Abstract
It has increasingly become clear over the last two decades that proteins can contain both globular domains and intrinsically unfolded regions that can both contribute to function. Although equally interesting, the disordered regions are difficult to study, because they usually do not crystallize unless bound to partners and are not easily amenable to cryo-electron microscopy studies. NMR spectroscopy remains the best technique to capture the structural features of intrinsically mixed folded proteins and describe their dynamics. These studies rely on the successful assignment of the spectrum, a task not easy per se given the limited spread of the resonances of the disordered residues. Here, we describe the structural properties of ataxin-3, the protein responsible for the neurodegenerative Machado-Joseph disease. Ataxin-3 is a 42-kDa protein containing a globular N-terminal Josephin domain and a C-terminal tail that comprises 13 polyglutamine repeats within a low complexity region. We developed a strategy that allowed us to achieve 87% assignment of the NMR spectrum using a mixed protocol based on high-dimensionality, high-resolution experiments and different labeling schemes. Thanks to the almost complete spectral assignment, we proved that the C-terminal tail is flexible, with extended helical regions, and interacts only marginally with the rest of the protein. We could also, for the first time to our knowledge, observe the structural propensity of the polyglutamine repeats within the context of the full-length protein and show that its structure is stabilized by the preceding region.
Introduction
Ataxin-3 is a human 42-kDa protein involved in the neurodegenerative disease spinocerebellar ataxia type-3, or Machado-Joseph disease (MJD) (1). The age of onset and the severity of this terminal and currently incurable disease depends on the length of a polymorphic tract of polyglutamine (polyQ) in the C-terminal half of ataxin-3. The disease is thought to be caused by the aggregation of the protein promoted by the polyQ tract (2, 3), which, in MJD, is expanded beyond the nonpathological level (ca. 42–50 repeats). Ataxin-3 is a deubiquitinating enzyme that recognizes and cleaves preferentially polyubiquitin chains of at least four subunits and is supposed to play an important role in the ubiquitin-proteasome pathway (4, 5). Several other cellular partners are recognized by ataxin-3 (4, 6, 7, 8, 9, 10), among which is the valosin-containing protein, an important component of the regulation of protein degradation by the proteasome (11, 12).
Ataxin-3 is an excellent example of a “mixed folded protein” (MFP), that is, a protein containing both globular and intrinsically unfolded regions. This class of proteins is distinct from that of globular proteins, which adopt a well-defined stable tertiary structure (13). It is also more complex than the so-called intrinsically unfolded proteins (IUPs or IDPs), which are devoid of a three-dimensional (3D) structure but can adopt it when and if needed (14). The architecture of ataxin-3 consists of an N-terminal 21-kDa domain, Josephin, followed by a region containing two or three (depending on the isoform) ubiquitin-interacting motifs (UIMs) and the polyQ repeats (15). Ubiquitin is recognized by ataxin-3 through multiple surfaces of the protein, distributed both along the Josephin domain and in the C-terminus (16, 17). Although the structure of the globular Josephin domain was solved early on after the identification of ataxin-3 (18), little is known about the intrinsically unfolded C-terminus. Solving the structure of ataxin-3 by crystallography has proven arduous, presumably because of the difficulty of obtaining ordered crystals of the full-length protein. Of the other two main methodologies able to describe structures at atomic resolution (NMR and, more recently, cryo-electron microscopy), NMR has proven most helpful in the characterization of IDPs and MFPs because it can operate directly in solution, capture molecular motions, and observe flexible structures. Despite the advantages mentioned, these studies have proven difficult because of inherent technical limitations. The first and possibly most important one is the fact that, as opposed to the generally good chemical shift dispersion that characterizes the NMR spectrum of folded proteins, IDPs and intrinsically unfolded regions suffer from an inevitable and inherent severe resonance overlap. Because the assignment of an NMR spectrum to each of the atoms along the sequence depends on the tracing of the magnetization transfer, that is, the transfer from one atom to the next as in a network, spectral overlap has the consequence of limiting the possibility of uniquely tracing the chain. Several strategies have been suggested to circumvent this problem. Yet no definite answer has been given to the question, and new ideas must be considered to eventually find, if possible, a unique and universal strategy. Additionally, ataxin-3 is prone to aggregate, and this makes it difficult to keep the protein in solution for the time (days or weeks) required for the formation of crystals or for the acquisition of complex NMR data sets.
Here, we describe a study of the structural properties of full-length ataxin-3 based on NMR. Despite the intrinsic difficulties, we successfully developed a strategy that allowed us to obtain the assignment of 87% of the spectrum and gain a clear map of ataxin-3 structure and flexibility. We could clearly observe the secondary structure propensity of the unfolded and flexible chain. Within this, the polyQ tract has a strong tendency toward helical conformation stabilized by the preceding region. We could also infer the dynamical properties of the C-terminal tail and compare them with those of Josephin. Our results provide a solid basis for a complete structural understanding of ataxin-3 and the essential prerequisite for any further study of the interactions between ataxin-3 and other cellular partners. In addition, the approaches developed provide, to our knowledge, new tools for the investigation of MFPs.
Materials and Methods
Construct choice and cloning
The plasmid used for ataxin-3(Q13) recombinant over-expression is a derivative of pMAL-c5× (New England Biolabs, Ipswich, MA), hereafter referred as pMht-atx3(Q13). The plasmid encodes maltose-binding protein (MBP) followed by a hexahistidine tag, a tobacco etch virus protease (TEVp) cleavage site, and the sequence encoding ataxin-3(Q13). The expression plasmid for the isolated Josephin domain (pMht-jos) was obtained by replacing the codon encoding residue 183 of ataxin-3 with a TAA stop codon via inverse polymerase chain reaction (PCR) site-directed mutagenesis. The mutants of ataxin-3(Q13) were obtained via inverse PCR site-directed mutagenesis using pMht-atx3(Q13) as a template. The desired mutation was inserted at the 5′-end of the forward mutagenic primer. After PCR amplification, the linearized plasmid was purified using Zyppy Plasmid Miniprep Kit (Zymo Research, Irvine, CA). The DNA was then incubated with DpnI (New England Biolabs), T4 polynucleotide kinase (New England Biolabs) and QuickLigase (New England Biolabs) to degrade the template, phosphorylate the 5′ ends, and recircularize the plasmid, respectively. The ligation mix was used to transform directly DH5α cells (New England Biolabs), and the cells were plated overnight on Luria-Bertani broth agar plates at 37°C. The outcome of the mutagenesis was checked by Sanger sequencing (GATC Biotech, Konstanz, Germany).
Protein production
The isolated Josephin domain, ataxin-3(Q13) wild type, and mutants were produced in a recombinant form using the pMht series plasmids described in the previous section and the Escherichia coli strain BL21 (New England Biolabs). Slightly different protocols were needed for producing the uniformly or selectively labeled samples. 15N-13C and 15N uniformly labeled proteins were expressed in M9 medium containing 15N ammonium sulfate and 13C-D-glucose (Cambridge Isotopes Laboratories, Tewksbury, MA) as the sole source of nitrogen and carbon, respectively. The cells were grown at 37°C until an OD600 of 0.8 was reached. Protein overexpression was induced by adding isopropyl β-D-1-thiogalactopyranoside (Generon, Slough, UK) to a final concentration of 1 mM. The cells were further incubated under shaking at 37°C for 3 h.
Selective labeling was achieved by first growing the cells in M9 medium supplemented with natural abundance ammonium sulfate, glucose, and amino acids. When the cells reached an OD of 0.8–1.0, 100 mg of the desired 15N labeled amino acid and 1 g of all other unlabeled amino acids were added to 1 L of culture. The cells were incubated under shaking. For 15N-glutamine selective labeling, 75 mg of 6-diazo-5-oxo-L-norleucine, 180 mg of L-methionine sulfoximine, and 180 mg of L-methionine sulfone (Sigma-Aldrich, St. Louis, MO) were also added to the culture. For 15N-glutamate selective labeling, 1 g of L-methionine sulfoximine, 1 g of L-methionine sulfone, 250 mg of disodium succinate, 250 mg of disodium maleate, and 250 mg of aminooxyacetate (Sigma-Aldrich) were added to the culture (19). After incubating the cells under shaking at 37°C for 15–20 min, isopropyl β-D-1-thiogalactopyranoside was added to a final concentration of 1 mM, and protein overexpression was allowed for 90 min.
All cultures were harvested by centrifugation at 6000 × g for 20 min on a Beckman Avanti centrifuge (Beckman Coulter Life Sciences, Indianapolis, IN). The cell pellet was transferred to 50 mL tubes and frozen at −20°C. The cell pellet was defrosted and resuspended in 20 mM sodium phosphate, 200 mM NaCl (pH 7.5) (buffer A) supplemented with DNase and lysozyme (Sigma-Aldrich). After sonication, ataxin-3 was purified by NiNTA-agarose (Generon) affinity chromatography using 20 mM sodium phosphate, 200 mM NaCl, 250 mM imidazole (pH 7.5) (buffer B) for elution. The 6×His-MBP tagged protein was then buffer-exchanged to buffer A, and hexahistidine-tagged TEVp was added to release untagged ataxin-3(Q13). The cleavage mixture was incubated with NiNTA-agarose resin to remove 6×His-MBP and 6×His-TEVp. The cleaved protein was concentrated with centrifugal filters and run on a Superdex 75 26/60 size-exclusion chromatography column (GE Healthcare, Little Chalfont, UK) to remove small traces of aggregates or impurities and exchange the protein into 20 mM sodium phosphate buffer, 2 mM TCEP (pH 6.5). The elution volumes obtained from size-exclusion chromatography confirmed that all the proteins were monomeric in solution. A final purity higher than 95% was assessed by SDS-PAGE.
