Abstract
Background
Children with acute lymphoblastic leukemia (ALL) often suffer from toxicity of chemotherapeutic drugs such as Methotrexate (MTX). Previously, we reported that 20% of patients receiving high-dose MTX developed oral mucositis. MTX inhibits folate metabolism, which is essential for DNA methylation. We hypothesize that MTX inhibits DNA methylation, which results into adverse effects. We studied DNA methylation markers during high-dose methotrexate treatment in pediatric acute lymphoblastic leukemia (ALL) in relation to developing oral mucositis.
Materials & methods
S-Adenosyl-Methionine (SAM) and S-Adenosyl-Homocysteine (SAH) levels and LINE1 DNA methylation were measured prospectively before and after high-dose methotrexate (HD-MTX 4 x 5g/m2) therapy in 82 children with ALL. Methotrexate-induced oral mucositis was registered prospectively. Oral mucositis (grade ≥ 3 National Cancer Institute Criteria) was used as clinical endpoint.
Results
SAM levels decreased significantly during methotrexate therapy (-16.1 nmol/L (-144.0 –+46.0), p<0.001), while SAH levels and the SAM:SAH ratio did not change significantly. LINE1 DNA methylation (+1.4% (-1.1 –+6.5), p<0.001) increased during therapy. SAM and SAH levels were not correlated to LINE1 DNA methylation status. No association was found between DNA methylation markers and developing oral mucositis.
Conclusions
This was the first study that assessed DNA methylation in relation to MTX-induced oral mucositis in children with ALL. Although global methylation markers did change during methotrexate therapy, methylation status was not associated with developing oral mucositis.
Introduction
Treatment outcome of pediatric acute lymphoblastic leukemia (ALL) has improved substantially over the past decades, with 5-year survival rates currently reaching 90% in developed countries [1–3]. We previously showed that 20% of children with ALL receiving high-dose MTX developed oral mucositis as an adverse effect despite folate rescue therapy [4]. It would be of clinical value to identify predictors of MTX-induced oral mucositis to select patients who could benefit from personalized intervention strategies [2].
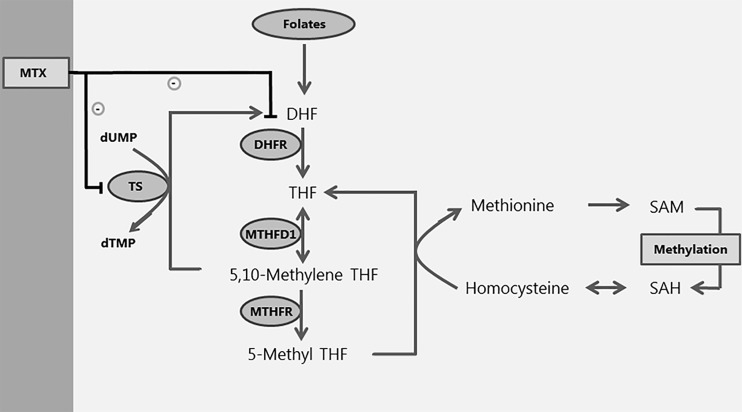
MTX is an inhibitor of methionine-adenosine transferase (MAT), resulting in lower levels of methionine with concomitant lower S-adenosyl-methionine (SAM) and is therefore expected to decrease DNA methylation [5, 6]. In a mouse neural tube defect model, MTX caused DNA hypomethylation [7]. In contrast, low-dose methotrexate caused global DNA hypermethylation in rheumatoid arthritis patients [8, 9]. DNA methylation is the process in which methyl-groups (-CH3) bind to Cytosine-phosphate-Guanine dinucleotides (CpG) in the DNA, by which it plays a role in ‘gene-silencing’ [10]. Methyl-groups are obtained from one-carbon metabolism, during which the methyl-group from SAM is donated to DNA, RNA and proteins, after which S-adenosyl-homocysteine (SAH) is formed (Fig 1). Plasma SAM and SAH metabolite levels and the SAM:SAH ratio reflect the global intracellular methylation status of the cell. Disturbances in SAM—SAH levels in combination with a decreased SAM:SAH ratio are associated with DNA hypomethylation [11]. Global DNA methylation status can be quantified by several methods, amongst which measuring DNA methylation status of Long Interspersed Nuclear 1 elements (LINE1). LINE1 elements occur frequently (~20.000 copies) in the human genome and DNA methylation status of these elements is therefore considered to be a proxy for global DNA methylation [12].
