Abstract
Mendelian disorders of cholesterol biosynthesis typically result in multi-system clinical phenotypes, underlining the importance of cholesterol in embryogenesis and development. FDFT1 encodes for an evolutionarily conserved enzyme, squalene synthase (SS, farnesyl-pyrophosphate farnesyl-transferase 1), which catalyzes the first committed step in cholesterol biosynthesis. We report three individuals with profound developmental delay, brain abnormalities, 2-3 syndactyly of the toes, and facial dysmorphisms, resembling Smith-Lemli-Opitz syndrome, the most common cholesterol biogenesis defect. The metabolite profile in plasma and urine suggested that their defect was at the level of squalene synthase. Whole-exome sequencing was used to identify recessive disease-causing variants in FDFT1. Functional characterization of one variant demonstrated a partial splicing defect and altered promoter and/or enhancer activity, reflecting essential mechanisms for regulating cholesterol biosynthesis/uptake in steady state.
Keywords: FDFT1, cholesterol biosynthesis, syndactyly, dysmorphism
Main Text
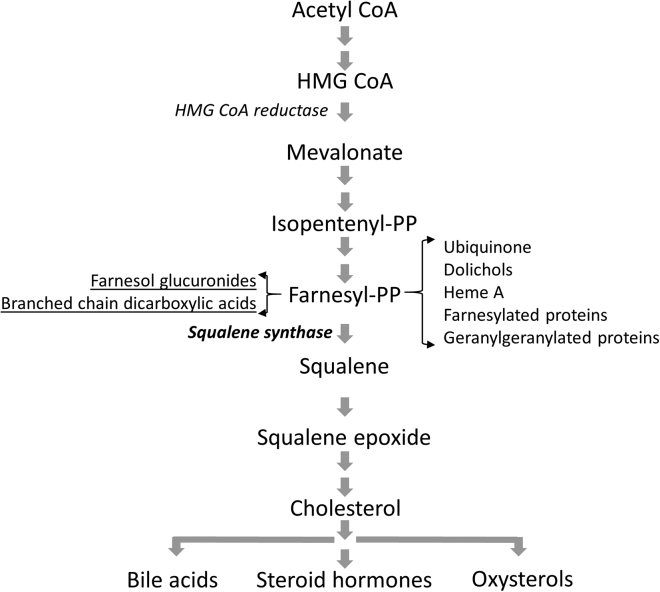
The isoprenoid biosynthesis pathway (Figure 1), essential for numerous biological processes, can be divided into a pre-squalene (also named the mevalonate pathway) and a post-squalene pathway, the latter specifically involved in the synthesis of sterol isoprenoids, including cholesterol. Cholesterol serves as an important structural component of the cell membrane and myelin but is also the precursor of oxysterols, steroid hormones, and bile acids.1 The two-step enzymatic conversion of farnesyl-pyrophosphate (FPP) to squalene by squalene synthase is the first committed step in cholesterol biosynthesis.2, 3
Figure 1.
Schematic Representation of the Isoprenoid/Cholesterol Biosynthesis Pathway
Metabolites that accumulate as a result of squalene synthase deficiency are underlined (pathway re-drawn based on Tansey and Shechter13).