NMR spectroscopy and sequential assignment
All NMR experiments were performed in 20 mM sodium phosphate buffer at pH 6.5 and 2 mM TCEP at 25°C. A protein concentration of 250 μM was used for all the experiments. Data were acquired at 600, 700, 800, and 950 MHz on Bruker Avance instruments (Bruker, Karlsruhe, Germany) equipped with cryogenic probes. HNCO, HNcaCONH, and HabCabCONH experiments were acquired with four scans per increment. The HNCO was acquired with an interscan delay of 1.5 s and spectral widths of 10416.7 Hz (acquisition dimension), 2594.7 Hz (15N), and 1810.94 Hz (13C′). Nonuniform sampling of the indirectly detected dimensions was applied. A five-dimensional (5D) HNcaCONH experiment was acquired with an interscan delay of 1.5 s and spectral widths of 10416.7 Hz (acquisition dimension), 2594.8 Hz (15N), 1811.1 Hz (13C′), 2594.8 Hz (15N), and 960.2 Hz (1H). The maximal evolution times were 50 ms for t1, t2, and t4 (1H and 15N) and 26.5 ms for t3 (13C′). The 5D HabCabCONH experiment was acquired with an interscan delay of 1.5 s and with spectral widths of 10416.7 Hz (acquisition dimension), 2594.8 Hz (15N), 1810.9 Hz (13C′), 12474.8 Hz (13Caliph), and 6401.2 (1Haliph). The maximal evolution times were 10 ms for the 1Haliph, 6.7 ms for 13Caliph, 26.4 ms for 13C′, and 49.3 ms for 15N indirect dimensions. In both experiments, a total number of 1910 points was acquired in the indirect dimensions. Nonuniform sampling was used for the indirectly detected dimensions. The time schedule was generated using a Poisson disk sampling (20). The whole data set of HNCO, HNcaCONH, and HabCabCONH experiments was acquired on the same protein sample in ∼65 h. Additional 15N-1H heteronuclear single quantum spectroscopy (HSQC) experiments were recorded immediately before and after each higher-dimensionality experiment to assess the spectral integrity of the protein. In all cases, the two HSQC spectra did not show discrepancies, suggesting sample stability, which is necessary to acquire high-quality data of high dimensionality.
The 3D nuclear Overhauser effect spectroscopy (NOESY)-15NHSQC experiment was acquired with an interscan delay of 1.0 s and a mixing time of 150 ms. The direct acquisition time (1H) was 85 ms, and the indirect evolution times were 15 ms for 1H and 18 ms for 15N. The 3D band-selective excitation short-transient (BEST)-transverse relaxation-optimized spectroscopy (TROSY) HNCACB experiment was acquired with an interscan delay of 0.12 s. The direct acquisition time (1H) was 91 ms, and the indirect evolution times were 7.5 ms for 13Cα,β and 23 ms for 15N.
HSQC-based interleaved relaxation data were acquired as pseudo-3D experiments at 800 MHz. For the isolated Josephin domain, delays of 100, 200, 400, 700, 1200, 1800, and 2500 ms and 16, 32, 64, 96, 144, and 192 ms were used for T1 and T2 measurements, respectively. For ataxin-3(Q13), delays of 100, 200, 400, 700, 1000, 1500, 2500, and 4000 ms and 16, 32, 48, 64, 96, 128, 160, 192, and 240 ms were used for T1 and T2 measurements, respectively. Average T1 and T2 values for Josephin and for ataxin-3(Q13) were calculated by fitting the area under the spectra in the range 7.7–10.5 ppm to an exponential function. The relaxation rates for Josephin residues within ataxin-3(Q13) were obtained by fitting the HSQC peak intensities. The correlation times were estimated using the formula (21).
The spectra were processed with NMRPipe based scripts and NMRDraw (22). The HNCO and the 5D experiments were processed with the multidimensional Fourier transform (23, 24) and the sparse multidimensional Fourier transform (25) algorithms. The assignment of the NMR spectra was performed with the software Sparky 3.115 (T. D. Goddard and D. G. Kneller, University of California, San Francisco, CA) and CcpNmr Analysis 2.4 (26). The relaxation rates were calculated and analyzed using nmrglue (27), nmrPipe, and CcpNmr Analysis 2.4. The assignment was submitted to the Biological Magnetic Resonance Bank database (accession number 27380).
Results
Strategy for spectral assignment of ataxin-3
Assignment of full-length ataxin-3 was performed on isoform 2 (UniProt: P54252-2) with an interrupted tract of 13 polyQ repeats, Q3KQ10—hereafter referred to as ataxin-3(Q13). This isoform contains three UIMs in the C-terminus at variance with isoform 1 (used in previous studies of our group (15, 28)), which has only two UIMs. We first reasoned that the problem of overlap would be significantly reduced by producing the recombinant C-terminal tail in isolation, assigning it, and then transferring the information to full-length ataxin-3. Unfortunately, we realized that the isolated tail easily degrades in the absence of the N-terminus (data not shown). We also tried to purify the tail using chimeric constructs in which the C-terminus could be released by protease cleavage. Once free in solution, the C-terminus aggregated rapidly. We thus had to work straightaway on full-length ataxin-3(Q13).
Two features of ataxin-3 had to be considered when choosing a suitable assignment strategy. Since ataxin-3 tends to aggregate with time, 15N-1H HSQC spectra were recorded before and after each experiment to assess whether the protein remained mainly in a monomeric state throughout the experiment. The spectra were assessed for variations of the peak volumes and of the resonance positions. Typically, a fresh sample of ataxin-3(Q13) at 250 μM could be used for at least 3 days in the NMR tube at 25°C. No significant changes in both peak intensity and position could be detected over this time frame. The sequence of the C-terminus of ataxin-3(Q13) (residues 183–361) contains multiple repetitive stretches, including three UIMs and the polyQ tract. The HSQC spectrum of ataxin-3(Q13) clearly shows the coexistence of well-dispersed resonances, with regions characterized by poor dispersion and high resonance overlap (Fig. 1 A). Spectral overlap and crowding were encompassing the frequency range 117–126 ppm (in the 15N dimension) and 7.9–8.7 ppm (in the 1H dimension). This is a hallmark of the intrinsic high flexibility of the C-terminus, as expected from the sequence. Another important feature is that the intensities of the resonances are highly uneven, with many resonances being extremely broad. This suggests different dynamical properties and/or solvent exchange regimes. Taken together, these data show that the tail of ataxin-3 has features of an intrinsically unfolded or partially structured system.
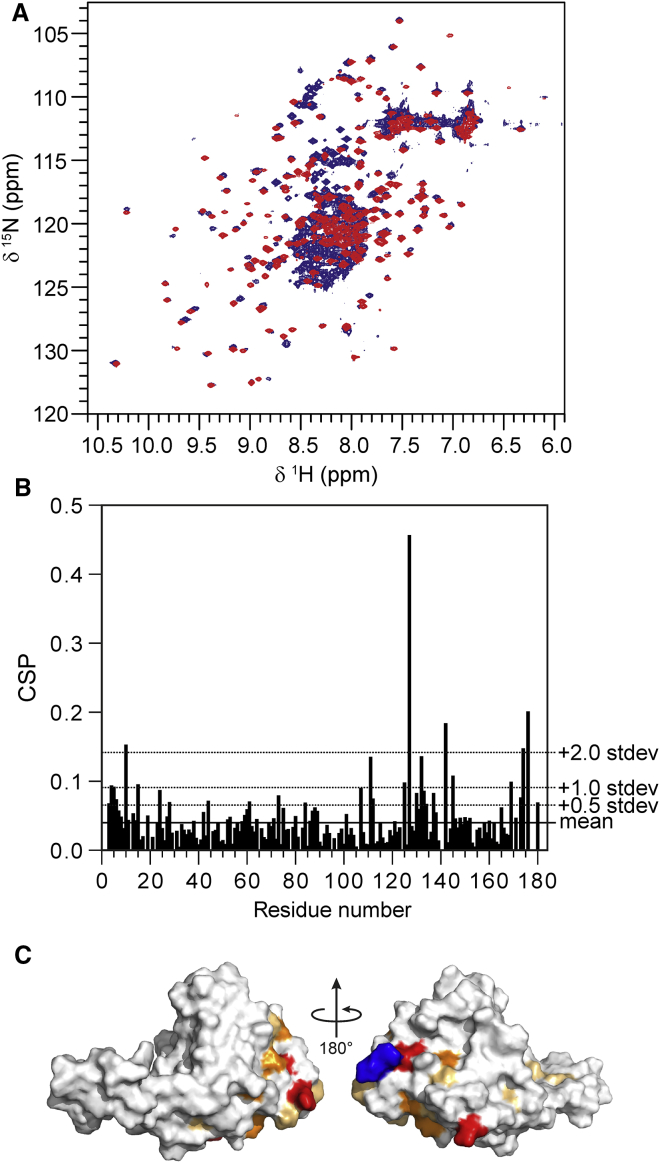
Figure 1.