Fig 1. Role of MTX in relation to one-carbon metabolism.
SAM: S-adenosylmethionine; SAH: S-adenosylhomocysteine; DHF: dihydrofolate; THF: tetrahydrofolate; TS: thymidylate synthase; DHFR: dihydrofolate reductase; MTHFD1: methylenetetrahydrofolate dehydrogenase 1; MTHFR: methylenetetrahydrofolate reductase; MTX: methotrexate. Folic acid donates a methyl-group to the one-carbon metabolism pathway. Through several steps methionine is transformed into SAM, which then donates the methyl-group for the DNA methylation process, and is transformed into SAH and homocysteine. MTX inhibits DHFR and TS. By inhibiting DHFR, MTX inhibits the pathway leading to methylation.
Currently, no studies on changes in DNA methylation in relation to the development of chemotherapy-related oral mucositis exist. However, DNA methylation has been implicated as a possible biomarker of treatment-related toxicity in other malignancies and rheumathoid arthritis treatment [13–16].
In the current prospective study we explored the hypothesis, that high-dose MTX therapy inhibits global DNA methylation, which is associated with the development of MTX-induced oral mucositis in children with ALL.
Materials & methods
Patients, treatment protocol and toxicity evaluation
The patient cohort and treatment protocol have been previously reported [4]. Briefly, patients between 1 and 18 years treated according to the standard and medium risk arms of the Dutch Childhood Oncology ALL-10 protocol (2004–2012) were eligible for the current study [17]. The study was approved by the Medical Ethical Committee (MEC-2005-358). Written informed consent was obtained before data- and sample collection. An overview of protocol M (HD-MTX phase; 5 gram/m2/course) is shown in S1 Fig. A modified version of The National Cancer Institute (NCI) Common Terminology Criteria for Adverse Events v.3.0 score system was used to score and document toxicity (S1 Table) [18]. Clinically relevant oral mucositis, defined as NCI grade ≥ 3, was used as endpoint in the analyses [4]. The highest grade of toxicity observed in each patient during protocol M was documented.
Sample collection
Peripheral EDTA blood samples were collected before the first HD-MTX course (T0) as well as two weeks after discontinuation of protocol M (T1) and were stored at -80°C (S1 Fig) [17]. DNA was isolated from whole blood using the MagNA Pure Compact Nucleid Acid isolation kit (Roche Molecular Biochemicals®) according to the manufacturer's instructions.
Cellular- and global DNA-methylation status
We measured plasma SAM and SAH levels as a proxy for cellular methylation status and LINE1 DNA methylation as a proxy for global DNA methylation status at T0 and T1.
SAM and SAH plasma levels were measured using a liquid chromatography tandem-mass spectrometry (LC-MS/MS) method using solid-phase extraction columns as previously described [19]. The SAM:SAH ratio was calculated. The LINE1 global DNA methylation assay was performed using primers as previously reported [20]. The LINE1 assay measured the methylation percentage at 12 CpG sites. Primers were designed using EpiTYPER Designer software (http://www.epidesigner.com/). We used a primer melting temperature of 64 °C, a primer size of 25 bp and an amplicon length of 300 bp. Primer sequences are depicted in S2 Table.
