Abstract
Next-generation sequencing has been invaluable in the elucidation of the genetic etiology of many subtypes of intellectual disability in recent years. Here, using exome sequencing and whole-genome sequencing, we identified three de novo truncating mutations in WAS protein family member 1 (WASF1) in five unrelated individuals with moderate to profound intellectual disability with autistic features and seizures. WASF1, also known as WAVE1, is part of the WAVE complex and acts as a mediator between Rac-GTPase and actin to induce actin polymerization. The three mutations connected by Matchmaker Exchange were c.1516C>T (p.Arg506Ter), which occurs in three unrelated individuals, c.1558C>T (p.Gln520Ter), and c.1482delinsGCCAGG (p.Ile494MetfsTer23). All three variants are predicted to partially or fully disrupt the C-terminal actin-binding WCA domain. Functional studies using fibroblast cells from two affected individuals with the c.1516C>T mutation showed a truncated WASF1 and a defect in actin remodeling. This study provides evidence that de novo heterozygous mutations in WASF1 cause a rare form of intellectual disability.
Keywords: WASF1, WAVE1 complex, neurodevelopmental disorder, autism, seizures, lamellipodia, actin cytoskeleton, recurrent de novo truncating mutations, developmental delay
Main Text
Neurodevelopmental disorders (NDDs), which include intellectual disability (ID), epilepsy, and autism spectrum disorder, are a heterogeneous group of disorders caused by abnormal development of the central nervous system (CNS). The complexity of CNS development is reflected in the fact that over 700 genes to date have been associated with ID, and very few occur at high prevalence.1, 2 Because of the extreme genetic heterogeneity of ID, the utilization of next-generation sequencing (NGS) technology provides an efficient method of determining the genetic cause of ID in individuals and discovering ID-associated genes. In addition, NGS of trios enables detection of de novo mutations,3 including single-nucleotide variants (SNVs) and small indels, which are a major contributing factor to the genetic etiology of moderate to severe ID and NDDs.4, 5, 6, 7
In this study, we used NGS approaches to identify three de novo variants in WAS protein family member 1 (WASF1 [MIM: 605035]), which encodes WASF1 (also known as WAVE1), in five unrelated individuals with overlapping neurodevelopmental abnormalities, including severe ID with autistic features and seizures. We used Matchmaker Exchange (MME)8 to connect the four international centers, which had each independently identified WASF1 as a candidate gene. All three de novo variants, including a recurrent truncating variant, cluster within the C-terminal actin-binding WCA domain of WASF1 and are predicted to result in a truncated protein.
The five affected individuals described in this report are from non-consanguineous families and are unrelated. All participants and parents gave informed consent, and the studies were approved by the appropriate institutional research ethics boards (Children’s Hospital of Eastern Ontario, Ottawa, Canada; IWK Health Centre, Halifax, Canada; Groupe Hospitalier Pitié-Salpêtrière, Paris, France; East of England Cambridge South, Cambridge, UK; Santa Maria Hospital, Lisbon, Portugal; and Radboud University Medical Center, Nijmegen, the Netherlands [2011-188]). The five affected individuals (P1–P5) have moderate to profound ID with autistic features, seizures, severe impairments in speech, gross motor delay, and a paucity of significant congenital abnormalities. A detailed clinical overview is provided in Table 1. The affected individuals have midfacial hypoplasia but lack a recognizable dysmorphic facial phenotype (Figures S1A–S1D). P5 started walking at 25 months, P1 and P2 began walking at age 3–4 years, and P4 did not walk until age 10 years. P1 requires wheelchair assistance when traveling out of his home. P3 has never achieved independent ambulation. All affected individuals either are non-verbal or have limited speech with a few or single words. All affected individuals except P5 have seizures, although these include a range of seizure types, including generalized and focal seizures; all require antiepileptic therapy. Four of the affected individuals (P1, P2, P4 and P5) had significant hypotonia in infancy, and two (P1 and P4) were described as having a wide-based gait, poor balance, and hyperactivity of movements. Musculoskeletal findings included joint hyperflexibility, ankle valgus, and pes planus in the more severely affected individuals. P5 presented with upper-limb dystonia in the first year of life. A high pain tolerance was observed in P1 and P3, whereas P4 and P5 exhibited automutilation, which is observed in those with an abnormal response to pain. Computed tomography of P1 showed mild atrophy near the Sylvian fissures, magnetic resonance imaging (MRI) of P2 and P3 was normal, and MRI of P4 revealed abnormalities of the periventricular white matter, although this individual also suffered a traumatic birth. MRI of P5 showed enlarged ventricles. Toe abnormalities (short third and fourth toes) were noted in three of the four affected individuals (Figures S1E–S1G). Testing for a range of other genetic conditions was undertaken in the affected individuals but resulted in no alternate diagnoses. Specific gene testing included MECP2, ATRX, UBE3A, CDKL5, MEF2C, FOXG1, TCF4, and NRXN1, reflecting the differential diagnosis and developmental severity of the condition. All had a normal result on diagnostic microarray testing. Metabolic testing was normal, as was a muscle biopsy of P3.
Table 1.