Assignment of the NH backbone resonances of the Josephin domain within the ataxin-3(Q13) construct. (A) Superimposition of the 15N-1H HSQC spectrum of the isolated Josephin domain (red) and ataxin-3(Q13) (blue). The spectra were recorded at 800 MHz and 25°C. (B) Chemical shift perturbation of Josephin NH backbone resonances in ataxin-3(Q13) with respect to the isolated domain. (C) Mapping of the chemical shift perturbations on the structure of the Josephin domain. Light orange, chemical shift perturbation > average + 0.5 SD; orange, chemical shift perturbation > average + 1 SD; red: chemical shift perturbation > average + 2 SD. The C-terminal arginine is colored in blue. The chemical shift perturbation (ΔδH-N) was calculated according to the formula , where α = 0.14 for glycine and α = 0.20 for any other residue. To see this figure in color, go online.
The Josephin domain does not significantly interact with the C-terminus
To identify the already assigned resonances of the smaller Josephin domain (residues 1–182) within the much more crowded spectrum of ataxin-3, we superimposed the 15N-1H HSQC spectra of isolated Josephin with that of full-length ataxin-3(Q13) (Fig. 1 A). Most of the well-dispersed Josephin resonances in the spectrum of full-length ataxin-3 overlap with those of the isolated domain. This allowed facile identification of several resonances. Some peaks, however, shift because of a change in the chemical environment. This could be a consequence of either interaction between the structured and unstructured regions or of minor changes in the experimental conditions. Although the latter effect can be minimal in a well-dispersed protein spectrum, unambiguous assignment of a spectrum of this complexity becomes prohibitive without further support. Initially, we could tentatively assign 124 out of the 174 nonproline resonances of Josephin (70%). Each of the NH correlations in the spectrum of the isolated Josephin domain was used to guide the search for the corresponding resonance of the full-length protein. The identity of the NH resonances of Josephin in ataxin-3(Q13) was further confirmed by inspection of the HNCACB spectrum (29). Given the unfavorable tumbling time of the Josephin domain in the context of the full-length protein, this experiment yielded weak signals for the Cα and Cβ resonances and no correlations with the preceding residue for most of the Josephin NH resonances. This partial information nevertheless proved very valuable for assigning the amino acid type, yielding unambiguous assignment in most cases. The assignment of the majority of the well-dispersed resonances of the Josephin domain could be achieved with confidence by superimposition of the spectrum of the isolated domain. The Cα and Cβ resonances proved helpful in confirming the amino acid type. The assignment of most of the residues resulted overall straightforward despite the limited chemical shift perturbation induced by the presence of the C-terminal tail. In contrast, the resonances of Josephin located in the highly degenerate central area of the HSQC spectrum demanded the use of information on the amino acid type from Cα and Cβ carbons and careful inspection of the carbon planes to avoid misassignments. This task was nevertheless facilitated by the low degree of overlap of the carbon resonances associated to Josephin and to the C-terminal tail, respectively.
To extend further the assignment of the Josephin resonances within the full-length ataxin-3(Q13) spectrum, we compared the 3D NOESY-HSQC spectra of ataxin-3(Q13) and of the isolated Josephin domain (Fig. S1). This allowed the assignment of 20 additional resonances, reaching a sequence coverage of 83% of the Josephin resonances. A few Josephin resonances in the HSQC spectrum of ataxin-3(Q13) showed a low signal-to-noise ratio and did not yield carbon correlations in the HNCACB spectrum or NOESY-HSQC amide or aliphatic proton patterns that could be identified in the spectrum of the isolated Josephin domain. To identify these resonances, we acquired a BEST-TROSY HSQC (30) and overlapped it with the conventional HSQC spectra of ataxin-3(Q13) and of the isolated Josephin domain. The overall increase in the signal-to-noise ratio of the BEST-TROSY HSQC spectrum made it possible to detect the previously invisible NH Josephin resonances and assign them. The combination of these experiments helped us to obtain an assignment coverage of 91% of the Josephin spectrum, which corresponds to an ∼55% coverage of full-length ataxin-3. These results strongly indicate that the assignment of difficult proteins can be achieved only by exploiting a broad repertoire of experiments, each designed to fulfill different purposes. This includes experiments that would not usually be considered as a first choice for protein assignment, such as the NOESY-HSQC.
The chemical shift perturbations observed in Josephin within ataxin-3(Q13) as compared to the isolated domain are small (overall within a standard deviation below or equal to 0.5) (Fig. 1 B). The only resonances with more appreciable variations are in the N- and C-termini and at residue G127. This is reasonable, because the N- and C-termini of Josephin are close in space. G127 is spatially close. This evidence demonstrates convincingly lack of tight interactions between the Josephin domain and the C-terminal tail of ataxin-3, which thus behaves semi-independently.
Walking through the assignment of ataxin-3 C-terminus
Triple resonance experiments (HNCA, HNCACB, CBCAcoNH) were initially performed to assign the ataxin-3(Q13) C-terminus (Fig. S2). The low dispersion of the 1H/15N/13C resonances associated to residues within repetitive regions impeded progress in the assignment, suggesting the necessity of ultra-high-resolution techniques. We exploited a strategy developed for intrinsically disordered proteins that uses 5D pulse sequences (20). The high resolution resulting from the higher dimensionality of these experiments has been shown to be particularly successful for systems that are not only intrinsically unfolded but also highly degenerate in sequence (31, 32, 33). In this strategy, a HN(CA)CONH experiment is used to provide sequential connectivity by linking the 15N-1H chemical shifts of consecutive residues, whereas a HabCabCONH spectrum is used to identify amino acid types from their Cα and Cβ resonances (20). The data are acquired using a nonuniform sampling scheme in which only 2% of the total data points are collected. The spectra are subsequently reconstructed using the sparse multidimensional Fourier transform algorithm (25). This strategy resulted in a reduction of the total acquisition time needed for a full set of assignment experiments from weeks to ∼2.5 days, a time frame in which the ataxin-3 spectral properties are unperturbed, indicating that the protein remains mainly monomeric. We first acquired a reference high-resolution HNCO spectrum with nonuniform sampling, which was used to extract the chemical shift values for each 1H/15N/13C′ resonance from the parent CON and CabHab two-dimensional planes of the 5D HNcaCONH and HabCabCONH data sets, respectively. The CON plane contains a weak resonance related to the HNCO peak of residue i and a stronger resonance related to the HNCO peak of residue i−1. The CabHab planes contain the Cα, Cβ, Hα, and Hβ resonances of residue i−1. The 5D spectra were of excellent quality and confirmed the initial assignment obtained via triple resonance experiments. Additional residues could be assigned, resulting in the coverage of ∼50% of the resonances of ataxin-3 C-terminus (Fig. 2). Assignment of the 1H, 15N, and 13C′ resonances was possible for residues 214–226, 240–246, 263–274, 298–312, and 319–360. Within these regions, the Cα chemical shifts were assigned for all the residues with the exception of residues 214, 218, 220, 227, 246, 268, 273, 274, 306, 310, 312, 324, 327, 331, 339, 359, and 360, for which it was not possible to detect a C′ resonance in the sequential assignment. The Cβ chemical shifts could be assigned for all the residues with the exception of residues 227, 246, 274, 312, and 360, for which no signals could be detected in the CabHab planes of the 5D HabCabCONH experiment.
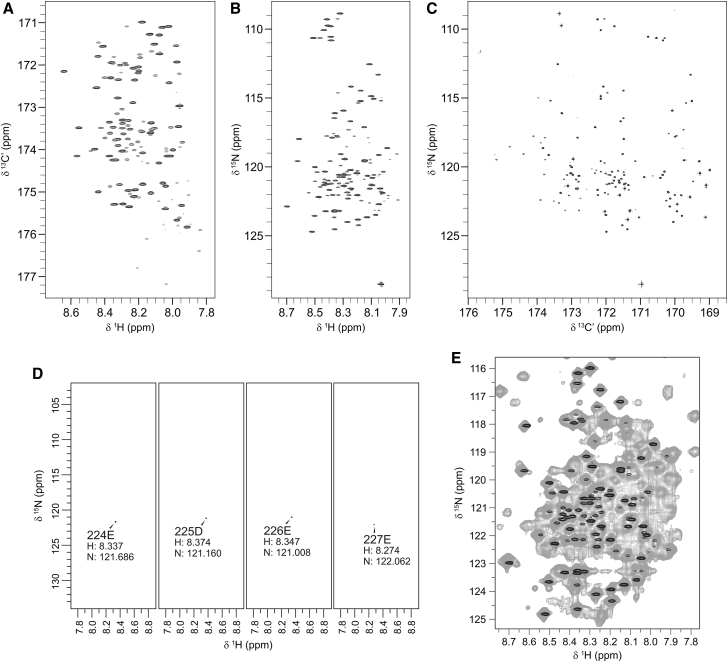
Figure 2.
Assisting assignment with 5D HNcaCONH experiments. (A) 13C-1H, (B) 15N-1H, and (C) 15N-13C projections of the HNCO experiment. (D) 15N-1H planes of consecutive residues associated to HNCO peaks in the 5D HNcaCONH experiment. (E) Superimposition of the HSQC spectrum of ataxin-3(Q13) (gray) with the 15N-1H projection of the HNCO spectrum used as reference for the 5D HNcaCONH (black).