For global LINE1 DNA methylation assay, isolated DNA (500 ng) was treated with sodium bisulphite to discriminate between methylated and unmethylated cytosines using the EZ DNA Methylation™ Kit (Zymo Research®) according to the manufacturer's instructions. Bisulphite-treated DNA was stored at +4°C and processed within 1 week according to the manufacturer’s instructions. The assay was performed in triplets per patient at T0 and T1 and mean DNA methylation values were calculated from these triplets when the variation coefficient was <10%. A PCR to amplify bisulphite-treated DNA was performed using the C-1000 Touch Thermal Cycler™ (Bio-Rad). Two μl of sodium bisulphite-treated DNA was added to each reaction (total volume reaction: 12 μl). The PCR master mix consisted of 1.2 μl 10x Buffer, 1.2 μl 2mM deoxyribonucleotide triphosphates (dNTPs), 0.7 μl 25mM MgCl2, 2 μl of the forward and reverse primer (1 pmol/μl), 0.1 μl AmpliTaq (5 U/L, Applied Biosystems, Waltham, MA, USA) and 2.8 μl H2O. A standard ‘step-down’ PCR thermal cycling protocol was performed: 10 minutes at 95°C; 5 cycles of 20 seconds (s) at 95°C, 30 s at 65°C and 1 minute at 72°C; 5 cycles of 20 s at 95°C, 30 s at 58°C and 1 minute at 72°C; 39 cycles of 20 s at 95°C, 30 s at 53°C, 1 minute at 72°C and a final elongation step for 3 min at 72°C followed by infinite hold at 12°C. After the PCR, a dilution of 10 μl H2O, 3 μl PCR product and 2 μl loading dye was loaded onto a 2% agarose gel to verify whether the PCR was successful. Unincorporated PCR primers and deoxynucleotide triphosphates in the samples were inactivated by using shrimp alkaline phosphatase (SAP) treatment as previously described [21]. Reverse transcription/RNase T cleavage was performed using the following conditions: 3.21 μl of RNase-free double-distilled H2O (ddH2O), 0.89 μl of 5x T7 polymerase buffer, 0.22 μl T Cleavage mix, 2.2 mM DTT, 20 U T7 DNA & RNA polymerase and 0.6 μg RNase A in 2 μl of purified DNA product. This mixture was incubated at 37°C for 3 hours. Thereafter, 6 mg of Clean Resin and 20 μl of milliQ H2O were added to each sample, rotated slowly for 20 minutes to mix reagents and then centrifuged down for 5 minutes at 3000rpm. Thereafter, 10–15 nL of the cleavage reaction was dispensed onto a SpectroCHIP array with a Nanodispenser RS1000 (Sequenom). The array was performed on a Matrix-assisted Laser Desorption/Ionization—Time Of Flight (MALDI-TOF) MassARRAY (Sequenom) analyzer according to the manufacturer’s instructions. LINE1 CpG4 could not be measured due to a silent signal. LINE1 CpG10 could not be analyzed due to a low mass fragment. LINE1 CpG6 and 7, CpG8 and 9 and CpG11 and 12 were each analyzed together as the two CpG sites were present in one fragment. A precision experiment was performed consisting of measuring 10 DNA controls, which should give DNA methylation percentages with a variation coefficient of <10%.
Statistical analysis
Statistical analyses were performed using SPSS Statistics Version 20.0.0.1 (SPSS, Chicago, IL, USA). For the LINE1 global DNA methylation assay the methylation percentage of individual CpG sites and the mean methylation percentage of all CpG sites measured in the assays was used for statistical analysis. Changes in SAM—SAH levels and the global LINE1 DNA methylation status between T0 and T1 were tested using a paired T test (mean ± standard deviation) or a Wilcoxon Rank Sum test (median, range), as appropriate, based on the normal distribution of data. The association between SAM—SAH levels and global LINE1 DNA methylation status at T0 and the change between T0 and T1 (delta T1 –T0) with the development of MTX-induced oral mucositis was tested using an independent T test (mean ± standard deviation) or a Mann Whitney U test (median, range) as appropriate. The correlation between SAM—SAH levels and LINE1 DNA methylation was tested using a Spearman’s rho coefficient. A correlation coefficient of >0.7 was considered relevant. In view of multiple comparisons the significance level was set at a p-value of 0.004 using a Bonferroni correction (p-value = 0.05 / 12). We tested the possible confounding effect of clinical characteristics (age at diagnosis, gender, ALL immunophenotype, ALL risk group) by testing whether these factors were significantly (p <0.05) related to both the determinant (DNA methylation) and the outcome (mucositis). If confounders were significant in both these univariate analyses, they were included in a multivariate regression model.
