primakoff P, Myles DG. Cell-cell membrane fusion during mammalian fertilization. FEBS Lett 2007; 581(11):2174–2180. [DOI] [PubMed] [Google Scholar]
- 6. Satouh Y, Inoue N, Ikawa M, Okabe M. Visualization of the moment of mouse sperm-egg fusion and dynamic localization of IZUMO1. J Cell Sci 2012; 125(21):4985–4990. [DOI] [PubMed] [Google Scholar]
- 7. Runge KE, Evans JE, He Z-Y, Gupta S, McDonald KL, Stahlberg H, Primakoff P, Myles DG. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev Biol 2007; 304(1):317–325. [DOI] [PubMed] [Google Scholar]
- 8. Nishimura H, Cho C, Branciforte DR, Myles DG, Primakoff P. Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin beta. Dev Biol 2001; 233(1):204–213. [DOI] [PubMed] [Google Scholar]
- 9. Ellerman DA, Pei J, Gupta S, Snell WJ, Myles D, Primakoff P. Izumo is part of a multiprotein family whose members form large complexes on mammalian sperm. Mol Reprod Dev 2009; 76(12):1188–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krauchunas AR, Marcello MR, Singson A. The molecular complexity of fertilization: Introducing the concept of a fertilization synapse. Mol Reprod Dev 2016; 83(5):376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klinovska K, Sebkova N, Dvorakova-Hortova K. Sperm-egg fusion: a molecular enigma of mammalian reproduction. Int J Mol Sci 2014; 15(12):10652–10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chu DS, Shakes DC. Spermatogenesis. Adv Exp Med Biol 2013; 757:171–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishimura H, L’Hernault SW. Spermatogenesis-defective (spe) mutants of the nematode Caenorhabditis elegans provide clues to solve the puzzle of male germline functions during reproduction. Dev Dyn 2010; 239:1502–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ward S, Argon Y, Nelson GA. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J Cell Biol 1981; 91(1):26–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ellis RE, Stanfield GM. The regulation of spermatogenesis and sperm function in nematodes. Semin Cell Dev Biol 2014; 0:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singaravelu G, Chatterjee I, Rahimi S, Druzhinina MK, Kang L, Xu XZS, Singson A. The sperm surface localization of the TRP-3/SPE-41 Ca2+-permeable channel depends on SPE-38 function in Caenorhabditis elegans. Dev Biol 2012; 365(2):376–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chatterjee I, Richmond A, Putiri E, Shakes DC, Singson A. The Caenorhabditis elegans spe-38 gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Dev Camb Engl 2005; 132:2795–2808. [DOI] [PubMed] [Google Scholar]
- 18. Xu X-ZS, Sternberg PW. A C. elegans sperm TRP protein required for sperm-egg interactions during fertilization. Cell 2003; 114(3):285–297. [DOI] [PubMed] [Google Scholar]
- 19. Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain P-O, Han J-DJ, Chesneau A, Hao T, Goldberg DS, Li N et al. A map of the interactome network of the metazoan C. elegans. Science 2004; 303(5657):540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simonis N, Rual J-F, Carvunis A-R, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, Cevik S, Klitgord N et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Meth 2009; 6(1):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Snider J, Stagljar I. Membrane yeast two-hybrid (MYTH) mapping of full-length membrane protein interactions. Cold Spring Harb Protoc 2016; 2016(1):pdb.top077560. [DOI] [PubMed] [Google Scholar]
- 22. Snider J, Kotlyar M, Saraon P, Yao Z, Jurisica I, Stagljar I. Fundamentals of protein interaction network mapping. Mol Syst Biol 2015; 11(12):848–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003; 4(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pavlopoulos GA, Secrier M, Moschopoulos CN, Soldatos TG, Kossida S, Aerts J, Schneider R, Bagos PG. Using graph theory to analyze biological networks. BioData Mining 2011; 4(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoon J, Blumer A, Lee K. An algorithm for modularity analysis of directed and weighted biological networks based on edge-betweenness centrality. Bioinformatics 2006; 22(24):3106–3108. [DOI] [PubMed] [Google Scholar]
- 27. Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science 2002; 296(5569):910–913. [DOI] [PubMed] [Google Scholar]
- 28. Achanzar WE, Ward S. A nematode gene required for sperm vesicle fusion. J Cell Sci 1997; 110(Pt 9):1073–1081. [DOI] [PubMed] [Google Scholar]