In the course of this analysis, we noticed that only a subset of the expected resonances could be detected. 5D experiments are optimized for highly disordered systems. The long polarization transfer pathway used in these experiments allows ultra-high resolution but also results in a considerable loss of signal-to-noise ratio. Consequently, fast-relaxing resonances located in rigid or semirigid portions of the polypeptide chain are usually nondetectable, whereas the effect is much less pronounced for the slowly relaxing resonances from highly flexible regions. Therefore, the mere fact that only a subset of the expected resonances could be detected in the 5D experiments indicates that the residues in the C-terminal tail of ataxin-3 have a wide range of dynamical properties. In particular, the ultra-high resolution of high-dimensionality experiments allowed sequential assignment of 8 out of 13 glutamine residues in the polyQ tract (residues 298–305). The signal-to-noise ratio of the carbon resonances in this region progressively increases from the N-terminus to the C-terminus. This observation suggests a higher flexibility of the polyQ tract at the C-terminus.
Systematic mutation of key residues
Additional resonances could be assigned by integrating the information obtained from the 5D experiments and from triple resonance experiments. Despite this, the information was still insufficient to obtain a satisfactory assignment coverage. For some amide resonances, a correlation with the previous or following residue was impeded by the absence of the corresponding Cαi−1 in the HNCACB spectra. An additional problem was posed by the ambiguity of many Cα-Cαi−1 correlations, leading to multiple equally likely candidates for sequential assignment. To overcome these problems, a systematic mutation strategy was exploited. Mutations were introduced at different sites within both assigned and unassigned regions. HSQC spectra were acquired for each mutant that could be purified and was stable. This assignment strategy relies on the fact that the resonance in the spectrum of the wild-type protein that is associated to the mutated residue will not be detectable in the spectrum of the mutant, whereas a resonance with different 1H and 15N chemical shifts, associated with the new residue, is expected to appear. Because the C-terminal tail of ataxin-3 is flexible and does not seem to form stable tertiary structures, a mutation at one site of the protein is expected to cause a chemical shift perturbation only for the amide NH resonances of residues proximal to the mutation. It is reasonable to expect that this perturbation is roughly inversely correlated with the distance in sequence from the mutated site in a flexible and largely solvent exposed chain. The mutants G193A, T207A, L213I, D228E, S256A, S260A, R282H, R284H, R285H, and C316A were successfully used to assign resonances for which a through-bond correlation could not be detected in triple resonance experiments. For example, the analysis of the HSQC spectrum of the mutant T207A in conjunction with the triple resonance experiments of the wild-type protein allowed unambiguous assignment of residues 206–208, based on the combination of the chemical shift perturbation observed and on information about the amino acid type from Cα and Cβ resonances obtained from the HNCACB experiment. Analogously, the spectrum of the mutant D228E helped to validate the assignment of a number of residues obtained both through triple resonance and 5D experiments (Fig. S3).
Further assignment using amino acid selective labeling
An amino acid selective labeling approach was used to resolve the ambiguities caused by the overlap of resonances associated with residues whose β carbons share similar chemical shifts. 14N-1H-labeled ataxin-3(Q13) samples were produced that incorporated a single 15N-labeled amino acid (15N-alanine, 15N-isoleucine, 15N-leucine, 15N-lysine, 15N-methionine, 15N-valine, 15N-arginine, 15N-glutamine, 15N-glutamate, 15N-tyrosine, and 15N-phenylalanine) (19). The samples resulted in an overall high labeling yield and selectivity, suggesting that amino acid metabolic scrambling was suppressed efficiently. Only the spectrum of selectively labeled 15N-glutamate showed considerable scrambling from glutamate to glutamine, aspartate, and alanine and some isotope dilution (Fig. S4). Despite this, a critical analysis of the spectrum, helped by comparison with the spectra of the other amino acid selectively labeled spectra, aided in discerning selective labeling from scrambling. This allowed unambiguous identification of amino acid types throughout the spectra (Table S1).
The assignment of overlapping resonances for which the chemical shifts of Cαi−i−1 and Cβi−i−1 connectivity could be detected in HNCACB spectra was straightforward. When the Cαi−1 and Cβi−1 were not detectable, assignment was obtained by producing amino acid selectively labeled mutants. For each labeling, the HSQC spectrum of wild-type ataxin-3 was compared with the HSQC of the corresponding point mutant. In this way, we could identify unambiguously V183, H187, L191, E194–A197, Q202–V204, K206, E210, R231, A232, R237, Q238, R251, A252, Q254, L255, M257, Q258, R262, E279–F289, and R318 (Fig. 3; Fig. S4).
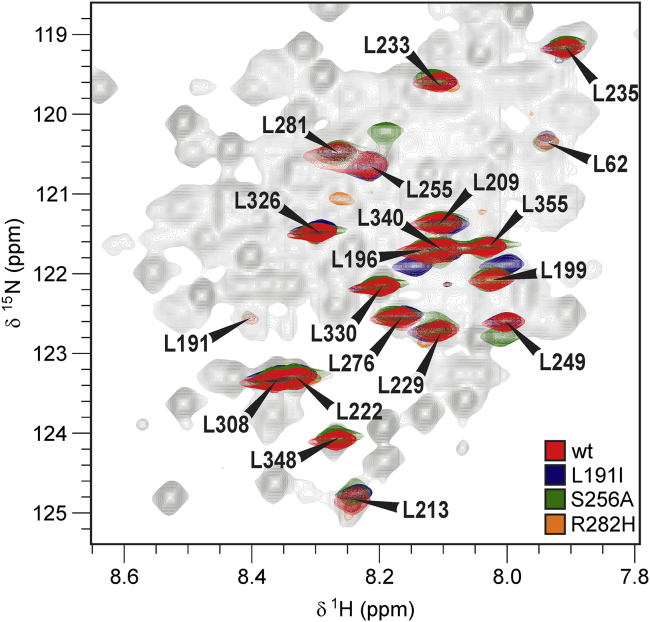
Figure 3.
15N-1H HSQC spectra of 15N-Leu selectively labeled wild-type and mutated ataxin-3(Q13). The spectrum of the wild-type protein (red) is superimposed to the spectra of three mutants (L191I: blue, S256A: green, R282H: orange). The counter levels were adjusted to show only the sharper resonances related mainly to residues in the C-terminal tail. The resonances of at least eight more leucines were also detectable in the experiment but with lower intensities. They all correspond to Josephin leucines. Minor scrambling was observed at the noise level. To see this figure in color, go online.
The NH resonances from most of the residues in the tract 183–192 could not be detected in the spectra of selective amino acid labeled samples. This fact and the lack of NH resonances of residues in the spatially close N- and C-termini of the Josephin domain suggest that this region is in an intermediate exchange regime.
The use of BEST-TROSY HNCACB to assist assignment
In an attempt to increase the signal-to-noise ratio further, BEST experiments were performed in association with TROSY using the BEST-TROSY HNCACB pulse sequence (34). This experiment achieves a longitudinal relaxation optimization, allowing fast pulsing and rapid acquisition of spectra with higher resolution and signal-to-noise ratio. The combination of BEST with TROSY enhances further these effects. It has been shown that the BEST sequence enhances preferentially resonances from transiently structured regions, which usually have a low signal-to-noise ratio. The increased signal-to-noise ratio of the BEST-TROSY-HNCACB experiment was used to confirm weak carbon resonances throughout the sequence of ataxin-3. This technique proved especially helpful in the sequential assignment of residues E279 and E280. These residues could not be assigned sequentially through the HNCACB experiment, because carbon correlations were below the detection limit. Conversely, BEST-TROSY HNCACB allowed us to detect the Cαi−1 and Cβi−1 correlations of residues E279 and E280 to the preceding serine and glutamate, respectively (Fig. S5). The combination of this information with glutamate 15N-selective labeling of wild-type ataxin-3 and of the R282H mutant led to unambiguous assignment of these residues.
Almost all the detectable resonances in the HSQC spectrum could be assigned unambiguously, reaching a coverage of the C-terminus of ataxin-3(Q13) of 158 out of 176 (90%) nonproline residues and coverage of 87% of the full-length protein (Figs. 4 and S6).