We identified three individuals, a sibship and an unrelated child, whose urine metabolic profiles were suggestive of a cholesterol biosynthesis defect. (For all individuals, informed consent for genetic testing was obtained. For individuals 1 and 2, additional written consent was obtained for photo publication. Ethics approval was provided by Royal Children’s Hospital and Health Service Ethics Committee and Sydney Children’s Hospitals Network Human Research Ethics Committee.) Salient clinical features include facial dysmorphisms (Figure 2), dry skin with photosensitivity, generalized tonic-clonic seizures, structural brain malformations, cortical visual impairment, profound global developmental delay, and genital malformations in the two males (Table 1, see Supplemental Note). The gas chromatography-mass spectrometry and nuclear magnetic resonance spectroscopy profiles yielded a consistent and complex pattern of abnormal metabolites including the accumulation of methylsuccinic acid, mevalonate lactone, mesaconic acid, and 3-methyladipic acid. Additionally, we observed saturated and unsaturated branched-chain dicarboxylic acids and glucuronides derived from farnesol (Figure 3). A similar metabolite profile has previously been observed in the urine of animal models and humans treated with pharmacological inhibitors of squalene synthase, as well as in animals loaded with farnesol.4, 5, 6 To confirm that the abnormal organic acids observed in the affected individuals derive from farnesol, we analyzed urine from a control subject after an oral farnesol load, which resulted in a similar metabolite profile (Figure S1). Plasma cholesterol in the three affected individuals was decreased (2.5–2.8 mmol/L; reference 3.0–5.5), and both HDL and LDL cholesterol levels were decreased or low normal (Table 1). Plasma total farnesol levels (the sum of free farnesol and farnesyl-pyrophosphate) in affected individuals were, however, significantly increased (1.5–3.9 μmol/L; reference < 0.12; Table 1) while plasma squalene levels were reduced or normal (0.17–0.93 μmol/L; reference 0.36–1.04). Thus, the body fluid metabolite profile of the affected individuals was strongly suggestive of a cholesterol biosynthesis defect at the level of squalene synthase, converting farnesyl-pyrophosphate into squalene.
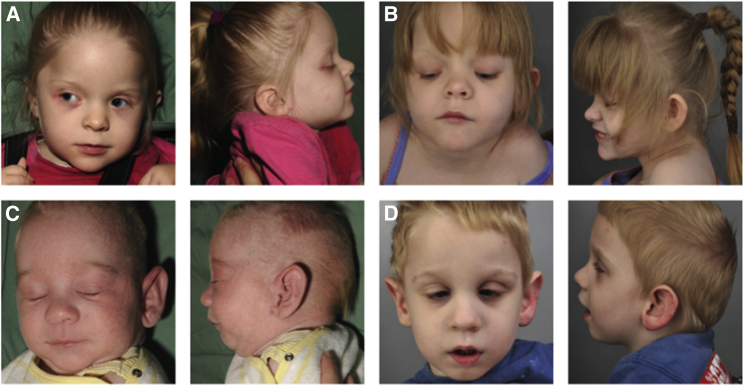
Figure 2.
Facial Characteristics
Individual 1 at 3 years of age (A) and 6 years of age (B) and individual 2 at 5 weeks of age (C) and 3 years of age (D). Dysmorphic features include depressed nasal bridge, low-set posteriorly rotated ears, square nasal tip, epicanthic folds, mild micrognathia, and retrognathia.
Table 1.
Summary of Key Clinical, Biochemical, and Molecular Features
| Individual 1 | Individual 2 | Individual 3 | |
|---|---|---|---|
| Age at presentation | day 5 with seizures | day 5 with seizures | first week of life with seizures |
| Current age | 10 years old | 7 years old | 9 years old |
| Gender | Female | Male | Male |
| Gestation | 37/40 | 39/40 | 39/40 |
| Birth weight | 2,600 g (40th percentile) | 2,750 g (10th percentile) | 3,033 g (25th percentile) |
| Birth length | 47 cm (50th percentile) | 49 cm (30th percentile) | 50 cm (50th percentile) |