Key Clinical Features of Affected Individuals
| Detail | P1 | P2 | P3 | P4 | P5 |
|---|---|---|---|---|---|
| General | |||||
| Age (years) | 21 | 23 | 23 | 30 | 23 |
| Sex | male | male | male | female | male |
| Birth | |||||
| Gestation (weeks) | 40 | 41 | 39 | NR | 41 |
| Weight (g) | 3,800 | 4,100 | 3,370 | 4,020 | 4,020 |
| Head circumference (cm) | NR | 35.5 | 35.5 | NR | 35.5 |
| Neurological | |||||
| Intellectual disability | severe to profound | moderate to severe | severe | profound | moderate to severe |
| Seizures | onset at 8 years; focal with occasional GTC | onset at 6 years; absence and GTC | onset at 8 months; infantile spasms initially, now GTC | onset NR; temporal-lobe epilepsy with partial seizures | none |
| Speech | single words | simple sentences | non-verbal | NR | single words |
| Hypotonia | yes | yes | no | yes (axial with hypertonia of extremities) | yes (head control achieved at 11 months) |
| History of regression | no | no | yes (8 months) | arrested development at age 1 year, 10 months | no |
| Wide-based gait with poor balance | yes | no | non-ambulant | yes | yes |
| High pain tolerance | yes | no | yes | possible (automutilation) | yes (automutilation) |
| Head imaging | MRI: scarce periventricular white matter, enlarged ventricles | MRI: normal | MRI: normal | CT: mild atrophy near Sylvian fissures | MRI: enlarged ventricles |
| Current Measurements | |||||
| Head circumference (cm) | 50.4 (<P1; −3.2 SD) | 58 (P98; +2 SD) | 53.2 (P25; −1.3 SD) | 54 (P25; −0.3 SD) | 57 (P99; +2.4 SD) |
| Weight (kg) | 40.8 (<P1) | 82 (P80) | 40.2 (P25) | unknown | 65 (P70) |
| Height (cm) | 156.7 (<P1; −2.8 SD) | 183 (P80; +1 SD) | 168 (P10; −1.2 SD) | 150 (P2; −2.8 SD) | 175 (P97; +1.8 SD) |
| Motor Development | |||||
| Age at unsupported sitting | 18 months | 9 months | 6 months | 22 months | NR |
| Age at walking | 4 years | 3 years | non-ambulant | 10 years | 25 months |
| Craniofacial | |||||
| Midface hypoplasia | yes | yes | no | yes | NR |
| Eyes | deep set, strabismus, gray sclera | exophthalmia | strabismus, gray sclera | strabismus, vision loss, upslanted palpebral fissures | strabismus |
| Musculoskleletal | |||||
| Joint hyperflexibility | yes | no | no | yes | yes |
| Ankle valgus | yes | no | no | yes | knee recurvatum |
| Long tapered fingers | yes | no | yes | no | NR |
| Feet | narrow, pes planus, short forth toes | short third toes | normal | short, pes planus, short third toes | pes planus |
| Other | |||||
| Nipples | widely spaced | normal | widely spaced | inverted | NR |
| Café au lait macules | yes | no | yes | no | NR |
| Feeding problems | trouble sucking, reflux, easy choking | no | cyclic vomiting resolved at age 16 years | no | feeding difficulties, reflux |
| Genitourinary | no | no | renal stones, recurrent UTIs | small kidneys, mildly dilated pyelum, recurrent UTIs | NR |
| Constipation | yes | no | yes | yes | yes |
| HGVSg variant | chr6: g.110422797G>A | chr6: g.110422797G>A | chr6: g.110421847G>A | chr6: g.110422831delinsCCTGGC | chr6: g.110422797G>A |
| HGVSc variant | c.1516C>T | c.1516C>T | c.1558C>T | c.1482delinsGCCAGG | c.1516C>T |
| HGVSp variant | p.Arg506Ter | p.Arg506Ter | p.Gln520Ter | p.Ile494MetfsTer23 | p.Arg506Ter |
| Genotype | heterozygous | heterozygous | heterozygous | heterozygous | heterozygous |
| Inheritance | de novo | de novo | de novo | de novo | de novo |
Abbreviations are as follows: CT, computed tomography; GTC, generalized tonic clonic seizure; MRI, magnetic resonance imaging; NR, not recorded; P, patient; and UTI, urinary tract infection.
Because the initial genetic tests were negative, all affected individuals had either exome sequencing or whole-genome sequencing (WGS) performed at their respective centers. Details of the methods used for each affected individual are provided in Table S1. Genomic coordinates throughout this report refer to GRCh37, and coding sequence and protein coordinates refer to the canonical transcript (Ensembl: ENST00000392589; GenBank: NM_003931.2).
Trio exome sequencing was performed on individual P1 and his parents as part of the Care4Rare Canada research program according to our standard approach as previously described.9 After filtering for rare variants (with a frequency less than 0.1% in gnomAD and present fewer than six times in our in-house controls), all variants in known disease-related genes were assessed, but no variants that could explain this individual’s phenotype were identified. In the search for potential novel genes, possible bi-allelic or X-linked recessive variants were examined, but there were no rare homozygous or hemizygous variants. Compound-heterozygous variants were identified in CROCC (MIM: 615776), but this gene was ruled out as a likely candidate because it has many loss-of-function variants in control databases (Table S2). Finally, de novo variants in WASF1, ATP5J (MIM: 603152), SLC38A4 (MIM: 608065), and ZNF175 (MIM: 601139) were identified (Table S2). Assessment of protein localization patterns and function and in silico mutation predictions determined that ATP5J, SLC38A4, and ZNF175 were unlikely to be responsible for this condition (refer to Table S2 for further details). Given the role of WASF1 in actin polymerization and the importance of actin regulation in achieving synaptic plasticity, the de novo heterozygous variant in WASF1 (c.1516C>T [p.Arg506Ter]) was judged to be the strongest candidate for causing this individual’s condition and was entered into MME.
Individuals P2 and P5 underwent trio exome sequencing as part of routine diagnostic testing at the Département de Génétique of Hôpital Pitié-Salpêtrière (Paris, France). After filtering for rare variants (with a frequency less than 0.1% in the ExAC Browser), no pathogenic variants, likely pathogenic variants, or variants of unknown significance (VUSs) were identified in known developmental-disease-associated genes. Next, rare variants in genes not previously known to be associated with disease were considered. A heterozygous de novo stop-gain variant in WASF1 (c.1516C>T [p.Arg506Ter]), the same variant identified in P1, was identified in both P2 and P5. A de novo missense variant in CDCA7L (MIM: 609685) was also identified in P2 but was not considered likely to be pathogenic (Table S2). No additional variants that required consideration of pathogenicity were identified in P5.
Individual P3 and his mother underwent WGS as part of the National Institute for Health Research (NIHR) BioResource study (UK) as previously described.10 No pathogenic or likely pathogenic variants were found in known developmental-disease-associated genes, but a heterozygous stop-gain variant in WASF1 (c.1558C>T [p.Gln520Ter]), which was not present in the unaffected mother, was identified. Sanger sequencing of P3 and his parents confirmed that the variant occurs de novo in the affected individual (Figure S2B). A hemizygous missense variant in X-linked ACSL4 (MIM: 300157), in which variants can cause X-linked ID (MIM: 300387), was also identified in P3 and was heterozygous in the mother. This was classified as a VUS because the variant was not previously associated with disease (Table S2).
Individual P4 underwent trio exome sequencing as part of routine diagnostic testing (Groningen, the Netherlands). No pathogenic variants, likely pathogenic variants, or VUSs in known developmental-disease-associated genes were identified. Next, de novo variants in genes not previously known to be associated with disease were considered. A heterozygous de novo frameshift variant in WASF1 (c.1482delinsGCCAGG [p.Ile494MetfsTer23]) was identified. No other coding variants that occurred de novo were identified.
Initially, the four groups independently identified WASF1 as a strong candidate because of features consistent with those of developmental-disorder-associated genes. This gene is constrained for loss-of-function variation in the ExAC Browser (pLi = 0.91)11 and is highly and specifically expressed in the adult human brain.12 All three WASF1 variants are absent from 1000 Genomes, the ExAC Browser, and gnomAD.11, 13 The variants in individuals P1 and P3–P5 were confirmed to be de novo by Sanger sequencing of the trio (Figure S2B). The read depths for P2 and his mother and father were 127 (with 69 read counts for the alternate allele), 143, and 124, respectively. MME connected three of the groups, and the fourth was connected by personal correspondence with the UK group.