Results
Plasma SAM—SAH levels and LINE1 DNA methylation were measured before start of MTX therapy (T0) and two weeks after end of MTX therapy (T1) in 82 pediatric ALL patients (Table 1). LINE1 methylation percentages were measured at all CpG sites with a variation coefficient of <10%. In total, 17 patients (21%) patients developed MTX-induced oral mucositis ≥ NCI grade 3. Baseline characteristics are summarized in Table 1.
Table 1. Baseline characteristics (n = 82).
| Patient characteristics | |
|---|---|
| Age at diagnosis | |
| median (range in years) | 5.4 (1–18) |
| Sex, n (%) | |
| Female | 46 (56) |
| Male | 36 (44) |
| Immunophenotype ALL, n (%) | |
| B-lineage | 71 (87) |
| T-lineage | 11 (13) |
| Risk group ALL-10 protocol, n(%) | |
| Standard risk | 23 (28) |
| Medium risk | 59 (72) |
| Mucositis, n (%)* | |
| No | 65 (79) |
| Yes | 17 (21) |
*Clinically relevant mucositis is defined as ≥ grade 3 according to the National Cancer Criteria v.3.0. [18].
Methylation markers—Changes during MTX therapy
Methylation marker levels at T0 and T1 are described in Table 2. Plasma SAM levels were significantly (mean -16.1 nmol/L [-144.0 –+46.0]) lower after MTX therapy (p-value < 0.001), whereas SAH levels and SAM:SAH ratio did not change significantly (Table 2). LINE1 DNA methylation increased (mean +1.4% [-1.1 –+6.5]) during MTX therapy (p-value <0.001, Table 2 + S3 Table). SAM—SAH levels and LINE1 DNA methylation status at T0 and T1 were not correlated (S4 Table).
Table 2. Methylation before and after stop of MTX therapy.
| T0 (before start MTX) | T1 (after stop MTX) | p-value | ||
|---|---|---|---|---|
| Cellular methylation | ||||
| SAM (nmol/L), median (range) | (n = 77) | 109.0 (71.0–245.0) | 99.0 (44.0–151.0) | <0.001* |
| SAH (nmol/L), median (range) | (n = 77) | 13.5 (8.1–78.2) | 12.9 (6.4–56.2) | 0.234 |
| SAM:SAH ratio, mean ± SD | (n = 77) | 8.0 ± 2.8 | 7.4 ± 3.1 | 0.207 |
| Global DNA methylation | ||||
| LINE1 –methylation (%), mean ± SD | (n = 80) | 65.1 ± 1.8 | 66.5 ± 1.9 | <0.001* |
T0: before start MTX; T1: after stop MTX. Mean percentage methylation of CpG sites in LINE1 (%) and plasma SAM—SAH levels (nmol/L) at T0 vs. T1; mean ± SD or median (range) based on normal distribution of data.
Methylation status in relation to MTX-induced oral mucositis
SAM and SAH levels and the SAM:SAH ratio at T0 were not associated with the occurrence of MTX-induced oral mucositis (Table 3). LINE1 DNA methylation at T0 was not significantly associated with the development of MTX-induced oral mucositis (Table 3 + S5 Table). In addition, changes in the methylation markers between T0 and T1 were not significantly associated with the development of MTX-induced oral mucositis (Table 3 + S6 Table). None of the tested clinical confounders (age at diagnosis, gender, ALL immunophenotype, ALL risk group) significantly affected these analyses.
Table 3. SAM and SAH levels and LINE1 DNA methylation in relation to MTX-induced oral mucositis.