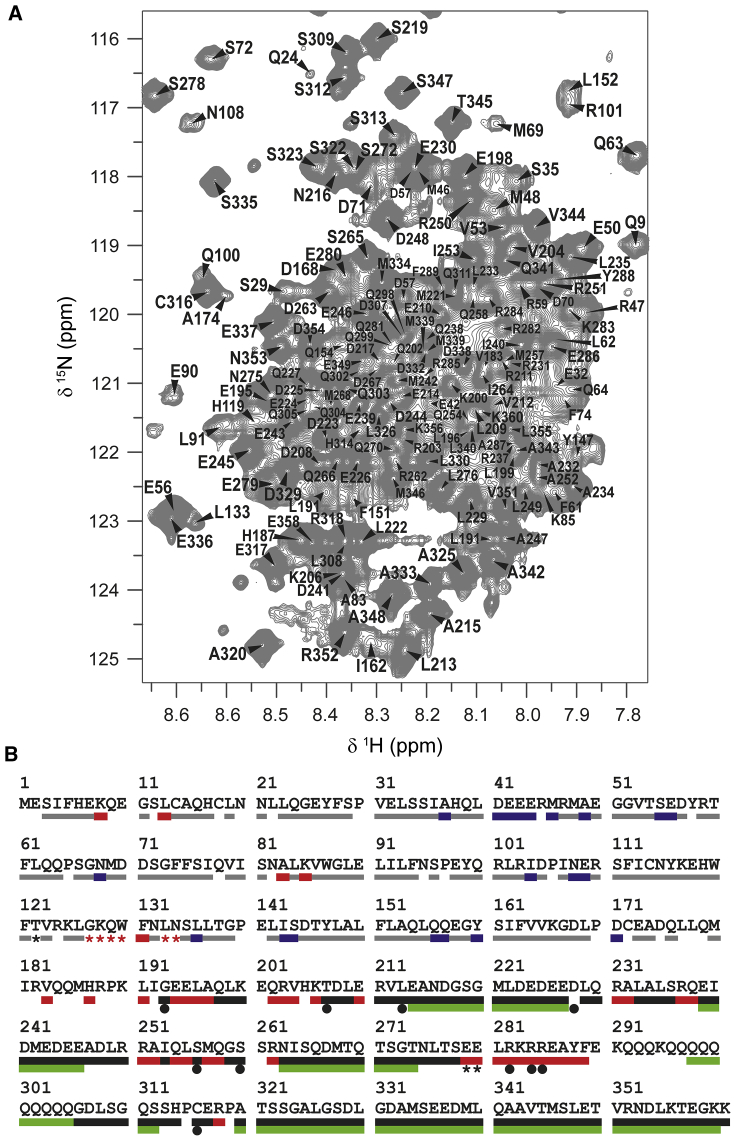
Figure 4.
Summary of the assignment. (A) Highly degenerate areas of the 1H-15NHSQC spectrum of ataxin-3(Q13) annotated with the assignment. (B) Final achieved assignment reported on thesequence of ataxin-3(Q13). The bar colors are associated to the technique used for assignment. The backbone NH pairs of the Josephin domain (residues 1–182) were assigned by comparison with the spectrum of the isolated domain without (gray bars) or with the aid of 15N-amino acid selective labeling (red bars) or BEST-TROSY HSQC (red stars). Residues assigned by comparison of the NOESY-HSQC of ataxin-3(Q13) with that of the isolated Josephin domain are indicated by blue bars. The C-terminal tail was assigned via HNCACB and CBCAcoNH (black bars), 5D HNcaCONH and HabCabCONH (green bars), selective 15N-amino acid labeling (red bars), BEST-TROSY-HNCACB (black stars), and single-point mutations (black circles). To see this figure in color, go online
Structure of the C-terminal tail of ataxin-3
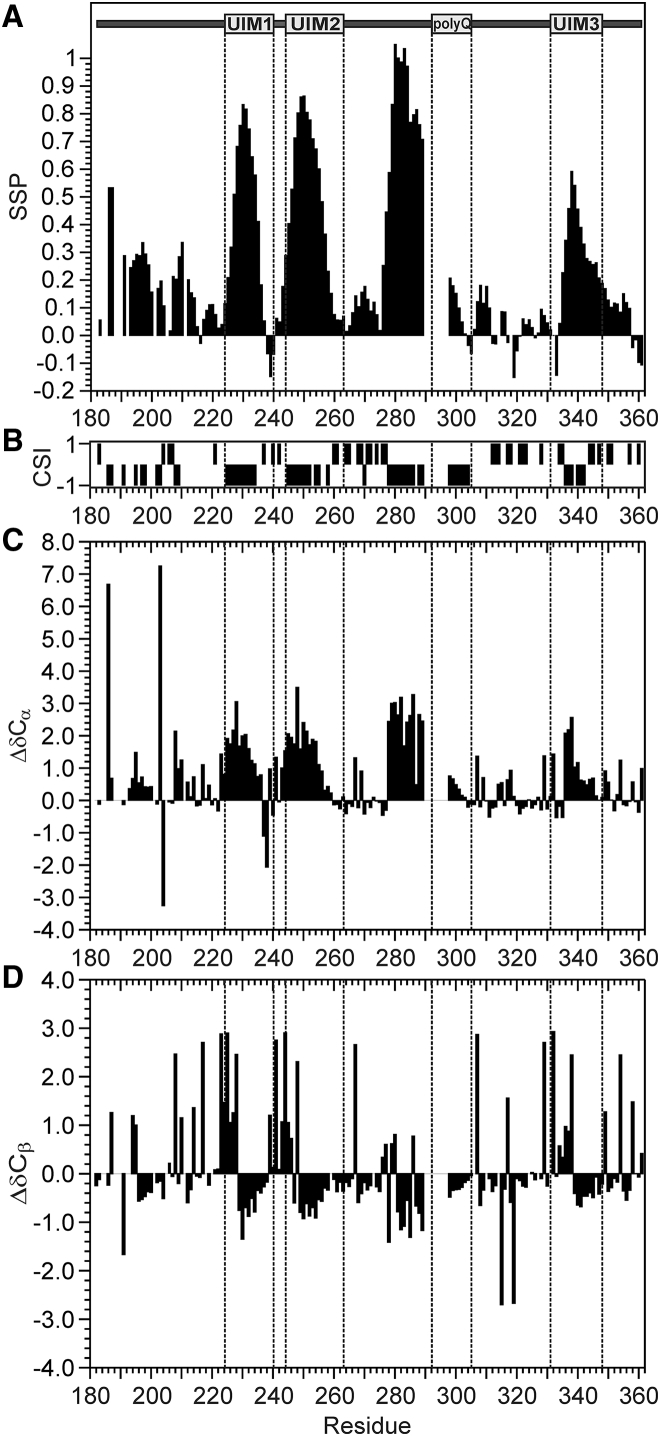
We exploited the chemical shifts obtained from our assignment strategy to gain structural information on the C-terminal tail within the context of the full-length protein. We used the algorithms SSP (35) and CSI (36) to generate scores indicating the propensity of each residue to populate helical, extended, or random coil secondary structure motifs (Fig. 5, A and B). The secondary chemical shifts of the α and β carbons, that is, the differences between the observed chemical shifts and the reference values in a random coil conformation, confirm and strengthen the conclusions obtained by the other two methods (Fig. 5, C and D). Overall, the C-terminal tail is mostly unstructured but with regions of strong helical propensity throughout, with maximal values in the three UIMs (residues 224–240, 244-263, and 331–348), in agreement with reports on peptides spanning the isolated UIMs (17). Interestingly, also the residues in the polyQ tract that could be assigned (298–305) exhibit some helical propensity. This supports the structure of a complex of a polyQ peptide with an antibody (37) but disagrees with our previous data showing that polyQ repeats C-terminally fused to a carrier protein (glutathione-S-transferase) are random coils independently of their length (38). Because, however, residues 278–289, which precede the polyQ repeats, have a helical propensity even higher than that of the UIMs, we can reasonably assume that the polyQ helicity is induced and stabilized by the preceding region. In agreement with this hypothesis is the fact that the α-helicity of the polyQ tract progressively decreases, moving from Q298 to Q305.
Figure 5.
Secondary structure of the C-terminus of ataxin-3(Q13). (A) SSP algorithm. Values around zero indicate random coil regions; positive and negative values indicate α-helical or β-sheet conformations, respectively. (B) CSI values (−1 indicates α-helix, 0 random coil, and +1 β-strand). (C) Secondary Cα chemical shifts. (D) Secondary Cβ chemical shifts.