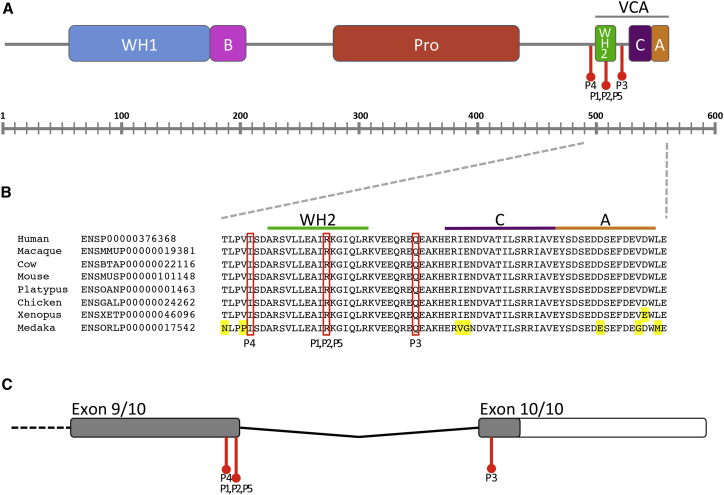
Interestingly, the three de novo variants appear to cluster around the WASP-homology 2 (WH2) domain of WASF1 (Figure 1A). A previously published method was used to determine that the clustering is statistically significant (p = 1.31 × 10−6).15 The C-terminal actin-binding WCA region, which includes the WH2 domain, is highly conserved throughout evolution (Figure 1B). The WCA region plays an important role in regulating WASF116, 17 so that actin and the Arp2/3 complex can bind to the WCA domain to promote actin polymerization.18 All three variants identified in the affected individuals fall either within the last 50 bp of the penultimate exon or within the last exon (Figure 1C) and are therefore predicted to result in the generation of a truncated protein that partially or fully eliminates this WCA domain.19
Figure 1.
Schematic Diagrams Showing Structure of WASF1 and WASF1
(A) Schematic diagram showing full-length WASF1 (also known as WAVE1 [Ensembl: ENSP00000376368]). Variants in the five individuals (indicated in red) cluster around the WH2 domain (domain coordinates are from Stradal et al.14). P1, P2, and P5 have p.Arg506Ter, P3 has p.Gln520Ter, and P4 has p.Ile494MetfsTer23. Abbreviations are as follows: WH1, WASP homology 1 domain; B, basic domain; Pro, proline-rich region; WH2, WASP homology 2 domain (also known as the verprolin homology domain); C, cofilin homology domain; A, acidic domain; WCA, collective name for the WH2, C, and A domains.
(B) Schematic diagram showing the amino acid sequence of part of WASF1. The WCA region of WASF1 is conserved throughout evolution. Yellow highlights residues that differ from the human protein sequence.
(C) Schematic diagram showing the 3′ part of WASF1, including locations of the participants’ variants in red. The gray boxes represent the coding sequence, and the white box represents the 3′ UTR. The variant in P1, P2, and P5 is 6 bps from the end of exon 9 (the penultimate exon). The variant in P4 is 40 bps from the end of exon 9. The variant in P3 is within exon 10 (the final exon).
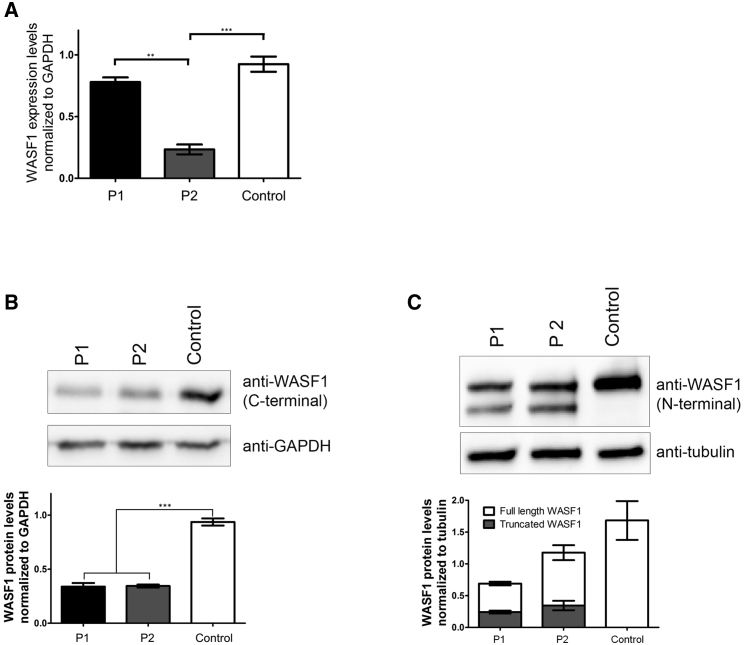
Next, the potential effect of the identified WASF1 variants on protein function was determined. Primary fibroblasts were obtained from individuals P1 and P2, who carry the same c.1516C>T variant (predicted to introduce a premature stop codon at amino acid 506). Amounts of WASF1 mRNA and WASF1 were examined. Real-time PCR showed variable levels of mRNA between the two affected individuals and control individuals (Figure 2A). For western blot analysis of WASF1, total protein extracts were probed with either a C-terminal antibody (epitope located after amino acid 506; Abcam, ab50356) or an N-terminal antibody (Sigma-Aldrich, W0267) against WASF1. Comparison of control and affected individuals revealed that the cells from affected individuals had both the full-length WASF1 (75 kDa) and a truncated ∼70 kDa protein that was not observed in control cells (Figures 2B and 2C). Densitometry quantification of these bands showed that the full-length protein was present at approximately 50% of the control levels, reflecting the presence of one wild-type allele, whereas the truncated protein was present at 14%–25% of control levels (Figures 2B and 2C). This suggests that although a truncated isoform is produced, it is unstable at either the mRNA or protein level such that the amount of protein is reduced. Therefore, the WASF1 c.1516C>T variant causes the production of a shorter mutant protein rather than the absence of a protein due to complete nonsense-mediated decay of the primary transcript.
Figure 2.
Amounts of WASF1 mRNA and WASF1 in Fibroblasts Derived from Affected Individuals with the c.1516C>T Variant
(A) RT-qPCR shows variable amounts of WASF1 mRNA between primary fibroblasts derived from individuals P1 and P2 and healthy control fibroblasts.
(B) Western blot analysis using an antibody with an epitope downstream of Arg506 showed that the amount of full-length WASF1 was approximately 50% lower in affected fibroblasts than in control fibroblasts.
(C) Western blot analysis using an antibody with an epitope in the N-terminal region of WASF1 showed the presence of the full-length and truncated WASF1 in affected fibroblasts. The truncated WASF1 was not present in control fibroblasts. All experiments were performed with fibroblasts derived from three healthy control individuals. Western blots were performed in triplicate, and band intensity was quantified with Image Lab Software (Bio-Rad).
Error bars indicate the range of measurement of triplicate samples.