| T0 | p-value | Change T0-T1 | p-value | ||||
|---|---|---|---|---|---|---|---|
| Cellular methylation | |||||||
| SAM (nmol/L), median (range) | SAM (nmol/L), median (range) | ||||||
| No Mucositis | n = 62 (78) | 109.5 (72.0–245.0) | 0.788 | No Mucositis | n = 62 (81) | -12.5 (-144.0 –+46.0) | 0.338 |
| Mucositis | n = 17 (22) | 107.0 (71.0–151.0) | Mucositis | n = 15 (19) | -9.0 (-46.0 –+31.0) | ||
| SAH (nmol/L), median (range) | SAH (nmol/L), median (range) | ||||||
| No Mucositis | n = 62 (78) | 13.7 (8.1–58.8) | 0.407 | No Mucositis | n = 62 (81) | -1.0 (-33.8 –+46.0) | 0.979 |
| Mucositis | n = 17 (22) | 11.4 (6.3–78.2) | Mucositis | n = 15 (19) | 0.4 (-53.6 –+11.1) | ||
| SAM:SAH ratio, mean ± SD | SAM:SAH ratio, mean ± SD | ||||||
| No Mucositis | n = 62 (78) | 7.9 ± 2.8 | 0.405 | No Mucositis | n = 62 (81) | -0.7 ± 3.6 | 0.501 |
| Mucositis | n = 17 (22) | 8.6 ± 3.1 | Mucositis | n = 15 (19) | 0.0 ± 3.3 | ||
| Global DNA methylation | |||||||
| LINE1 –total methylation (%), mean ± SD | LINE1 –total methylation (%), mean ± SD | ||||||
| No Mucositis | n = 65 (79) | 65.0 (± 1.9) | 0.339 | No Mucositis | n = 63 (79) | 1.5 ± 1.4 | 0.290 |
| Mucositis | n = 17 (21) | 65.5 (± 1.3) | Mucositis | n = 17 (21) | 1.1 ± 1.2 | ||
Mean percentage methylation of LINE1 and plasma SAM—SAH levels in nmol/L before start of MTX (T0) and the change (T0 –T1) during MTX therapy in relation to MTX-induced oral mucositis; mean ± SD or median based on normal distribution of data.
Discussion
This is the first study on the role of cellular methylation status and global DNA methylation in relation to the development of MTX-induced oral mucositis in children with ALL. Although we showed that global DNA methylation markers were changed after MTX therapy, plasma SAM—SAH levels and LINE1 DNA methylation were not associated with developing oral mucositis due to high-dose MTX treatment.
We observed a decrease in SAM levels after high-dose MTX treatment in pediatric ALL patients, while the SAH levels and the SAM:SAH ratio did not show a significant change. This very likely means that the change in SAM was not large enough to affect the SAM:SAH ratio significantly. Inhibition of SAM levels has been shown previously in an in vitro methotrexate model and a methotrexate-induced neural tube defect mouse model [7, 22]. Our study in the pediatric ALL setting using a high-dose MTX regimen confirmed these results. However, this decrease in SAM seen at the end of MTX therapy can also be caused by other factors, such as environmental factors. More studies are necessary to assess the role of MTX in relation to plasma SAM.
We observed small increases of 1–2% in LINE1 DNA methylation status after high-dose MTX treatment, which are are in line with previous reports in other diseases [9, 23]. For example, in patients using selective serotonin reuptake inhibitors, global and gene-specific methylation differences of 1–5% were found to be associated with treatment response [23]. Furthermore, 5-methylcytosine levels, which is another global DNA methylation measure, differed between healthy controls and rheumathoid arthritis patients receiving MTX with 1–2% [9]. The observed increase in DNA methylation status after HD-MTX therapy should be replicated in an independent case-control setting or validated using another global DNA methylation assay than the LINE1 assay to assess whether the observed changes are due to HD-MTX and not due to other environmental factors, such as nutrition. The LINE1 hypermethylation we found was contradictory to what we hypothesized, as MTX is expected to inhibit DNA methylation. In line with our results, previous studies in rheumathoid arthritis patients showed that MTX treatment induced DNA hypermethylation [8, 9].
A possible explanation for the observed DNA hypermethylation after MTX therapy in our study is that concomitant therapy is administered, such as folate rescue therapy and 6-Mercaptopurine. Folates provide methyl-groups necessary for methylation reactions and increased DNA methylation status in several mouse tissues (liver, kidney, brain) as well as in peripheral mononuclear cells [24, 25]. In contrast, 6-Mercaptopurine causes DNA hypomethylation [26]. It is possible that the inhibitory effect of MTX and 6-Mercaptopurine on DNA methylation status is masked by folate rescue therapy, as folate increases DNA methylation status.