The C-terminal tail is highly flexible
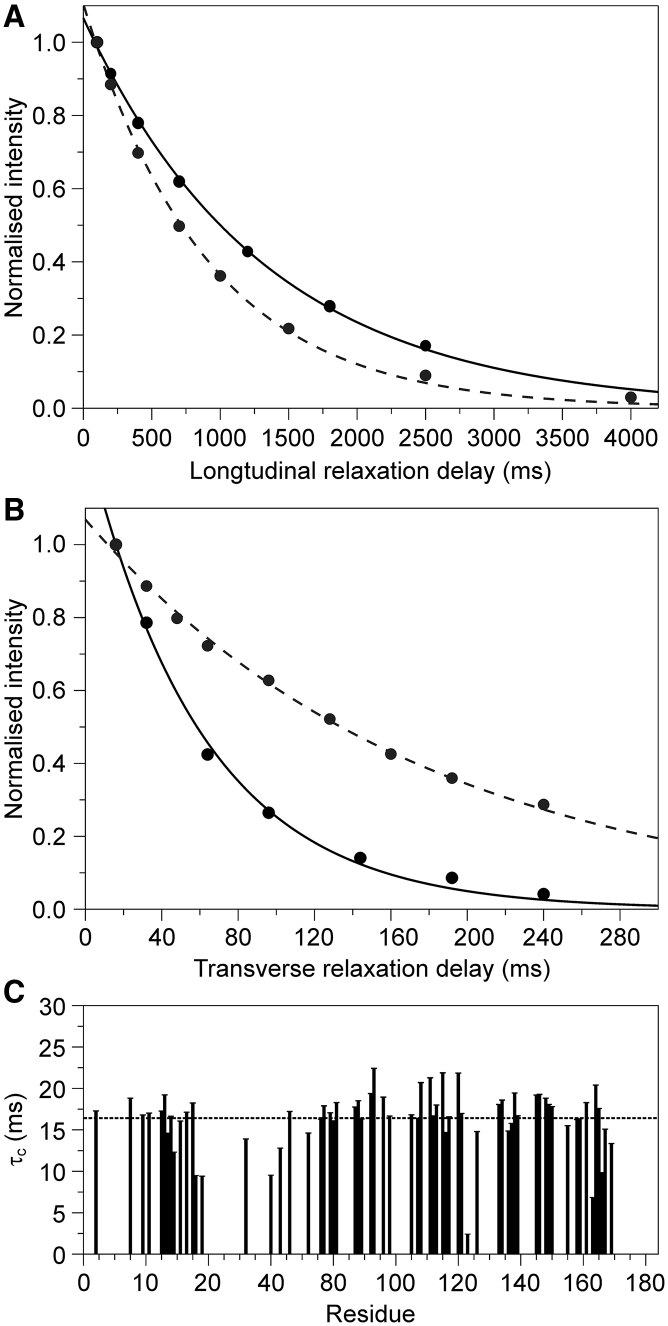
The 15N-1H relaxation values informed us of the dynamical properties of the C-terminal tail and allowed comparison with the properties of isolated Josephin. We reasoned that the strong spectral overlap would prevent us from gaining statistically significant per-residue relaxation times. We thus estimated the overall T1 and T2 relaxation times of full-length ataxin-3 by fitting the area of the resonance envelop over the 1H region 7.7–10.5 ppm of the ataxin-3(Q13) spectrum. Despite its obvious limitations, this approach can still provide valuable overall information. The T1 and T2 relaxation times are 902.2 and 176.1 ms, respectively, as opposed to values of 1322.8 and 61.3 ms for the isolated Josephin domain (Fig. 6, A and B). Assuming a spherical and rigid polypeptide chain, the averaged correlation time obtained from the T1/T2 ratios of the isolated Josephin domain is 10.9 ns. This value is in excellent agreement with the value previously reported (18) and corresponds to that expected for a protein with a molecular weight of ∼18 kDa (39). Under the same assumptions, we estimated an average value of 4.8 ns for full-length ataxin-3(Q13). This marked underestimation clearly indicates that the assumptions do not pertain to this case and that the C-terminal tail is flexible and contributes by significantly lowering the apparent correlation time. To estimate the correlation time of Josephin within ataxin-3(Q13), we selected a set of well-dispersed isolated resonances of Josephin and measured their relaxation rates by traditional experiments (Fig. 6 C). The resulting averaged correlation time (16.4 ns) is that expected for a protein with a molecular weight of ∼25 kDa (29). These data clearly show that the C-terminal tail has higher flexibility than the Josephin domain.
Figure 6.
Description of the dynamics of ataxin-3(Q13). (A) Longitudinal and (B) transverse relaxation of the spectrum of the isolated Josephin domain (continuous line) and ataxin-3(Q13) (dashed line) in the range 7.7–10.5 ppm. (C) Correlation times (τC) of well-dispersed resonances of Josephin within the spectrum of ataxin-3(Q13). The dashed line indicates the average τC.
Discussion
Ataxin-3 is an interesting protein, not only because it is associated to the neurodegenerative MJD, but even more because it is a deubiquitinating enzyme with very specific fold and features (18, 40, 41). The first biophysical studies on this protein date to around 2000, although much of the original interest was devoted toward the aggregation properties of the protein, because they seem to be directly related to disease (2, 42, 43). In 2003, we proved that ataxin-3 is an MFP that contains an N-terminal Josephin domain present in a restricted number of other eukaryotic species and a C-terminal tail (15). The first NMR structure of Josephin was published in 2005 and validated the year after (18, 44), whereas no direct information has been available for the ataxin-3 C-terminal tail other than the generic deduction that it should be more disordered based on the NMR spectrum (15). It is only now that we can report full assignment of the NMR spectrum of ataxin-3. Spectral assignment was indeed not trivial because of the several problems imposed by an MFP of this size: the NMR spectrum of ataxin-3 fully reflects a mixture of globular and unstructured regions. As such, the assignment strategy was far from standard, and we had to develop an ad hoc procedure that combined different independent routes. We found the application of 5D techniques indispensable to achieve the resolution necessary to assign highly degenerate regions. Despite this, the use of 5D experiments was limited by the high dynamic heterogeneity and the presence of resonances in or near an intermediate exchange regime, as supported by selective labeling. Judicious introduction of site-specific mutations helped us to push the assignment coverage and confirm the information obtained independently. This validation proved essential to exclude incorrect tracing given the high complexity of the system.
In addition to the techniques adopted in this study, carbon-detected experiments may have potentially been an excellent choice (45). These methods have several advantages over proton-detection methods, as they provide high-resolution spectra and permit the ability to overcome signal-to-noise ratio problems caused by unfavorable local proton solvent exchange regimes. However, the intrinsically low sensitivity of carbon detection compared to conventional proton detection usually demands protein concentrations above 0.5 mM for low molecular weight proteins, both structured and flexible (46, 47, 48), and of 1 mM or higher for medium molecular weight proteins (45). Small, well structured, well known aggregation-prone proteins were studied at lower concentrations by two-dimensional carbon detected experiments (49) but not by higher dimensionality experiments. For higher molecular weight proteins, only those with high solubility and stability over a sufficiently long experimental time-frame are amenable. This condition is generally difficult to achieve with aggregation-prone proteins such as ataxin-3(Q13), for which millimolar concentrations would be unattainable. The concentration used in this study (250 μM) was in fact carefully chosen to yield acceptable signal-to-noise ratios while delaying aggregation.
Our effort resulted in an overall 87% coverage assignment that extends over the whole protein. Although we cannot exclude the possibility that the unassigned residues have resonances perversely overlapping, independent evidence suggests the presence of conformational and/or solvent exchange of some residues, especially in the region between Josephin and the C-terminal tail. It was also paradoxically difficult to identify some of the Josephin resonances despite the almost complete assignment previously achieved (50). These resonances, which correspond to residues equally distributed along the Josephin domain, were genuinely not observed in the full-length ataxin-3 spectrum and undetectable in the residue-specific selectively labeled samples, indicating that the full-length protein has different exchange regimes as compared to the isolated domain.
This is the first time, to our knowledge, that we can assess directly both the structure in solution and the dynamical behavior of one of the nine polyQ-containing proteins associated with disease within the context of the full-length protein. Because of the difficulties in resolving polyQ protein structures at sufficient resolution, previous studies were all based on fragments (51, 52, 53, 54) or on anti-polyQ antibodies, such as the monoclonal antibodies MW1–5, 1F8, 3B5H10, and 1C2 that specifically bind to polyQ (37, 55, 56, 57, 58, 59, 60). Despite being overall highly flexible, the C-terminal tail of ataxin-3 contains several stretches of well-defined secondary structure. The three UIMs are helical, in agreement with a previous study of a construct containing UIM1 and 2 (17). In addition to these regions, a helical tract was observed also between residues 278 and 289, which directly precede the polyQ tract. The identification of a stable α-helix in this region was recently suggested by a crystallographic study on a construct containing ataxin-3 residues 278–305 fused to MBP and a short C-terminal crystallization tag (61). Interestingly, two different crystals were obtained in this study. One included the monomeric MBP with the ataxin-3 region 278–305 exposed to the solvent. In this structure, residues 278–284 form two helical turns, whereas the other residues could not be modeled because of absence of electron density. In the second crystal, the protein forms a dimer in which the tract 278–304 is helical, partially shielded from the solvent and stabilized by contacts with MBP. We performed our structural analysis of full-length ataxin-3 in solution, under near-physiological conditions, and without using any tag that could potentially interfere with the structure of the protein. We can thus conclusively affirm that the 278–289 tract forms a helical conformation in solution when in a monomeric state.
We should also put our observations on the polyQ tract (residues 292–305) within the context of other studies. The importance of the regions flanking the polyQ tract has increasingly been stressed (62, 63, 64). A number of studies have shown that the regions immediately N- and C-terminal to the polyQ repeats play an important role in modulating protein self-assembly and aggregate morphology (51, 53, 62, 63, 64, 65, 66). A recent bioinformatics study based on the analysis of the currently available structures has, for instance, revealed that the secondary structure in polyQ vicinity is predominantly random coil or helical and found that the helix is preferentially N-terminal to the polyQ middle position, whereas the random coil is preferentially C-terminal (65). Based on this analysis, the authors suggested that the polyQ function could be linked to a helix/random coil structure context and that even short stretches of repeats can serve this function. We could not obtain sequential assignment and hence estimate the structure of residues 292–297. It is likely that these residues have a shorter T2, as also suggested by the fact that their resonances are below the detection threshold of 5D experiments. This could be caused by several effects, as for instance intramolecular interactions or chemical exchange, but not by an intermediate exchange regime, because the associated H, N, Cα, and Cβ resonances were detectable in the HNCACB experiment, albeit not sequentially assignable because of severe overlap. The minor helical tendency observed within the following polyQ repeats suggests anyway that the whole polyQ has an appreciable tendency to adopt a helical conformation stabilized by the preceding residues. This hypothesis is further supported by the crystal structure of exon I of huntingtin (54), another well-characterized member of the polyQ family, in which the whole region is helical. Another study on an intrinsically unfolded fragment of the androgen receptor also showed that the polyglutamine tract and a stretch of four N-terminally flanking residues (55LLLL58) have a propensity to form a helical structure. The helicity of both the polyQ tract and the LLLL region increases remarkably upon expansion from 4 to 25 glutamines. Deletion of the LLLL stretch led to a decrease in polyQ helicity and increase in disorder. Interestingly, the authors showed that both the helicity and the flexibility of the polyQ tract decrease progressively when moving from the LLLL region toward the C-terminus in a way reminiscent of our observations. In the case of ataxin-3, it could be speculated that the 278–289 tract has a role in propagating its helicity to the polyQ tract in a repeat-dependent fashion.