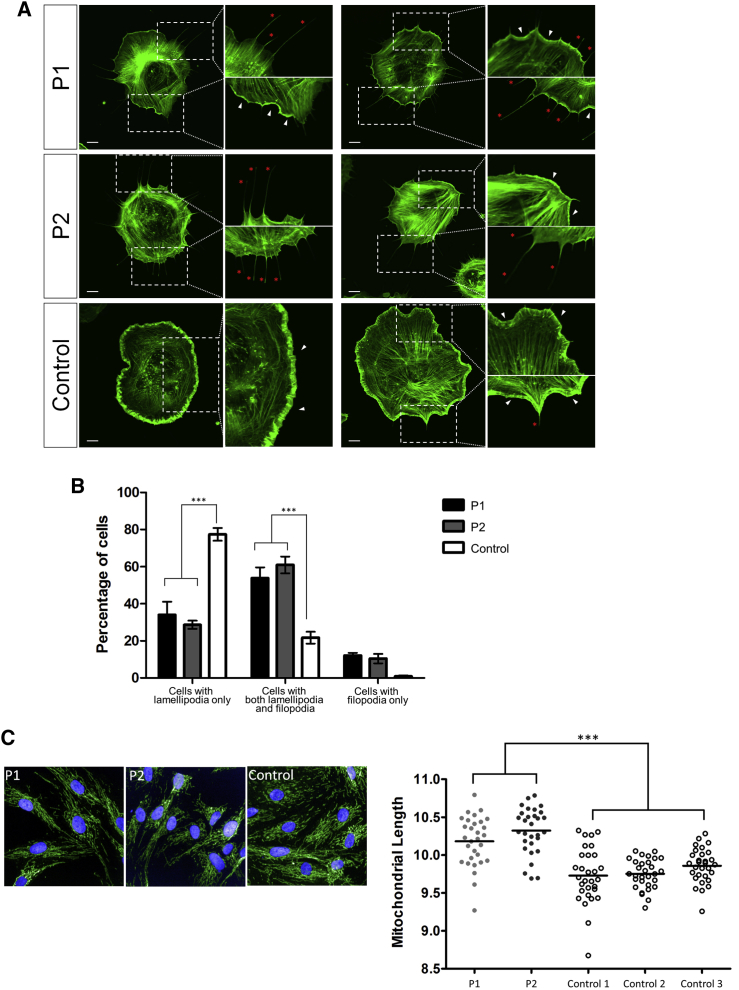
WASF1 plays a critical role in binding actin to initiate actin polymerization. Examination of the reorganization of the actin cytoskeleton during lamellipodia formation in fibroblasts was used for testing this role.20, 21, 22 Serum-starved fibroblasts were trypsinized, re-plated onto poly-l-lysine-coated coverslips, and stimulated with platelet-derived growth factor (PDGF; Sigma-Aldrich, P3201) for inducing the formation of lamellipodia, as previously described.20 Then cells were fixed, filamentous actin was visualized by labeling with phalloidin (Thermo Fisher Scientific, A12349), and the actin phenotype was quantified in each genotype. In the majority of control cells (77%), actin at the cell periphery formed well-organized, sheet-like lamellipodia structures (Figures 3A and 3B, white arrowhead; Figure S3). This was interspersed with cells in which the actin sheets were interjected by filopodia, which are finger-like actin projections (Figures 3A and 3B, red asterisk; Figure S3). We next assessed fibroblasts from affected individuals and found that although a sheet-like lamellipodia structure was observed along the periphery of 34% and 24% of P1 and P2 cells, respectively, the actin bundles were thinner and less organized than in the control cells (Figures 3A and 3B). We also noted that a portion of cells from P1 and P2 had severe disruptions in actin organization such that no lamellipodia delineated the cell periphery and only filopodial projections were present (12% and 11% for P1 and P2, respectively; Figures 3A and 3B). This phenotype was not seen in control cells. Therefore, cells from affected individuals have an alteration in actin organization, suggesting that the presence of a truncated WASF1 results in defective actin remodeling during the formation of lamellipodia.
Figure 3.
Lamellipodia Formation and Mitochondrial Morphology in Fibroblasts Derived from Individuals with the c.1516C>T Variant
(A) Primary fibroblasts were treated with PDGF for inducing the formation of lamellipodia. Visualization of the filamentous actin by phalloidin staining revealed the disruption of actin in the cell periphery of P1 and P2 fibroblasts. In the insets, lamellipodia and filopodia are marked by white arrowheads and red asterisks, respectively. Scale bars represent 10 μm.
(B) Cells were categorized into three groups on the basis of the predominant actin phenotype present: cells displaying lamellipodia only, cells displaying a mixture of lamellipodia and filopodia, and cells displaying filopodia only. Quantification based on these three categories indicates that significantly fewer affected fibroblasts than control fibroblasts are able to form solely lamellipodia.
(C) Confocal microscopic analysis of TOMM-20-immunostained mitochondria (in green) indicated that both affected fibroblasts have significantly elongated mitochondria. The nuclei were visualized by DAPI staining (in blue).
Finally, WASF1-dependent actin polymerization has been shown to mediate mitochondrial trafficking into dendritic spines in primary neurons;23 therefore, we assessed mitochondrial morphology in fibroblasts with the c.1516C>T variant. Mitochondria were visualized and the average length was quantified as previously described.24 As expected, a dense and complex network of mitochondria was present in both control and affected fibroblasts. Quantification revealed that mitochondria in the cells from affected individuals were significantly longer than those in control fibroblasts (Figure 3C). This result suggests that the presence of the c.1516C>T variant in WASF1 disrupts the regulation of mitochondrial dynamics and alters the normal balance between fission and fusion in affected fibroblasts.
This report provides evidence that de novo truncating variants in WASF1 in five unrelated individuals cause a NDD comprising severe ID with autistic features, seizures, and developmental delay. Interestingly, three of the five individuals in this study have the same de novo variant (c.1516C>T [Ensembl: ENST00000392589]). Three of the four individuals have VUSs in other genes in addition to the WASF1 variants. Population-level sequencing initiatives have enabled increased recognition of the prevalence of recurrent benign de novo mutations.25 Although it is unlikely, the possibility that they contribute to the respective individuals’ phenotypes cannot be excluded.
The variants described as associated with this NDD are all stop-gain or frameshift variants and significantly cluster around the C-terminal WH2 domain in the WCA region of WASF1. The truncated protein observed for c.1516C>T (p.Arg506Ter) suggests that all three variants are likely to lead to altered function of the mutant protein rather than complete protein loss or haploinsufficiency from degradation through nonsense-mediated decay. In a disease context, recurrent de novo events are known to be associated with specific dominant-negative or gain-of-function effects, such as FGFR3 (MIM: 134934) variants causing achondroplasia (MIM: 100800), and are usually missense variants.26 Clustering and recurrence of de novo protein-truncating mutations also do occur, albeit less frequently because the genic localization of a pathogenic mutation resulting in haploinsufficiency is generally not critical.15, 27, 28 Additional individuals with rare WASF1 variants are required for determining whether any pathogenic variants lie outside of this WCA region and/or whether a spectrum of phenotypes is perhaps associated with different variants in this gene.