- 29. Kroft TL, Gleason EJ, L’Hernault SW. The spe-42 gene is required for sperm-egg interactions during C. elegans fertilization and encodes a sperm-specific transmembrane protein. Dev Biol 2005; 286(1):169–181. [DOI] [PubMed] [Google Scholar]
- 30. Singson A, Mercer KB, L’Hernault SW. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell 1998; 93(1):71–79. [DOI] [PubMed] [Google Scholar]
- 31. Arduengo PM, Appleberry OK, Chuang P, L’Hernault SW. The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J Cell Sci 1998; 111(Pt 24):3645–3654. [DOI] [PubMed] [Google Scholar]
- 32. Beel AJ, Sanders CR. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell Mol Life Sci 2008; 65:1311–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gleason EJ, Lindsey WC, Kroft TL, Singson AW, L’hernault SW. spe-10 encodes a DHHC-CRD zinc-finger membrane protein required for endoplasmic reticulum/Golgi membrane morphogenesis during Caenorhabditis elegans spermatogenesis. Genetics 2006; 172:145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel 2008; 21:639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miyamoto T. The dendritic cell-specific transmembrane protein DC-STAMP is essential for osteoclast fusion and osteoclast bone-resorbing activity. Mod Rheumatol 2006; 16:341–342. [DOI] [PubMed] [Google Scholar]
- 36. Yagi M, Miyamoto T, Toyama Y, Suda T. Role of DC-STAMP in cellular fusion of osteoclasts and macrophage giant cells. J Bone Miner Metab 2006; 24:355–358. [DOI] [PubMed] [Google Scholar]
- 37. Ma X, Zhu Y, Li C, Xue P, Zhao Y, Chen S, Yang F, Miao L. Characterisation of Caenorhabditis elegans sperm transcriptome and proteome. BMC Genomics 2014; 15:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ortiz MA, Noble D, Sorokin EP, Kimble J. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 2014; 4:1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Allen AK, Nesmith JE, Golden A. An RNAi-based suppressor screen identifies interactors of the Myt1 ortholog of Caenorhabditis elegans. G3 2014; 4:2329–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caufield JH, Sakhawalkar N, Uetz P. A comparison and optimization of yeast two-hybrid systems. Methods 2012; 58:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Y-C, Rajagopala SV, Stellberger T, Uetz P. Exhaustive benchmarking of the yeast two-hybrid system. Nat Methods 2010; 7:667–668; author reply 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Putiri E, Zannoni S, Kadandale P, Singson A. Functional domains and temperature-sensitive mutations in SPE-9, an EGF repeat-containing protein required for fertility in Caenorhabditis elegans. Dev Biol 2004; 272:448–459. [DOI] [PubMed] [Google Scholar]
- 43. Washington NL, Ward S. FER-1 regulates Ca2+-mediated membrane fusion during C. elegans spermatogenesis. J Cell Sci 2006; 119:2552–2562. [DOI] [PubMed] [Google Scholar]
- 44. Nixon B, Bromfield EG, Dun MD, Redgrove KA, McLaughlin EA, Aitken RJ. The role of the molecular chaperone heat shock protein A2 (HSPA2) in regulating human sperm-egg recognition. Asian J Androl 2015; 17:568–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bromfield EG, Nixon B. The function of chaperone proteins in the assemblage of protein complexes involved in gamete adhesion and fusion processes. Reproduction 2013; 145:R31–R42. [DOI] [PubMed] [Google Scholar]
- 46. Ikawa M, Wada I, Kominami K, Watanabe D, Toshimori K, Nishimune Y, Okabe M. The putative chaperone calmegin is required for sperm fertility. Nature 1997; 387:607–611. [DOI] [PubMed] [Google Scholar]
- 47. Ikawa M, Nakanishi T, Yamada S, Wada I, Kominami K, Tanaka H, Nozaki M, Nishimune Y, Okabe M. Calmegin is required for fertilin alpha/beta heterodimerization and sperm fertility. Dev Biol 2001; 240:254–261. [DOI] [PubMed] [Google Scholar]
- 48. Ikawa M, Tokuhiro K, Yamaguchi R, Benham AM, Tamura T, Wada I, Satouh Y, Inoue N, Okabe M. Calsperin is a testis-specific chaperone required for sperm fertility. J Biol Chem 2011; 286:5639–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cho C. Testicular and epididymal ADAMs: expression and function during fertilization. Nat Rev Urol 2012; 9:550–560. [DOI] [PubMed] [Google Scholar]
- 50. Nishimura H, Myles DG, Primakoff P. Identification of an ADAM2-ADAM3 complex on the surface of mouse testicular germ cells and cauda epididymal sperm. J Biol Chem 2007; 282:17900–17907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.