| Birth head circumference | 32.5 cm (50th percentile) | 34 cm (50th percentile) | 33 cm (10th percentile) |
| Ethnicity | European | European | European |
| Parental relationship | non-consanguineous | non-consanguineous | non-consanguineous |
| MRI brain abnormalities | hypoplastic corpus callosum, white matter loss | hypoplastic corpus callosum, white matter loss | diffuse polymicrogyria, central white matter and cortical volume loss |
| Seizures | yes, neonatal onset | yes, neonatal onset | yes, neonatal onset |
| Global developmental delay | profound across all developmental modalities | profound across all developmental modalities | profound across all developmental modalities |
| Irritability | yes | yes | yes |
| Optic nerve hypoplasia | yes | yes | no |
| Cataracts | no | no | no |
| Visual impairment | cortical VI | cortical VI | cortical VI |
| Dysmorphism | depressed nasal bridge low set posteriorly rotated ears square nasal tip epicanthic folds mild micrognathia mild retrognathia 2-3 toe syndactyly |
depressed nasal bridge large ears square nasal tip epicanthic folds mild micrognathia mild retrognathia 2-3 toe syndactyly dorsal foot fat pads |
coarse facial features bitemporal narrowing prominent ears triangular facies |
| Genitals | normal | bilateral cryptorchidism | hypospadias |
| Cardiac malformations | no | no | bicuspid aortic valve |
| Gastrointestinal | IUGR with FTT PEG feeds constipation |
IUGR with FTT PEG feeds constipation |
IUGR with FTT PEG feeds constipation |
| Skeletal | thin gracile bones fixed flexion deformity knees |
thin gracile bones fixed flexion deformity knees |
fixed flexion deformities in elbows |
| Skin | dry skin photosensitivity lack of hair pigment on LM/EM |
dry skin photosensitivity lack of hair pigment on LM/EM |
dry skin photosensitivity |
| Sleep cycle | poor sleep initiation | poor sleep initiation | poor sleep initiation and maintenance |
| Cholesterol | 2.5 (3–5.5 mmol/L) | 2.8 (3–5.5 mmol/L) | 2.7 (3–5.5 mmol/L) |
| Triglyceride | 1.1 (0.55–2.0 mmol/L) | 1.6 (0.55–2.0 mmol/L) | 0.8 (0.55–2.0 mmol/L) |
| HDL-C | 0.7 (0.9–2.2 mmol/L) | 1.2 (0.9–2.2 mmol/L) | 1.06 (0.9–2.2 mmol/L) |
| LDL-C | 1.3 (2.0–3.4 mmol/L) | 0.8 (2.0–3.4 mmol/L) | 1.3 (2.0–3.4 mmol/L) |
| VLDL-C | 0.5 (0.1–20.65 mmol/L) | 0.7 (0.1–20.65 mmol/L) | not performed |
| Plasma total farnesol | 1.5, 1.6 (<0.12 μmol/L) | 1.7, 3.9 (<0.12 μmol/L) | not performed |
| Plasma squalene | 0.81, 0.93 (0.36–1.04 μmol/L) | 0.17, 0.37 (0.36–1.04 μmol/L) | not performed |
| Molecular | compound heterozygous mutations: chr8(GRCh37): g. 11667760-11787743, maternal; chr8(GRCh37): g.11689003_11689004delinsAG; NM_001287742.1 (FDFT1): c.880-24_880-23delinsAG, paternal | compound heterozygous mutations: chr8(GRCh37): g. 11667760-11787743, maternal; chr8(GRCh37): g.11689003_11689004delinsAG; NM_001287742.1 (FDFT1): c.880-24_880-23delinsAG, paternal | homozygous mutation: chr8(GRCh37): g.11660095_11660110del; NM_001287742.1: c.-75+131_-75+146del |
Abbreviations: IUGR, intra-uterine growth retardation; PEG, percutaneous endoscopic gastrostomy; FTT, failure to thrive; LM/EM, light microscopy and electroelectron microscopy.
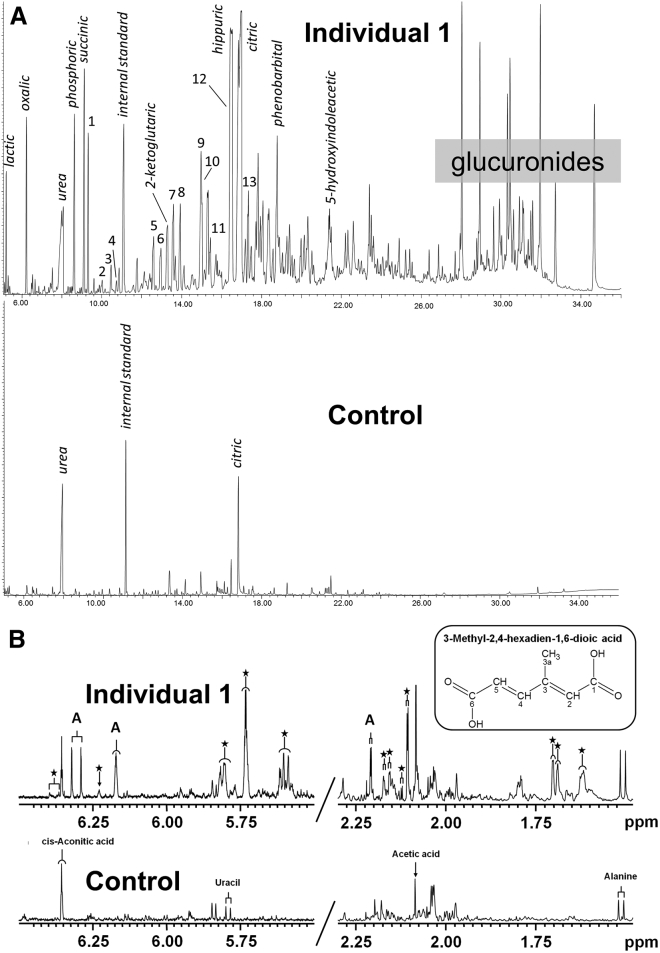
Figure 3.