WASF1 is an essential component of the actin pathway where RAC1 activation triggers a conformational change in WASF1 to allow binding of actin and ARP2/3 to the WCA domain to initiate actin polymerization.20, 21, 29, 30 The presence of a truncated protein that lacks the WCA region, as observed here, most likely disrupts the WASF1 complex itself, its interactions with CYFIP1, its proteasomal degradation, and the binding of actin (Figure 2C).16, 17, 31 Like mutations in WASF1, mutations in RAC1 similarly disrupt the formation of lamellipodia in fibroblasts,32 indicating that the organization and stabilization of actin bundles during the formation of lamellipodia is likely to be compromised by truncated WASF1.
WASF1-dependent actin polymerization is known to be important in CNS development and synaptic plasticity.18, 33, 34, 35, 36, 37, 38, 39 Two different WASF1-null mouse models demonstrate cognitive impairments, including deficits in sensorimotor function, learning, and memory.12, 40 In addition, mutations in a number of genes in the actin regulatory pathway, namely, formin 2 (FMN2 [MIM: 606373]),41 actin gamma-1 (ACTG1 [MIM: 102560]),42 rho guanine nucleotide exchange factor 6 (ARHGEF6 [MIM: 300267]),43, 44 and RAS-related C3 botulinum toxin substrate 1 (RAC1 [MIM: 602048]),32 are associated with ID.
WASF1 localizes to the outer mitochondrial membrane, where it has been shown to play a role in the trafficking of mitochondria to the dendritic spines.23, 44, 45 Actin itself has also been shown to be necessary for mediating mitochondrial fission.45 Given that fibroblasts derived from affected individuals with the c.1516C>T variant show elongated mitochondria (Figure 3C), WASF1 most likely plays additional roles in regulating mitochondrial dynamics, although how variants in WASF1 affect mitochondrial function in affected individuals remains to be elucidated.
In summary, de novo heterozygous truncating variants in WASF1 cause a NDD in individuals with ID associated with autistic features, seizures, and developmental delay. The three de novo variants, identified in five unrelated affected individuals, are all predicted to affect the actin-binding C-terminal WCA region of WASF1. The clustering of truncating pathogenic variants reported here and the presence of a truncated protein in cells from affected individuals imply either a gain-of-function or dominant-negative mechanism of disease. Because WASF1 functions within a large protein complex with ABI2, CYFIP1 or CYFIP2, BRK1, and NCKAP1, the hypothesis that these variants have a most likely dominant-negative effect remains to be tested. This study further expands the list of actin-regulatory-pathway genes associated with NDD and demonstrates the value of sharing genomic data through MME to identify the consequence of extremely rare mutational events.
Consortia
The NIHR BioResource consists of Timothy Aitman, David Bennett, Mark Caulfield, Patrick Chinnery, Daniel Gale, Ania Koziell, Taco W. Kuijpers, Michael A. Laffan, Eamonn Maher, Hugh S. Markus, Nicholas W. Morrell, Willem H. Ouwehand, David J. Perry, F. Lucy Raymond, Irene Roberts, Kenneth G.C. Smith, Adrian Thrasher, Hugh Watkins, Catherine Williamson, Geoffrey Woods, Sofie Ashford, John R. Bradley, Debra Fletcher, Tracey Hammerton, Roger James, Nathalie Kingston, Christopher J. Penkett, Kathleen Stirrups, Marijke Veltman, Tim Young, Matthew Brown, Naomi Clements-Brod, John Davis, Eleanor Dewhurst, Helen Dolling, Marie Erwood, Amy Frary, Rachel Linger, Jennifer M. Martin, Sofia Papadia, Karola Rehnstrom, Hannah Stark, David Allsup, Steve Austin, Tamam Bakchoul, Tadbir K. Bariana, Paula Bolton-Maggs, Elizabeth Chalmers, Janine Collins, Peter Collins, Wendy N. Erber, Tamara Everington, Remi Favier, Kathleen Freson, Bruce Furie, Michael Gattens, Johanna Gebhart, Keith Gomez, Daniel Greene, Andreas Greinacher, Paolo Gresele, Daniel Hart, Johan W.M. Heemskerk, Yvonne Henskens, Rashid Kazmi, David Keeling, Anne M. Kelly, Michele P. Lambert, Claire Lentaigne, Ri Liesner, Mike Makris, Sarah Mangles, Mary Mathias, Carolyn M. Millar, Andrew Mumford, Paquita Nurden, Jeanette Payne, John Pasi, Kathelijne Peerlinck, Shoshana Revel-Vilk, Michael Richards, Matthew Rondina, Catherine Roughley, Sol Schulman, Harald Schulze, Marie Scully, Suthesh Sivapalaratnam, Matthew Stubbs, R. Campbell Tait, Kate Talks, Jecko Thachil, Cheng-Hock Toh, Ernest Turro, Chris Van Geet, Minka De Vries, Timothy Q. Warner, Henry Watson, Sarah Westbury, Abigail Furnell, Rutendo Mapeta, Paula Rayner-Matthews, Ilenia Simeoni, Simon Staines, Jonathan Stephens, Christopher Watt, Deborah Whitehorn, Antony Attwood, Louise Daugherty, Sri V.V. Deevi, Csaba Halmagyi, Fengyuan Hu, Vera Matser, Stuart Meacham, Karyn Megy, Olga Shamardina, Catherine Titterton, Salih Tuna, Ping Yu, Julie von Ziegenweldt, William Astle, Marta Bleda, Keren J. Carss, Stefan Gräf, Matthias Haimel, Hana Lango-Allen, Sylvia Richardson, Paul Calleja, Stuart Rankin, Wojciech Turek, Julie Anderson, Christine Bryson, Jenny Carmichael, Coleen McJannet, Sophie Stock, Louise Allen, Gautum Ambegaonkar, Ruth Armstrong, Gavin Arno, Maria Bitner-Glindzicz, Angie Brady, Natalie Canham, Manali Chitre, Emma Clement, Virginia Clowes, Patrick Deegan, Charu Deshpande, Rainer Doffinger, Helen Firth, Frances Flinter, Courtney French, Alice Gardham, Neeti Ghali, Paul Gissen, Detelina Grozeva, Robert Henderson, Anke Hensiek, Simon Holden, Muriel Holder, Susan Holder, Jane Hurst, Dragana Josifova, Deepa Krishnakumar, Manju A. Kurian, Melissa Lees, Robert MacLaren, Anna Maw, Sarju Mehta, Michel Michaelides, Anthony Moore, Elaine Murphy, Soo-Mi Park, Alasdair Parker, Chris Patch, Joan Paterson, Julia Rankin, Evan Reid, Elisabeth Rosser, Alba Sanchis-Juan, Richard Sandford, Saikat Santra, Richard Scott, Aman Sohal, Penelope Stein, Ellen Thomas, Dorothy Thompson, Marc Tischkowitz, Julie Vogt, Emma Wakeling, Evangeline Wassmer, Andrew Webster, Sonia Ali, Souad Ali, Harm J. Boggard, Colin Church, Gerry Coghlan, Victoria Cookson, Paul A. Corris, Amanda