The evidence discussed here brings new light into the debate whether the polyQ toxicity follows the “Linear Lattice” or the “Structural Toxic Threshold” (or the “Conformation Emergence”) model (37, 57, 59, 67, 68). The first model states that polyQ toxicity is a linear function of the repeat length (57, 69). The latter postulates that polyQ can adopt several different conformations, among which is a toxic one (59, 67, 68). It is clear that polyQ is a chameleon sequence, as it can adopt different conformations according to the context. In 2002, we demonstrated conclusively that, when attached to a noninteracting carrier protein and in a monomeric form, polyQ is in a random coil conformation (38). Solid-state NMR data have instead indicated the presence of β-structures for polyQ aggregates (70, 71). Yet no conclusive evidence shows that this behavior is exclusive for polyQ in the pathological range. We could thus suggest that the two models are not mutually exclusive: the same toxic species may form in short and long polyQ without a clear threshold, but the longer the polyQ tract, the more stable that structure is, thus resulting in higher toxicity.
The inherent flexibility of the C-terminal tail and the lack of interactions with Josephin support the hypothesis that ataxin-3 is a “molecular ruler” able to select polyubiquitin chains with at least four repeats (16). The flexible C-terminus is likely to act as a “tentacle” that hooks the ubiquitin chain and brings it back to the catalytic site for cleavage. The intrinsically unfolded nature of the C-terminal tail may help tune the affinities of the interaction. The helicity of the region preceding the polyQ tract will instead promote interactions with other proteins such as valosin-containing protein, whose binding site is supposed to be located in the region that includes the basic motif RKRR in the C-terminal tail (72).
Conclusions
In conclusion, the data presented in this study allowed us to assess for the first time, to our knowledge, the structural behavior of full-length ataxin-3 in solution. This work paves the way to a better understanding of ataxin-3 normal and pathological functions and will help in future investigations of interactions of ataxin-3 with other proteins. In particular, the ability to obtain detailed structural information of the full-length protein rather than protein fragments will allow us to draw a complete picture of the complex multisite interactions and molecular mechanisms simultaneously involving the Josephin domain and the C-terminal tail. Furthermore, our work may potentially be important for the advancement of our understanding of ataxin-3 aggregation.
Author Contributions
A.S. collected the data and carried out most of the analysis. G.K. and A.O. provided technical support for recording and analyzing the data. J.N. and V.S. provided advice and help in the recording and processing of 5D experiments. A.P. supervised the research. A.P. and A.S. wrote the manuscript.
Acknowledgments
We are indebted to the Medical Research Council NMR facilities at The Francis Crick Institute, London.
The Medical Research Council (U117584256) and the Czech Infrastructure for Integrative Structural Biology research infrastructure (project LM2015043) funded by Ministry of Education, Youth, and Sports Czech Republic are also gratefully acknowledged.
Editor: David Eliezer.
Footnotes
Six figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30668-4.
Supporting Material
References
- 1.Paulson H.L., Perez M.K., Pittman R.N. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 2.Ellisdon A.M., Thomas B., Bottomley S.P. The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J. Biol. Chem. 2006;281:16888–16896. doi: 10.1074/jbc.M601470200. [DOI] [PubMed] [Google Scholar]
- 3.Saunders H.M., Gilis D., Bottomley S.P. Flanking domain stability modulates the aggregation kinetics of a polyglutamine disease protein. Protein Sci. 2011;20:1675–1681. doi: 10.1002/pro.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doss-Pepe E.W., Stenroos E.S., Madura K. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol. Cell. Biol. 2003;23:6469–6483. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scaglione K.M., Zavodszky E., Paulson H.L. Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell. 2011;43:599–612. doi: 10.1016/j.molcel.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu Y., Liu H., Tang T.S. Ataxin-3 promotes genome integrity by stabilizing Chk1. Nucleic Acids Res. 2017;45:4532–4549. doi: 10.1093/nar/gkx095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee A., Saha S., Hazra T.K. The role of the mammalian DNA end-processing enzyme polynucleotide kinase 3′-phosphatase in spinocerebellar ataxia type 3 pathogenesis. PLoS Genet. 2015;11:e1004749. doi: 10.1371/journal.pgen.1004749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jana N.R., Dikshit P., Nukina N. Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 2005;280:11635–11640. doi: 10.1074/jbc.M412042200. [DOI] [PubMed] [Google Scholar]
- 9.Bonanomi M., Mazzucchelli S., Tortora P. Interactions of ataxin-3 with its molecular partners in the protein machinery that sorts protein aggregates to the aggresome. Int. J. Biochem. Cell Biol. 2014;51:58–64. doi: 10.1016/j.biocel.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Ferro A., Carvalho A.L., Maciel P. NEDD8: a new ataxin-3 interactor. Biochim. Biophys. Acta. 2007;1773:1619–1627. doi: 10.1016/j.bbamcr.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Moir D., Stewart S.E., Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer H.H., Wang Y., Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelstein A.V. 50+ years of protein folding. Biochemistry (Mosc.) 2018;83(Suppl 1):S3–S18. doi: 10.1134/S000629791814002X. [DOI] [PubMed] [Google Scholar]
- 14.DeForte S., Uversky V.N. Order, disorder, and everything in between. Molecules. 2016;21::E1090. doi: 10.3390/molecules21081090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masino L., Musi V., Pastore A. Domain architecture of the polyglutamine protein ataxin-3: a globular domain followed by a flexible tail. FEBS Lett. 2003;549:21–25. doi: 10.1016/s0014-5793(03)00748-8. [DOI] [PubMed] [Google Scholar]
- 16.Nicastro G., Masino L., Pastore A. Josephin domain of ataxin-3 contains two distinct ubiquitin-binding sites. Biopolymers. 2009;91:1203–1214. doi: 10.1002/bip.21210. [DOI] [PubMed] [Google Scholar]
- 17.Song A.X., Zhou C.J., Hu H.Y. Structural transformation of the tandem ubiquitin-interacting motifs in ataxin-3 and their cooperative interactions with ubiquitin chains. PLoS One. 2010;5:e13202. doi: 10.1371/journal.pone.0013202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicastro G., Menon R.P., Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc. Natl. Acad. Sci. USA. 2005;102:10493–10498. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong K.I., Yamamoto M., Tanaka T. Selective isotope labeling of recombinant proteins in Escherichia coli. Methods Mol. Biol. 2012;896:439–448. doi: 10.1007/978-1-4614-3704-8_30. [DOI] [PubMed] [Google Scholar]
- 20.Motáčková V., Nováček J., Sklenář V. Strategy for complete NMR assignment of disordered proteins with highly repetitive sequences based on resolution-enhanced 5D experiments. J. Biomol. NMR. 2010;48:169–177. doi: 10.1007/s10858-010-9447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavanagh J., Fairbrother W.J., Skelton N.J. Academic Press; Cambridge, MA: 2006. Protein NMR Spectroscopy. [Google Scholar]
- 22.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Kazimierczuk K., Koźmiński W., Zhukov I. Two-dimensional fourier transform of arbitrarily sampled NMR data sets. J. Magn. Reson. 2006;179:323–328. doi: 10.1016/j.jmr.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Kazimierczuk K., Zawadzka A., Zhukov I. Random sampling of evolution time space and Fourier transform processing. J. Biomol. NMR. 2006;36:157–168. doi: 10.1007/s10858-006-9077-y. [DOI] [PubMed] [Google Scholar]
- 25.Kazimierczuk K., Zawadzka A., Koźmiński W. Narrow peaks and high dimensionalities: exploiting the advantages of random sampling. J. Magn. Reson. 2009;197:219–228. doi: 10.1016/j.jmr.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Vranken W.F., Boucher W., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 27.Helmus J.J., Jaroniec C.P. Nmrglue: an open source Python package for the analysis of multidimensional NMR data. J. Biomol. NMR. 2013;55:355–367. doi: 10.1007/s10858-013-9718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masino L., Nicastro G., Pastore A. The Josephin domain determines the morphological and mechanical properties of ataxin-3 fibrils. Biophys. J. 2011;100:2033–2042. doi: 10.1016/j.bpj.2011.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubey A., Kadumuri R.V., Atreya H.S. Rapid NMR assignments of proteins by using optimized combinatorial selective unlabeling. ChemBioChem. 2016;17:334–340. doi: 10.1002/cbic.201500513. [DOI] [PubMed] [Google Scholar]