Urine Metabolite Profiles
(A) Urine organic acid GC-MS profiles of individual 1 (top) and a control subject (bottom). Normal urine components are indicated in italics. Numbered peaks indicate abnormal metabolites: (1) methylsuccinic, (2) mevalonic lactone, (3) mesaconic acid, (4) 2-methylgluatic acid, (5) 3-methyladipic acid, (6) 3-methylhex-3-enedioic acid, (7) 3-methylhex-2-endioic acid, (8) 2,6-dimethylheptanedioic acid, (9) unknown, (10) 3-methylhex-2,4-dienedioic, (11) 2,6-dimethylhept-2-enedioic acid, (12) 3,7-dimethyloctanedioic, and (13) 3,7-dimethyl-2,6-dienedioic. Control values are given in Figure S1.
(B) One-dimensional 500 MHz 1H-NMR spectra of urine of individual 1 and age-matched control subject measured at pH 2.5 (regions 5.60–6.50 ppm and 1.50–2.25 ppm). The insert shows the structure of 3-methylhex-2,4-dienedioic acid. Proton assignments of 3-methylhex-3,4-dienedioic acid (A) and other farnesol-derived dicarboxylic acids (asterisk).
The affected individuals and their unaffected parents were subjected to whole-exome sequencing (WES). Informed consent for genetic testing was obtained for all individuals under protocols approved by the Royal Children’s Hospital and Health Service Ethics Committee and Sydney Children’s Hospitals Network Human Research Ethics Committee. Assuming a recessive mode of inheritance, prioritization of variants revealed compound heterozygous variants affecting FDFT1, encoding the SS enzyme, in the sibship (individuals 1 and 2): a maternally inherited 120 kb deletion (chr8(GRCh37):g.11667760–11787743), resulting in loss of exons 6–10 of FDFT1 and the entire coding sequence of the neighboring CTSB gene (encoding cathepsin B [MIM: 116810], haploinsufficiency for which is asymptomatic in the mother), and a paternally inherited variant GenBank: NM_001287742.1(FDFT1); c.880−24_880−23delinsAG, predicted to create a novel splice acceptor site. The latter prediction was functionally tested using a minigene splice assay, which indeed showed the retention of 22 bp of intron 8 sequence (Figure S2). FDFT1cDNA analysis using RNA isolates generated from fibroblasts of the sibship confirmed the partial splicing defect, showing both normally spliced FDFT1 cDNA and mis-spliced FDFT1 cDNA (Figure S2). Subsequent western blot analysis demonstrated a marked reduction in squalene synthase protein in cultured lymphoblasts and fibroblasts in individuals 1 and 2 (Figures 4B and S2). An even greater reduction was observed when fibroblasts were cultured in lipid-depleted media. While the FDFT1 level is further reduced relative to the control in the LD-FBS (compared to FBS), we hypothesize that the absolute level may not be decreased, but FDFT1 expression in the control is upregulated in LD-FBS. We hypothesize that the residual squalene synthase protein remaining is likely to arise from the correctly spliced paternal allele, which would also explain our observation at cDNA level. In individual 3, no pathogenic bi-allelic variants affecting the coding sequence of FDFT1 could be identified. Interestingly, however, some of the FDFT1 deeper intronic sequence was interpretable from WES data, in allowing the identification of a rare homozygous intronic 16 bp deletion (Chr8(GRCh37):g.11660095_11660110del). The variant was validated using Sanger sequencing in individual 3 and confirmed to be heterozygous in both non-consanguineous parents (Figure S3). Notably, the deletion is absent from our in-house database containing WES data of >15,000 individuals, nor has it been reported in GnomAD (access date October 25, 2017),7 suggesting that this is a private mutation (Figure S3). The residual variation intolerance score (RVIS) of FDFT1 shows that it is among the 6.5% of human genes most intolerant to functional variation.8 We next determined the functional consequence of the 16 bp intronic deletion in individual 3.