Creaser-Myers, Rosa DaCosta, Natalie Dormand, Mélanie Eyries, Henning Gall, Pavandeep K. Ghataorhe, Stefano Ghio, Ardi Ghofrani, J. Simon R. Gibbs, Barbara Girerd, Alan Greenhalgh, Charaka Hadinnapola, Arjan C. Houweling, Marc Humbert, Anna Huis in’t Veld, Fiona Kennedy, David G. Kiely, Gabor Kovacs, Allan Lawrie, Rob V. Mackenzie Ross, Rajiv Machado, Larahmie Masati, Sharon Meehan, Shahin Moledina, David Montani, Shokri Othman, Andrew J. Peacock, Joanna Pepke-Zaba, Val Pollock, Gary Polwarth, Lavanya Ranganathan, Christopher J. Rhodes, Kevin Rue-Albrecht, Gwen Schotte, Debbie Shipley, Florent Soubrier, Laura Southgate, Laura Scelsi, Jay Suntharalingam, Yvonne Tan, Mark Toshner, Carmen M. Treacy, Richard Trembath, Anton Vonk Noordegraaf, Sara Walker, Ivy Wanjiku, John Wharton, Martin Wilkins, Stephen J. Wort, Katherine Yates, Hana Alachkar, Richard Antrobus, Gururaj Arumugakani, Chiara Bacchelli, Helen Baxendale, Claire Bethune, Shahnaz Bibi, Claire Booth, Michael Browning, Siobhan Burns, Anita Chandra, Nichola Cooper, Sophie Davies, Lisa Devlin, Elizabeth Drewe, David Edgar, William Egner, Rohit Ghurye, Kimberley Gilmour, Sarah Goddard, Pavel Gordins, Sofia Grigoriadou, Scott Hackett, Rosie Hague, Lorraine Harper, Grant Hayman, Archana Herwadkar, Aarnoud Huissoon, Stephen Jolles, Peter Kelleher, Dinakantha Kumararatne, Sara Lear, Hilary Longhurst, Lorena Lorenzo, Jesmeen Maimaris, Ania Manson, Elizabeth McDermott, Sai Murng, Sergey Nejentsev, Sadia Noorani, Eric Oksenhendler, Mark Ponsford, Waseem Qasim, Isabella Quinti, Alex Richter, Crina Samarghitean, Ravishankar Sargur, Sinisa Savic, Suranjith Seneviratne, Carrock Sewell, Emily Staples, Hans Stauss, James Thaventhiran, Moira Thomas, Steve Welch, Lisa Willcocks, Nigel Yeatman, Patrick Yong, Phil Ancliff, Christian Babbs, Mark Layton, Eleni Louka, Simon McGowan, Adam Mead, Noémi Roy, Jenny Chambers, Peter Dixon, Cecelia Estiu, Bill Hague, Hanns-Ulrich Marschall, Michael Simpson, Sam Chong, Ingrid Emmerson, Lionel Ginsberg, David Gosal, Rob Hadden, Rita Horvath, Mohamed Mahdi-Rogers, Adnan Manzur, Andrew Marshall, Emma Matthews, Mark McCarthy, Mary Reilly, Tara Renton, Andrew Rice, Andreas Themistocleous, Tom Vale, Natalie Van Zuydam, Suellen Walker, Liz Ormondroyd, Gavin Hudson, Wei Wei, Patrick Yu Wai Man, James Whitworth, Maryam Afzal, Elizabeth Colby, Moin Saleem, Omid S. Alavijeh, H. Terry Cook, Sally Johnson, Adam P. Levine, Edwin K.S. Wong, and Rhea Tan.
The project was selected for analysis by the Care4Rare Consortium Gene Discovery Steering Committee, consisting of Kym Boycott, Alex MacKenzie, Jacek Majewski, Michael Brudno, Dennis Bulman, and David Dyment.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the four affected individuals involved in this study and their families. This work was supported by the Cambridge Biomedical Research Centre and the National Institute for Health Research (NIHR) for the NIHR BioResource (grant number RG65966). This work was supported by the Care4Rare Canada Consortium (Enhanced Care for Rare Genetic Diseases in Canada), which is funded by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, the Ontario Research Fund, Genome Quebec, and the Children’s Hospital of Eastern Ontario Foundation.
Published: June 28, 2018
Footnotes
Supplemental Data include three figures and two tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.06.001.
Contributor Information
F. Lucy Raymond, Email: flr24@cam.ac.uk.
NIHR BioResource:
Timothy Aitman, David Bennett, Mark Caulfield, Patrick Chinnery, Daniel Gale, Ania Koziell, Taco W. Kuijpers, Michael A. Laffan, Eamonn Maher, Hugh S. Markus, Nicholas W. Morrell, Willem H. Ouwehand, David J. Perry, F. Lucy Raymond, Irene Roberts, Kenneth G.C. Smith, Adrian Thrasher, Hugh Watkins, Catherine Williamson, Geoffrey Woods, Sofie Ashford, John R. Bradley, Debra Fletcher, Tracey Hammerton, Roger James, Nathalie Kingston, Christopher J. Penkett, Kathleen Stirrups, Marijke Veltman, Tim Young, Matthew Brown, Naomi Clements-Brod, John Davis, Eleanor Dewhurst, Helen Dolling, Marie Erwood, Amy Frary, Rachel Linger, Jennifer M. Martin, Sofia Papadia, Karola Rehnstrom, Hannah Stark, David Allsup, Steve Austin, Tamam Bakchoul, Tadbir K. Bariana, Paula Bolton-Maggs, Elizabeth Chalmers, Janine Collins, Peter Collins, Wendy N. Erber, Tamara Everington, Remi Favier, Kathleen Freson, Bruce Furie, Michael Gattens, Johanna Gebhart, Keith Gomez, Daniel Greene, Andreas Greinacher, Paolo Gresele, Daniel Hart, Johan W.M. Heemskerk, Yvonne Henskens, Rashid Kazmi, David Keeling, Anne M. Kelly, Michele P. Lambert, Claire Lentaigne, Ri Liesner, Mike Makris, Sarah Mangles, Mary Mathias, Carolyn M. Millar, Andrew Mumford, Paquita Nurden, Jeanette Payne, John Pasi, Kathelijne Peerlinck, Shoshana Revel-Vilk, Michael Richards, Matthew Rondina, Catherine Roughley, Sol Schulman, Harald Schulze, Marie Scully, Suthesh Sivapalaratnam, Matthew Stubbs, R. Campbell Tait, Kate Talks, Jecko Thachil, Cheng-Hock Toh, Ernest Turro, Chris Van Geet, Minka De Vries, Timothy Q. Warner, Henry Watson, Sarah Westbury, Abigail Furnell, Rutendo Mapeta, Paula Rayner-Matthews, Ilenia Simeoni, Simon Staines, Jonathan Stephens, Christopher Watt, Deborah Whitehorn, Antony Attwood, Louise Daugherty, Sri V.V. Deevi, Csaba Halmagyi, Fengyuan Hu, Vera Matser, Stuart Meacham, Karyn Megy, Olga Shamardina, Catherine Titterton, Salih Tuna, Ping Yu, Julie von Ziegenweldt, William Astle, Marta Bleda, Keren J. Carss, Stefan Gräf, Matthias Haimel, Hana Lango-Allen, Sylvia Richardson, Paul Calleja, Stuart Rankin, Wojciech Turek, Julie Anderson, Christine Bryson, Jenny Carmichael, Coleen McJannet, Sophie Stock, Louise Allen, Gautum Ambegaonkar, Ruth Armstrong, Gavin Arno, Maria Bitner-Glindzicz, Angie Brady, Natalie Canham, Manali Chitre, Emma Clement, Virginia Clowes, Patrick Deegan, Charu Deshpande, Rainer Doffinger, Helen Firth, Frances Flinter, Courtney French, Alice Gardham, Neeti Ghali, Paul Gissen, Detelina Grozeva, Robert Henderson, Anke Hensiek, Simon Holden, Muriel Holder, Susan Holder, Jane