Our study showed no association between global methylation markers at start and at the end of MTX therapy in relation to MTX-induced oral mucositis. These results do not confirm the hypothesis that global methylation is associated with MTX-induced mucositis. However, a recent study showed that the hypermethylation in CpG1 and CpG2 of the promotor of the ɣ-Glutamyl Hydrolase (GGH) gene, which is involved in polyglutamating MTX, can significantly reduce GGH mRNA expression in pediatric ALL [27]. Therefore, performing genome-wide DNA methylation analyses using an Illumina methylation EPIC array could be relevant in future studies to assess whether gene-specific DNA methylation status could be used as a possible biomarker in predicting MTX-induced toxicity, such as oral mucositis.
Finally, in our study DNA methylation status was measured in DNA isolated from whole blood leucocytes. At this point in therapy, patients are considered to be in complete remission, and therefore the DNA methylation profile of leukemic blasts, which are known to be different from normal leucocytes, should not interfere with our analysis. DNA methylation status differs per tissue [28, 29]. In future studies, it would be interesting to look into DNA methylation changes in the oral mucosa in relation to MTX-induced oral mucositis.
Strengths of our study are the prospective collection of toxicity data and the fact that all patients were treated according to the same standardized treatment protocol. A limitation is the relatively small sample size of this study.
Conclusion
This study is the first report to study methylation status in relation to the development of MTX-induced oral mucositis in children with ALL. Global methylation markers changed after high-dose MTX therapy, but we could not demonstrate that global methylation markers are associated with the development of MTX-induced oral mucositis in pediatric ALL.
Supporting information
Protocol M consists of a 57-day period in which patients receive 6-Mercaptopurine (6-MP) orally in a dose of 25 mg/m2/day. Patients receive 2-weekly high-dose methotrexate (HD-MTX) intravenously in a dose of 5000 mg/m2/dose in 24 hours. Intrathecal infusions of methotrexate, cytarabine (ARA-C) and di-adreson F (DAF) are administered 2-weekly. Leucovorin is administered at 36 hours, 42 hours and 48 hours after start of the HD-MTX infusion at a dose of 15 mg/m2/dose. Peripheral EDTA blood samples were collected at day 1 of protocol M (T0)) as well as two weeks after discontinuation of protocol M (T1).
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Abbreviations
- ALL
Acute Lymphoblastic Leukemia
- CpG
Cytosine-phosphate-Guanine dinucleotide
- LINE1
Long Interspersed Nuclear Elements 1
- MAT
Methionine Adenosine Transferase
- MTX
Methotrexate
- NCI
National Cancer Institute
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- SAP
Shrimp Alkaline Phosphatase
Data Availability
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.t3g1vc3.
Funding Statement
This project was supported by Stichting Kinderen Kankervrij, Amstelveen, The Netherlands, grant number 197. This grant was received by SGH and MMvdHE. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children's oncology group. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30(14):1663–9. Epub 2012/03/14. doi: 10.1200/jco.2011.37.8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pui CH, Mullighan CG, Evans WE, Relling MV. Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood. 2012;120(6):1165–74. Epub 2012/06/26. doi: 10.1182/blood-2012-05-378943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamps WA, van der Pal-de Bruin KM, Veerman AJ, Fiocco M, Bierings M, Pieters R. Long-term results of Dutch Childhood Oncology Group studies for children with acute lymphoblastic leukemia from 1984 to 2004. Leukemia. 2010;24(2):309–19. Epub 2009/12/18. doi: 10.1038/leu.2009.258 . [DOI] [PubMed] [Google Scholar]