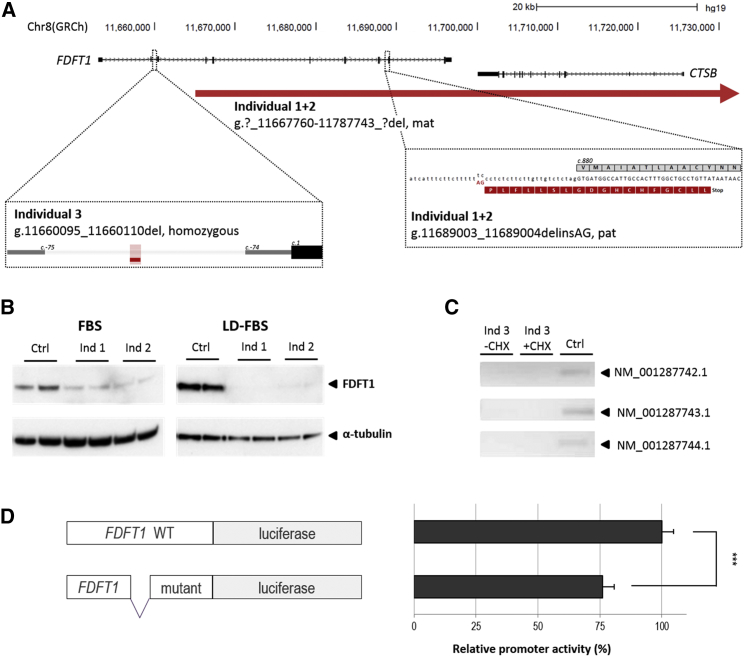
Figure 4.
Schematic Representation of FDFT1, the Variants Identified in Individuals 1, 2, and 3, and Functional Follow-up
(A) The maternal deletion identified in the sibship extends ∼50 kb more proximally, but no other genes are included. Genomic positions are based on hg19. Red horizontal lines indicate deleted sequence. For the paternal mutation in individuals 1 and 2, the wild-type protein amino acid sequence is provided in gray, whereas the predicted sequence of the splice mutation is shown in red.
(B) Western blot analysis of individuals 1 (Ind 1) and 2 (Ind 2), using two different culturing conditions (fetal bovine serum [FBS] and lipid depleted [LD-] FBS), using an antibody to detect FDFT1 in fibroblast of individuals 1 and 2. Alpha-tubulin was used as loading control. FDFT1 reduction is more prominent when cells were cultured under lipid depletion.
(C) Isoform-specific PCR using cDNA generated from EBV-transformed PBMCs of individual 3 (Ind 3) was used for the detection of FDFT1 isoforms. Three FDFT1 isoforms could not be detected in individual 3 whereas these were present in EBV-transformed PBMCs of a control individual. The addition of cycloheximide (CHX; - absent, + present) did not facilitate expression detection. Profile of all isoforms is presented in Figure S5.
(D) The genomic segment containing the homozygous 16 bp deletion in individual 3 was tested for promoter activity using a luciferase assay, which showed significantly reduced activity when compared to the control construct containing the wild-type sequence (two-tailed t test, p = 2.9 × 10−6). ∗∗∗p < 0.001. Error bars represent one SD.