Hurst, Dragana Josifova, Deepa Krishnakumar, Manju A. Kurian, Melissa Lees, Robert MacLaren, Anna Maw, Sarju Mehta, Michel Michaelides, Anthony Moore, Elaine Murphy, Soo-Mi Park, Alasdair Parker, Chris Patch, Joan Paterson, Julia Rankin, Evan Reid, Elisabeth Rosser, Alba Sanchis-Juan, Richard Sandford, Saikat Santra, Richard Scott, Aman Sohal, Penelope Stein, Ellen Thomas, Dorothy Thompson, Marc Tischkowitz, Julie Vogt, Emma Wakeling, Evangeline Wassmer, Andrew Webster, Sonia Ali, Souad Ali, Harm J. Boggard, Colin Church, Gerry Coghlan, Victoria Cookson, Paul A. Corris, Amanda Creaser-Myers, Rosa DaCosta, Natalie Dormand, Mélanie Eyries, Henning Gall, Pavandeep K. Ghataorhe, Stefano Ghio, Ardi Ghofrani, J. Simon R. Gibbs, Barbara Girerd, Alan Greenhalgh, Charaka Hadinnapola, Arjan C. Houweling, Marc Humbert, Anna Huis in’t Veld, Fiona Kennedy, David G. Kiely, Gabor Kovacs, Allan Lawrie, Rob V. Mackenzie Ross, Rajiv Machado, Larahmie Masati, Sharon Meehan, Shahin Moledina, David Montani, Shokri Othman, Andrew J. Peacock, Joanna Pepke-Zaba, Val Pollock, Gary Polwarth, Lavanya Ranganathan, Christopher J. Rhodes, Kevin Rue-Albrecht, Gwen Schotte, Debbie Shipley, Florent Soubrier, Laura Southgate, Laura Scelsi, Jay Suntharalingam, Yvonne Tan, Mark Toshner, Carmen M. Treacy, Richard Trembath, Anton Vonk Noordegraaf, Sara Walker, Ivy Wanjiku, John Wharton, Martin Wilkins, Stephen J. Wort, Katherine Yates, Hana Alachkar, Richard Antrobus, Gururaj Arumugakani, Chiara Bacchelli, Helen Baxendale, Claire Bethune, Shahnaz Bibi, Claire Booth, Michael Browning, Siobhan Burns, Anita Chandra, Nichola Cooper, Sophie Davies, Lisa Devlin, Elizabeth Drewe, David Edgar, William Egner, Rohit Ghurye, Kimberley Gilmour, Sarah Goddard, Pavel Gordins, Sofia Grigoriadou, Scott Hackett, Rosie Hague, Lorraine Harper, Grant Hayman, Archana Herwadkar, Aarnoud Huissoon, Stephen Jolles, Peter Kelleher, Dinakantha Kumararatne, Sara Lear, Hilary Longhurst, Lorena Lorenzo, Jesmeen Maimaris, Ania Manson, Elizabeth McDermott, Sai Murng, Sergey Nejentsev, Sadia Noorani, Eric Oksenhendler, Mark Ponsford, Waseem Qasim, Isabella Quinti, Alex Richter, Crina Samarghitean, Ravishankar Sargur, Sinisa Savic, Suranjith Seneviratne, Carrock Sewell, Emily Staples, Hans Stauss, James Thaventhiran, Moira Thomas, Steve Welch, Lisa Willcocks, Nigel Yeatman, Patrick Yong, Phil Ancliff, Christian Babbs, Mark Layton, Eleni Louka, Simon McGowan, Adam Mead, Noémi Roy, Jenny Chambers, Peter Dixon, Cecelia Estiu, Bill Hague, Hanns-Ulrich Marschall, Michael Simpson, Sam Chong, Ingrid Emmerson, Lionel Ginsberg, David Gosal, Rob Hadden, Rita Horvath, Mohamed Mahdi-Rogers, Adnan Manzur, Andrew Marshall, Emma Matthews, Mark McCarthy, Mary Reilly, Tara Renton, Andrew Rice, Andreas Themistocleous, Tom Vale, Natalie Van Zuydam, Suellen Walker, Liz Ormondroyd, Gavin Hudson, Wei Wei, Patrick Yu Wai Man, James Whitworth, Maryam Afzal, Elizabeth Colby, Moin Saleem, Omid S. Alavijeh, H. Terry Cook, Sally Johnson, Adam P. Levine, Edwin K.S. Wong, and Rhea Tan
Care4Rare Canada Consortium:
Kym M. Boycott, Alex MacKenzie, Jacek Majewski, Michael Brudno, Dennis Bulman, and David Dyment
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
Matchmaker Exchange, http://www.matchmakerexchange.org/
OMIM, http://omim.org/
Supplemental Data
References
- 1.Vissers L.E.L.M., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 2.Martínez F., Caro-Llopis A., Roselló M., Oltra S., Mayo S., Monfort S., Orellana C. High diagnostic yield of syndromic intellectual disability by targeted next-generation sequencing. J. Med. Genet. 2017;54:87–92. doi: 10.1136/jmedgenet-2016-103964. [DOI] [PubMed] [Google Scholar]
- 3.Veltman J.A., Brunner H.G. De novo mutations in human genetic disease. Nat. Rev. Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 4.Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T., Albrecht B., Bartholdi D., Beygo J., Di Donato N. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 5.Hamdan F.F., Srour M., Capo-Chichi J.-M., Daoud H., Nassif C., Patry L., Massicotte C., Ambalavanan A., Spiegelman D., Diallo O. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilfert A.B., Sulovari A., Turner T.N., Coe B.P., Eichler E.E. Recurrent de novo mutations in neurodevelopmental disorders: properties and clinical implications. Genome Med. 2017;9:101. doi: 10.1186/s13073-017-0498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaulieu C.L., Majewski J., Schwartzentruber J., Samuels M.E., Fernandez B.A., Bernier F.P., Brudno M., Knoppers B., Marcadier J., Dyment D., FORGE Canada Consortium FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am. J. Hum. Genet. 2014;94:809–817. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carss K.J., Arno G., Erwood M., Stephens J., Sanchis-Juan A., Hull S., Megy K., Grozeva D., Dewhurst E., Malka S., NIHR-BioResource Rare Diseases Consortium Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am. J. Hum. Genet. 2017;100:75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahl J.P., Wang-Dunlop J., Gonzales C., Goad M.E.P., Mark R.J., Kwak S.P. Characterization of the WAVE1 knock-out mouse: implications for CNS development. J. Neurosci. 2003;23:3343–3352. doi: 10.1523/JNEUROSCI.23-08-03343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stradal T.E.B., Rottner K., Disanza A., Confalonieri S., Innocenti M., Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14:303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Lelieveld S.H., Wiel L., Venselaar H., Pfundt R., Vriend G., Veltman J.A., Brunner H.G., Vissers L.E.L.M., Gilissen C. Spatial Clustering of de Novo Missense Mutations Identifies Candidate Neurodevelopmental Disorder-Associated Genes. Am. J. Hum. Genet. 2017;101:478–484. doi: 10.1016/j.ajhg.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., Borek D., Padrick S.B., Gomez T.S., Metlagel Z., Ismail A.M., Umetani J., Billadeau D.D., Otwinowski Z., Rosen M.K. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padrick S.B., Cheng H.-C., Ismail A.M., Panchal S.C., Doolittle L.K., Kim S., Skehan B.M., Umetani J., Brautigam C.A., Leong J.M., Rosen M.K. Hierarchical regulation of WASP/WAVE proteins. Mol. Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padrick S.B., Doolittle L.K., Brautigam C.A., King D.S., Rosen M.K. Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. USA. 2011;108:E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 20.Suetsugu S., Yamazaki D., Kurisu S., Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- 21.Yamazaki D., Fujiwara T., Suetsugu S., Takenawa T. A novel function of WAVE in lamellipodia: WAVE1 is required for stabilization of lamellipodial protrusions during cell spreading. Genes Cells. 2005;10:381–392. doi: 10.1111/j.1365-2443.2005.00845.x. [DOI] [PubMed] [Google Scholar]