- 4.den Hoed MA, Lopez-Lopez E, te Winkel ML, Tissing W, de Rooij JD, Gutierrez-Camino A, et al. Genetic and metabolic determinants of methotrexate-induced mucositis in pediatric acute lymphoblastic leukemia. The pharmacogenomics journal. 2015;15(3):248–54. Epub 2014/10/29. doi: 10.1038/tpj.2014.63 . [DOI] [PubMed] [Google Scholar]
- 5.Wang YC, Chiang EP. Low-dose methotrexate inhibits methionine S-adenosyltransferase in vitro and in vivo. Molecular medicine (Cambridge, Mass). 2012;18:423–32. Epub 2011/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broxson EH Jr., Stork LC, Allen RH, Stabler SP, Kolhouse JF. Changes in plasma methionine and total homocysteine levels in patients receiving methotrexate infusions. Cancer research. 1989;49(21):5879–83. Epub 1989/11/01. . [PubMed] [Google Scholar]
- 7.Wang X, Guan Z, Chen Y, Dong Y, Niu Y, Wang J, et al. Genomic DNA hypomethylation is associated with neural tube defects induced by methotrexate inhibition of folate metabolism. PloS one. 2015;10(3):e0121869 Epub 2015/03/31. doi: 10.1371/journal.pone.0121869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YI, Logan JW, Mason JB, Roubenoff R. DNA hypomethylation in inflammatory arthritis: reversal with methotrexate. The Journal of laboratory and clinical medicine. 1996;128(2):165–72. Epub 1996/08/01. . [DOI] [PubMed] [Google Scholar]
- 9.de Andres MC, Perez-Pampin E, Calaza M, Santaclara FJ, Ortea I, Gomez-Reino JJ, et al. Assessment of global DNA methylation in peripheral blood cell subpopulations of early rheumatoid arthritis before and after methotrexate. Arthritis research & therapy. 2015;17:233 Epub 2015/09/04. doi: 10.1186/s13075-015-0748-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13(7):484–92. Epub 2012/05/30. doi: 10.1038/nrg3230 . [DOI] [PubMed] [Google Scholar]
- 11.Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, et al. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. The Journal of nutrition. 2001;131(11):2811–8. Epub 2001/11/06. doi: 10.1093/jn/131.11.2811 . [DOI] [PubMed] [Google Scholar]
- 12.Lisanti S, Omar WA, Tomaszewski B, De Prins S, Jacobs G, Koppen G, et al. Comparison of methods for quantification of global DNA methylation in human cells and tissues. PloS one. 2013;8(11):e79044 Epub 2013/11/22. doi: 10.1371/journal.pone.0079044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Ede AE, Laan RF, Blom HJ, De Abreu RA, van de Putte LB. Methotrexate in rheumatoid arthritis: an update with focus on mechanisms involved in toxicity. Seminars in arthritis and rheumatism. 1998;27(5):277–92. Epub 1998/05/08. . [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Orozco LD, Wang J, Rau CD, Rubbi L, Ren S, et al. DNA Methylation Indicates Susceptibility to Isoproterenol-Induced Cardiac Pathology and Is Associated With Chromatin States. Circulation research. 2016;118(5):786–97. Epub 2016/02/04. doi: 10.1161/CIRCRESAHA.115.305298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flanagan JM, Wilhelm-Benartzi CS, Metcalf M, Kaye SB, Brown R. Association of somatic DNA methylation variability with progression-free survival and toxicity in ovarian cancer patients. Annals of oncology: official journal of the European Society for Medical Oncology. 2013;24(11):2813–8. Epub 2013/10/12. doi: 10.1093/annonc/mdt370 . [DOI] [PubMed] [Google Scholar]
- 16.Dexheimer GM, Alves J, Reckziegel L, Lazzaretti G, Abujamra AL. DNA Methylation Events as Markers for Diagnosis and Management of Acute Myeloid Leukemia and Myelodysplastic Syndrome. Disease markers. 2017;2017:5472893 Epub 2017/10/19. doi: 10.1155/2017/5472893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pieters R, de Groot-Kruseman H, Van der Velden V, Fiocco M, van den Berg H, de Bont E, et al. Successful Therapy Reduction and Intensification for Childhood Acute Lymphoblastic Leukemia Based on Minimal Residual Disease Monitoring: Study ALL10 From the Dutch Childhood Oncology Group. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2016;34(22):2591–601. Epub 2016/06/09. doi: 10.1200/jco.2015.64.6364 . [DOI] [PubMed] [Google Scholar]