FDFT1 exists in 11 different isoforms, encoding 5 different protein variants (Figure S4). Using fetal and adult organ-specific cDNA libraries, we determined the normal pattern for 10/11 isoforms and detected ubiquitous FDFT1 presence in all major organ tissues tested (Figure S5). Next, we tested cDNA generated from a fibroblast cell line of individual 3, as well as from a control individual, for the presence of these FDFT1 isoforms. Whereas all ten isoforms tested were present in the control fibroblast line, only seven of ten were present in individual 3 (Figure S5), and three isoforms remained undetected (GenBank: NM_001287742.1, NM001287743.1, and NM00128774.4; Figure 4C). The addition of cycloheximide (CHX) failed to result in isoform detection (Figure 4C), suggesting that the absence of these isoforms is a consequence of abnormal regulation rather than erroneous splicing degraded by nonsense-mediated RNA decay. Under normal conditions, the absent FDFT1 isoforms are observed in fetal and adult skeletal muscle, as well as in adult brain, spleen, testis, lung, and kidney (Figure S5). In line with the presence of the seven other FDFT1 isoforms, western blot analysis on fibroblasts from individual 3 identified squalene synthase at protein levels comparable to a normal control individual (Figure S6).
We next explored the possibility that the 16 bp intronic deletion in FDFT1 generates pathogenicity as a consequence of a regulatory disturbance. Annotation of the chromatic state for FDFT1 indicates that the region containing the 16 bp deletion is predicted to have promoter and/or enhancer effects (Figure S7). To assess the effect of the deletion on the putative promoter activity, two constructs were generated and tested in a luciferase assay: one construct containing the 1,024-bp wild-type promoter sequence and one containing the same fragment but with the 16 bp deletion. Analysis of the wild-type sequence showed that indeed the sequence has promoter capacity (Figure 4D, Table S1). Moreover, the normalized luciferase readout of the construct containing the 16-bp deletion shows a significantly reduced promoter activity sequence (two-tailed t test, p = 2.90 × 10−6), suggesting that the aberrant expression of FDFT1 isoforms in individual 3 may indeed be the consequence of diminished promoter activity due to the homozygous 16 bp deletion (Figure 4D, Table S1).
Farnesol and its products exhibit a wide variety of biological activities, including cell growth inhibition, induction of apoptosis, and regulation of bile acid secretion.9 The primitive streak gives rise to the endoderm and mesoderm in embryonic stem cells (ES). Mouse ES primitive streak development is inhibited by statin medication, which is driven by protein farnesylation.10 Farnesol-induced apoptosis is associated with ER stress and activation of the unfolded protein response, inhibition of the phosphatidylcholine system, activation of MAP-kinases, and activation of the apoptosome intrinsic mitochondrial-dependent caspases.9 Evidence is emerging that dysregulation of the mevalonate pathway may be involved in the progression of neurodegeneration in disorders such as Alzheimer disease (MIM: 104300).11
Cholesterol biosynthesis defects are noted for significant birth malformations.1 These are also evidenced in our individuals with FDFT1 disruptive variants (Table 1). Pathogenic mechanisms could involve direct toxicity of accumulated metabolites, abnormal processes that involve farnesyl-pyrophosphate (such as protein farnesylation, Figure 1), or reduced cholesterol moiety attachments to the hedgehog patterning gene family.1 Also, tissue-specific analyses of the various FDFT1 isoforms show that at least one isoform (GenBank: NM_001287743.1) is detected in brain, which is absent in individual 3 (Figure S5), and could potentially explain the severe neurodevelopmental disorder observed. Importantly, Fdft1-null mice demonstrate embryonic lethality at day 12.5 in conjunction with growth restriction and neurodevelopmental disorders.12 The fact that the FDFT1 variants in individuals 1, 2, and 3 are compatible with life may be explained by the fact that all individuals have some form of residual FDFT1 activity, either resulting from the diminished levels of correctly spliced FDFT1 in individuals 1 and 2, or alternatively, by functional compensation for disrupted regulation in individual 3.
In conclusion, we describe squalene synthase deficiency due to pathogenic variants in FDFT1 leading to altered splicing and transcriptional deregulation of the FDFT1 isoforms. The clinical phenotype resembles other known cholesterol biosynthesis defects. The accumulation of farnesyl pyrophosphate in this disorder initiates a complex metabolic cascade involving glucuronidation, hydroxylation, and oxidation to shorter chain molecules. Our cohort exhibits a urine metabolite profile which is an important diagnostic indicator and provides insight into the important role of FDFT1 in embryogenesis and morphogenesis.
Declaration of Interests
The authors declare no competing financial interests.