- 22.Takenawa T., Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- 23.Sung J.Y., Engmann O., Teylan M.A., Nairn A.C., Greengard P., Kim Y. WAVE1 controls neuronal activity-induced mitochondrial distribution in dendritic spines. Proc. Natl. Acad. Sci. USA. 2008;105:3112–3116. doi: 10.1073/pnas.0712180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanstone J.R., Smith A.M., McBride S., Naas T., Holcik M., Antoun G., Harper M.-E., Michaud J., Sell E., Chakraborty P., Care4Rare Consortium DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur. J. Hum. Genet. 2016;24:1084–1088. doi: 10.1038/ejhg.2015.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosmicki J.A., Samocha K.E., Howrigan D.P., Sanders S.J., Slowikowski K., Lek M., Karczewski K.J., Cutler D.J., Devlin B., Roeder K. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellus G.A., Hefferon T.W., Ortiz de Luna R.I., Hecht J.T., Horton W.A., Machado M., Kaitila I., McIntosh I., Francomano C.A. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am. J. Hum. Genet. 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- 27.Hood R.L., Lines M.A., Nikkel S.M., Schwartzentruber J., Beaulieu C., Nowaczyk M.J.M., Allanson J., Kim C.A., Wieczorek D., Moilanen J.S., FORGE Canada Consortium Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am. J. Hum. Genet. 2012;90:308–313. doi: 10.1016/j.ajhg.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamdan F.F., Myers C.T., Cossette P., Lemay P., Spiegelman D., Laporte A.D., Nassif C., Diallo O., Monlong J., Cadieux-Dion M., Deciphering Developmental Disorders Study High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am. J. Hum. Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miki H., Suetsugu S., Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B., Chou H.-T., Brautigam C.A., Xing W., Yang S., Henry L., Doolittle L.K., Walz T., Rosen M.K. Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. eLife. 2017;6:6. doi: 10.7554/eLife.29795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunda P., Craig G., Dominguez V., Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Reijnders M.R.F., Ansor N.M., Kousi M., Yue W.W., Tan P.L., Clarkson K., Clayton-Smith J., Corning K., Jones J.R., Lam W.W.K., Deciphering Developmental Disorders Study RAC1 Missense Mutations in Developmental Disorders with Diverse Phenotypes. Am. J. Hum. Genet. 2017;101:466–477. doi: 10.1016/j.ajhg.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaworski J., Kapitein L.C., Gouveia S.M., Dortland B.R., Wulf P.S., Grigoriev I., Camera P., Spangler S.A., Di Stefano P., Demmers J. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Fischer M., Kaech S., Knutti D., Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 35.van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu. Rev. Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- 36.Pilpel Y., Segal M. Rapid WAVE dynamics in dendritic spines of cultured hippocampal neurons is mediated by actin polymerization. J. Neurochem. 2005;95:1401–1410. doi: 10.1111/j.1471-4159.2005.03467.x. [DOI] [PubMed] [Google Scholar]
- 37.Soderling S.H., Guire E.S., Kaech S., White J., Zhang F., Schutz K., Langeberg L.K., Banker G., Raber J., Scott J.D. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J. Neurosci. 2007;27:355–365. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y., Sung J.Y., Ceglia I., Lee K.-W., Ahn J.-H., Halford J.M., Kim A.M., Kwak S.P., Park J.B., Ho Ryu S. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- 39.Hazai D., Szudoczki R., Ding J., Soderling S.H., Weinberg R.J., Sótonyi P., Rácz B. Ultrastructural abnormalities in CA1 hippocampus caused by deletion of the actin regulator WAVE-1. PLoS ONE. 2013;8:e75248. doi: 10.1371/journal.pone.0075248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soderling S.H., Langeberg L.K., Soderling J.A., Davee S.M., Simerly R., Raber J., Scott J.D. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl. Acad. Sci. USA. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law R., Dixon-Salazar T., Jerber J., Cai N., Abbasi A.A., Zaki M.S., Mittal K., Gabriel S.B., Rafiq M.A., Khan V. Biallelic truncating mutations in FMN2, encoding the actin-regulatory protein Formin 2, cause nonsyndromic autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2014;95:721–728. doi: 10.1016/j.ajhg.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivière J.-B., van Bon B.W.M., Hoischen A., Kholmanskikh S.S., O’Roak B.J., Gilissen C., Gijsen S., Sullivan C.T., Christian S.L., Abdul-Rahman O.A. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat. Genet. 2012;44:440–444, S1–S2. doi: 10.1038/ng.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kutsche K., Yntema H., Brandt A., Jantke I., Nothwang H.G., Orth U., Boavida M.G., David D., Chelly J., Fryns J.P. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat. Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- 44.Yan Y., Tsukamoto O., Nakano A., Kato H., Kioka H., Ito N., Higo S., Yamazaki S., Shintani Y., Matsuoka K. Augmented AMPK activity inhibits cell migration by phosphorylating the novel substrate Pdlim5. Nat. Commun. 2015;6:6137. doi: 10.1038/ncomms7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z., Okamoto K., Hayashi Y., Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.