- 18.Trotti A, Colevas AD, Setser A, Rusch V, Jaques D, Budach V, et al. CTCAE v3.0: development of a comprehensive grading system for the adverse effects of cancer treatment. Seminars in radiation oncology. 2003;13(3):176–81. Epub 2003/08/07. doi: 10.1016/S1053-4296(03)00031-6 . [DOI] [PubMed] [Google Scholar]
- 19.Gellekink H, van Oppenraaij-Emmerzaal D, van Rooij A, Struys EA, den Heijer M, Blom HJ. Stable-isotope dilution liquid chromatography-electrospray injection tandem mass spectrometry method for fast, selective measurement of S-adenosylmethionine and S-adenosylhomocysteine in plasma. Clinical chemistry. 2005;51(8):1487–92. Epub 2005/05/28. doi: 10.1373/clinchem.2004.046995 . [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Wang F, Guan J, Le J, Wu L, Zou J, et al. Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. The American journal of clinical nutrition. 2010;91(5):1359–67. Epub 2010/02/19. doi: 10.3945/ajcn.2009.28858 . [DOI] [PubMed] [Google Scholar]
- 21.Ehrich M, Zoll S, Sur S, van den Boom D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic acids research. 2007;35(5):e29 Epub 2007/01/30. doi: 10.1093/nar/gkl1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nesher G, Moore TL, Dorner RW. In vitro effects of methotrexate on peripheral blood monocytes: modulation by folinic acid and S-adenosylmethionine. Annals of the rheumatic diseases. 1991;50(9):637–41. Epub 1991/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viuff AC, Pedersen LH, Kyng K, Staunstrup NH, Borglum A, Henriksen TB. Antidepressant medication during pregnancy and epigenetic changes in umbilical cord blood: a systematic review. Clinical epigenetics. 2016;8(1):94 Epub 2016/09/10. doi: 10.1186/s13148-016-0262-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Molecular and cellular biology. 2003;23(15):5293–300. Epub 2003/07/16. doi: 10.1128/MCB.23.15.5293-5300.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingrosso D, Cimmino A, Perna AF, Masella L, De Santo NG, De Bonis ML, et al. Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinaemia in patients with uraemia. Lancet (London, England). 2003;361(9370):1693–9. Epub 2003/05/28. doi: 10.1016/s0140-6736(03)13372-7 . [DOI] [PubMed] [Google Scholar]
- 26.Hogarth LA, Redfern CP, Teodoridis JM, Hall AG, Anderson H, Case MC, et al. The effect of thiopurine drugs on DNA methylation in relation to TPMT expression. Biochemical pharmacology. 2008;76(8):1024–35. Epub 2008/08/19. doi: 10.1016/j.bcp.2008.07.026 . [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Liu S, Wang H, Mai H, Yuan X, Li C, et al. Methylation level of CpG islands in GGH gene promoter in pediatric acute leukemia. PloS one. 2017;12(3):e0173472 Epub 2017/03/10. doi: 10.1371/journal.pone.0173472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Bustos C, Ramos E, Young JM, Tran RK, Menzel U, Langford CF, et al. Tissue-specific variation in DNA methylation levels along human chromosome 1. Epigenetics & chromatin. 2009;2(1):7 Epub 2009/06/10. doi: 10.1186/1756-8935-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Day K, Waite LL, Thalacker-Mercer A, West A, Bamman MM, Brooks JD, et al. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome biology. 2013;14(9):R102 Epub 2013/09/17. doi: 10.1186/gb-2013-14-9-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protocol M consists of a 57-day period in which patients receive 6-Mercaptopurine (6-MP) orally in a dose of 25 mg/m2/day. Patients receive 2-weekly high-dose methotrexate (HD-MTX) intravenously in a dose of 5000 mg/m2/dose in 24 hours. Intrathecal infusions of methotrexate, cytarabine (ARA-C) and di-adreson F (DAF) are administered 2-weekly. Leucovorin is administered at 36 hours, 42 hours and 48 hours after start of the HD-MTX infusion at a dose of 15 mg/m2/dose. Peripheral EDTA blood samples were collected at day 1 of protocol M (T0)) as well as two weeks after discontinuation of protocol M (T1).
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.t3g1vc3.