Acknowledgments
The authors would, first and foremost, like to thank the families, without whose participation this work would not have been possible. This work was supported by the Kevin Milo Trust and internal research funds at the Children’s Hospital at Westmead. The authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about. The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program.
Published: June 14, 2018
Footnotes
Supplemental Data include seven figures, one table, Supplemental Case Reports, and Supplemental Methods and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.05.004.
Contributor Information
David Coman, Email: david.coman@health.qld.gov.au.
James Pitt, Email: james.pitt@vcgs.org.au.
Web Resources
DECIPHER, https://decipher.sanger.ac.uk/
gnomAD Browser, http://gnomad.broadinstitute.org/
MutationTaster, http://www.mutationtaster.org/
NIST Standard Reference Databases: Analytical Chemistry, http://www.nist.gov/srd/analy.cfm
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Primer3, http://bioinfo.ut.ee/primer3
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental Data
References
- 1.Waterham H.R. Defects of cholesterol biosynthesis. FEBS Lett. 2006;580:5442–5449. doi: 10.1016/j.febslet.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Schechter I., Conrad D.G., Hart I., Berger R.C., McKenzie T.L., Bleskan J., Patterson D. Localization of the squalene synthase gene (FDFT1) to human chromosome 8p22-p23.1. Genomics. 1994;20:116–118. doi: 10.1006/geno.1994.1135. [DOI] [PubMed] [Google Scholar]
- 3.Do R., Kiss R.S., Gaudet D., Engert J.C. Squalene synthase: a critical enzyme in the cholesterol biosynthesis pathway. Clin. Genet. 2009;75:19–29. doi: 10.1111/j.1399-0004.2008.01099.x. [DOI] [PubMed] [Google Scholar]
- 4.Jemal M., Ouyang Z. Gas chromatography-mass spectrometric method for quantitative determination in human urine of dicarboxylic (dioic) acids produced in the body as a consequence of cholesterol biosynthesis inhibition. J. Chromatogr. B Biomed. Sci. Appl. 1998;709:233–241. doi: 10.1016/s0378-4347(98)00080-2. [DOI] [PubMed] [Google Scholar]
- 5.Vaidya S., Bostedor R., Kurtz M.M., Bergstrom J.D., Bansal V.S. Massive production of farnesol-derived dicarboxylic acids in mice treated with the squalene synthase inhibitor zaragozic acid A. Arch. Biochem. Biophys. 1998;355:84–92. doi: 10.1006/abbi.1998.0704. [DOI] [PubMed] [Google Scholar]
- 6.Bostedor R.G., Karkas J.D., Arison B.H., Bansal V.S., Vaidya S., Germershausen J.I., Kurtz M.M., Bergstrom J.D. Farnesol-derived dicarboxylic acids in the urine of animals treated with zaragozic acid A or with farnesol. J. Biol. Chem. 1997;272:9197–9203. doi: 10.1074/jbc.272.14.9197. [DOI] [PubMed] [Google Scholar]
- 7.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joo J.H., Jetten A.M. Molecular mechanisms involved in farnesol-induced apoptosis. Cancer Lett. 2010;287:123–135. doi: 10.1016/j.canlet.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto-Uchida Y., Yu R., Miyamura N., Arima N., Ishigami-Yuasa M., Kagechika H., Yoshida S., Hosoya T., Nawa M., Kasama T. The mevalonate pathway regulates primitive streak formation via protein farnesylation. Sci. Rep. 2016;6:37697. doi: 10.1038/srep37697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hottman D.A., Li L. Protein prenylation and synaptic plasticity: implications for Alzheimer’s disease. Mol. Neurobiol. 2014;50:177–185. doi: 10.1007/s12035-013-8627-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tozawa R., Ishibashi S., Osuga J., Yagyu H., Oka T., Chen Z., Ohashi K., Perrey S., Shionoiri F., Yahagi N. Embryonic lethality and defective neural tube closure in mice lacking squalene synthase. J. Biol. Chem. 1999;274:30843–30848. doi: 10.1074/jbc.274.43.30843. [DOI] [PubMed] [Google Scholar]
- 13.Tansey T.R., Shechter I. Structure and regulation of mammalian squalene synthase. Biochim. Biophys. Acta. 2000;1529:49–62. doi: 10.1016/s1388-1981(00)00137-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.