- 30.Lescop E., Kern T., Brutscher B. Guidelines for the use of band-selective radiofrequency pulses in hetero-nuclear NMR: example of longitudinal-relaxation-enhanced BEST-type 1H-15N correlation experiments. J. Magn. Reson. 2010;203:190–198. doi: 10.1016/j.jmr.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 31.MacRaild C.A., Zachrdla M., Norton R.S. Conformational dynamics and antigenicity in the disordered malaria antigen merozoite surface protein 2. PLoS One. 2015;10:e0119899. doi: 10.1371/journal.pone.0119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyarko A., Song Y., Barbar E. Multiple recognition motifs in nucleoporin Nup159 provide a stable and rigid Nup159-Dyn2 assembly. J. Biol. Chem. 2013;288:2614–2622. doi: 10.1074/jbc.M112.432831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orbán-Németh Z., Henen M.A., Konrat R. Backbone and partial side chain assignment of the microtubule binding domain of the MAP1B light chain. Biomol. NMR Assign. 2014;8:123–127. doi: 10.1007/s12104-013-9466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solyom Z., Schwarten M., Brutscher B. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR. 2013;55:311–321. doi: 10.1007/s10858-013-9715-0. [DOI] [PubMed] [Google Scholar]
- 35.Marsh J.A., Singh V.K., Forman-Kay J.D. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart D.S., Sykes B.D., Richards F.M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 37.Li P., Huey-Tubman K.E., Bjorkman P.J. The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model. Nat. Struct. Mol. Biol. 2007;14:381–387. doi: 10.1038/nsmb1234. [DOI] [PubMed] [Google Scholar]
- 38.Masino L., Kelly G., Pastore A. Solution structure of polyglutamine tracts in GST-polyglutamine fusion proteins. FEBS Lett. 2002;513:267–272. doi: 10.1016/s0014-5793(02)02335-9. [DOI] [PubMed] [Google Scholar]
- 39.Maciejewski M.W., Liu D., Mullen G.P. Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity. J. Mol. Biol. 2000;296:229–253. doi: 10.1006/jmbi.1999.3455. [DOI] [PubMed] [Google Scholar]
- 40.Burnett B., Li F., Pittman R.N. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- 41.Chai Y., Berke S.S., Paulson H.L. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J. Biol. Chem. 2004;279:3605–3611. doi: 10.1074/jbc.M310939200. [DOI] [PubMed] [Google Scholar]
- 42.Chow M.K., Mackay J.P., Bottomley S.P. Structural and functional analysis of the Josephin domain of the polyglutamine protein ataxin-3. Biochem. Biophys. Res. Commun. 2004;322:387–394. doi: 10.1016/j.bbrc.2004.07.131. [DOI] [PubMed] [Google Scholar]
- 43.Chow M.K., Paulson H.L., Bottomley S.P. Destabilization of a non-pathological variant of ataxin-3 results in fibrillogenesis via a partially folded intermediate: a model for misfolding in polyglutamine disease. J. Mol. Biol. 2004;335:333–341. doi: 10.1016/j.jmb.2003.08.064. [DOI] [PubMed] [Google Scholar]
- 44.Nicastro G., Habeck M., Pastore A. Structure validation of the Josephin domain of ataxin-3: conclusive evidence for an open conformation. J. Biomol. NMR. 2006;36:267–277. doi: 10.1007/s10858-006-9092-z. [DOI] [PubMed] [Google Scholar]
- 45.Bermel W., Felli I.C., Zawadzka-Kazimierczuk A. High-dimensionality 13C direct-detected NMR experiments for the automatic assignment of intrinsically disordered proteins. J. Biomol. NMR. 2013;57:353–361. doi: 10.1007/s10858-013-9793-z. [DOI] [PubMed] [Google Scholar]
- 46.O’Hare B., Benesi A.J., Showalter S.A. Incorporating 1H chemical shift determination into 13C-direct detected spectroscopy of intrinsically disordered proteins in solution. J. Magn. Reson. 2009;200:354–358. doi: 10.1016/j.jmr.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 47.Sahu D., Bastidas M., Showalter S.A. Generating NMR chemical shift assignments of intrinsically disordered proteins using carbon-detected NMR methods. Anal. Biochem. 2014;449:17–25. doi: 10.1016/j.ab.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence C.W., Showalter S.A. Carbon-detected (15)N NMR spin relaxation of an intrinsically disordered protein: FCP1 dynamics unbound and in complex with RAP74. J. Phys. Chem. Lett. 2012;3:1409–1413. doi: 10.1021/jz300432e. [DOI] [PubMed] [Google Scholar]
- 49.Davies H.A., Rigden D.J., Madine J. Probing medin monomer structure and its amyloid nucleation using 13C-direct detection NMR in combination with structural bioinformatics. Sci. Rep. 2017;7:45224. doi: 10.1038/srep45224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicastro G., Masino L., Pastore A. Assignment of the 1H, 13C, and 15N resonances of the Josephin domain of human ataxin-3. J. Biomol. NMR. 2004;30:457–458. doi: 10.1007/s10858-004-4343-3. [DOI] [PubMed] [Google Scholar]
- 51.Eftekharzadeh B., Piai A., Salvatella X. Sequence context influences the structure and aggregation behavior of a PolyQ tract. Biophys. J. 2016;110:2361–2366. doi: 10.1016/j.bpj.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baias M., Smith P.E., Frydman L. Structure and dynamics of the huntingtin exon-1 N-terminus: A solution NMR perspective. J. Am. Chem. Soc. 2017;139:1168–1176. doi: 10.1021/jacs.6b10893. [DOI] [PubMed] [Google Scholar]
- 53.Thakur A.K., Jayaraman M., Wetzel R. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim M.W., Chelliah Y., Bezprozvanny I. Secondary structure of huntingtin amino-terminal region. Structure. 2009;17:1205–1212. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Legleiter J., Lotz G.P., Muchowski P.J. Monoclonal antibodies recognize distinct conformational epitopes formed by polyglutamine in a mutant huntingtin fragment. J. Biol. Chem. 2009;284:21647–21658. doi: 10.1074/jbc.M109.016923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daldin M., Fodale V., Caricasole A. Polyglutamine expansion affects huntingtin conformation in multiple Huntington’s disease models. Sci. Rep. 2017;7:5070. doi: 10.1038/s41598-017-05336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennett M.J., Huey-Tubman K.E., Bjorkman P.J. A linear lattice model for polyglutamine in CAG-expansion diseases. Proc. Natl. Acad. Sci. USA. 2002;99:11634–11639. doi: 10.1073/pnas.182393899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Persichetti F., Trettel F., MacDonald M.E. Mutant huntingtin forms in vivo complexes with distinct context-dependent conformations of the polyglutamine segment. Neurobiol. Dis. 1999;6:364–375. doi: 10.1006/nbdi.1999.0260. [DOI] [PubMed] [Google Scholar]
- 59.Miller J., Arrasate M., Finkbeiner S. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat. Chem. Biol. 2011;7:925–934. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trottier Y., Lutz Y., Tora L. Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature. 1995;378:403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 61.Zhemkov V.A., Kulminskaya A.A., Kim M. The 2.2-Angstrom resolution crystal structure of the carboxy-terminal region of ataxin-3. FEBS Open Bio. 2016;6:168–178. doi: 10.1002/2211-5463.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhattacharyya A., Thakur A.K., Wetzel R. Oligoproline effects on polyglutamine conformation and aggregation. J. Mol. Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 63.Darnell G., Orgel J.P., Meredith S.C. Flanking polyproline sequences inhibit beta-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J. Mol. Biol. 2007;374:688–704. doi: 10.1016/j.jmb.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 64.Shen K., Calamini B., Frydman J. Control of the structural landscape and neuronal proteotoxicity of mutant Huntingtin by domains flanking the polyQ tract. eLife. 2016;5:e18065. doi: 10.7554/eLife.18065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Totzeck F., Andrade-Navarro M.A., Mier P. The protein structure Context of PolyQ Regions. PLoS One. 2017;12:e0170801. doi: 10.1371/journal.pone.0170801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crick S.L., Ruff K.M., Pappu R.V. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA. 2013;110:20075–20080. doi: 10.1073/pnas.1320626110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagai Y., Inui T., Toda T. A toxic monomeric conformer of the polyglutamine protein. Nat. Struct. Mol. Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 68.Peters-Libeu C., Miller J., Finkbeiner S. Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure. J. Mol. Biol. 2012;421:587–600. doi: 10.1016/j.jmb.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klein F.A., Zeder-Lutz G., Trottier Y. Linear and extended: a common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10. Hum. Mol. Genet. 2013;22:4215–4223. doi: 10.1093/hmg/ddt273. [DOI] [PubMed] [Google Scholar]
- 70.Hoop C.L., Lin H.K., van der Wel P.C. Huntingtin exon 1 fibrils feature an interdigitated β-hairpin-based polyglutamine core. Proc. Natl. Acad. Sci. USA. 2016;113:1546–1551. doi: 10.1073/pnas.1521933113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider R., Schumacher M.C., Baldus M. Structural characterization of polyglutamine fibrils by solid-state NMR spectroscopy. J. Mol. Biol. 2011;412:121–136. doi: 10.1016/j.jmb.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 72.Rao M.V., Williams D.R., Loll P.J. Interaction between the AAA+ ATPase p97 and its cofactor ataxin3 in health and disease: Nucleotide-induced conformational changes regulate cofactor binding. J. Biol. Chem. 2017;292:18392–18407. doi: 10.1074/jbc.M117.806281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.