Abstract
The application of non-targeted metabolomic profiling has recently become a powerful non-invasive tool to discover new clinical biomarkers. This study aimed to identify metabolic pathways that could be exploited for prognostic and therapeutic purposes in hepatorenal dysfunction in cirrhosis. One hundred three subjects with cirrhosis had glomerular filtration rate (GFR) measured using iothalamate plasma clearance, and were followed until death, transplantation, or the last encounter. Concomitantly, plasma metabolomic profiling was performed using ultrahigh performance liquid chromatography-tandem mass spectrometry to identify preliminary metabolomic biomarker candidates. Among the 1028 metabolites identified, 34 were significantly increased in subjects with high liver and kidney disease severity compared with those with low liver and kidney disease severity. The highest average fold-change (2.39) was for 4-acetamidobutanoate. Metabolite-based enriched pathways were significantly associated with the identified metabolomic signature (P values ranged from 2.07E-06 to 0.02919). Ascorbate and aldarate metabolism, methylation, and glucuronidation were among the most significant protein-based enriched pathways associated with this metabolomic signature (P values ranged from 1.09E-18 to 7.61E-05). Erythronate had the highest association with measured GFR (R-square=0.571, P<0.0001). Erythronate (R=0.594, P<0.0001) and N6-carbamoylthreonyladenosine (R=0.591, P<0.0001) showed stronger associations with measured GFR compared to Cr (R=0.588, P<0.0001) even after controlling for age, gender and race. The 5 most significant metabolites that predicted mortality independent of kidney disease and demographics were S-adenosylhomocysteine (P=0.0003), glucuronate (P=0.0006), trans-aconitate (P=0.0018), 3-ureidopropionate (P=0.0021), and 3-(4-hydroxyphenyl)lactate (P=0.0047). A unique metabolomic signature associated with hepatorenal dysfunction in cirrhosis was identified for further investigations that provide potentially important mechanistic insights into cirrhosis-altered metabolism.
INTRODUCTION
The many successes realized in the fields of liver and kidney transplantation have prolonged the duration of life for many patients, but demand for donor organs remains high. Donor organ allocation and distribution is imperfect and presents important issues regarding how these organs can be equitably distributed. As such, there is a particular need for developing accurate biomarkers to conveniently assess a level of renal function and extent of kidney damage in patients with cirrhosis, because serum creatinine (Cr) concentration alone is insufficient to give true clinical assessments.(1)
In recent years, progress has been made toward a better estimation of GFR in cirrhosis. Serum Cr, which is heavily weighted in the Model for End-Stage Liver Disease-Sodium (MELD-Na) score(2, 3), inaccurately reflects glomerular filtration rate (GFR) in cirrhosis due to reduced muscle mass and increased tubular secretion of Cr.(4, 5) The Cr-based Modification of Diet in Renal Disease Study equation-6 (MDRD-6) used for simultaneous liver-kidney transplantation listing underestimates measured GFR(6) and increases the probability of unnecessary kidney transplantation. Conversely, severe renal disease, with significant increase in morbidity and mortality, can occur after liver transplantation when pre-transplant GFR is overestimated.(6, 7) Though not ideal or widespread in clinical use, the GFR equations combining serum Cr and cystatin C including the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) Cr-Cystatin C equation(8) and Cr-Cystatin C GFR Equation for Cirrhosis(9) have shown significantly higher accuracy in predicting measured GFR compared to the conventional Cr-based GFR-estimating equations in cirrhosis.(5, 8–10) These tests have not gained widespread clinical use.
The application of non-targeted serum metabolomic profiling has evolved as a powerful non-invasive tool to identify new clinical biomarkers.(11–13) Select metabolites can provide early diagnostic differentiation and insight into the pathological mechanisms underlying a variety of liver diseases and can also be exploited for drug development. In this preliminary study, we aimed to explore novel pathophysiological metabolomic pathways that could be subsequently validated and exploited for prognostic (e.g., to predict response to hepatorenal syndrome or predict native kidney recovery after liver transplantation) and therapeutic purposes (e.g. development of new drugs targeting specific enriched pathways associated with a metabolomic signature) in kidney disease in cirrhosis.
MATERIALS AND METHODS
Study Population
After obtaining approval from the University of Maryland, Baltimore Institutional Review Board between 2010 and 2016, we conducted this study using the General Clinical Research Center and outpatient clinics of the University of Maryland Medical Center. Written informed consent was obtained from all study subjects. This study was also approved by Baylor College of Medicine Biomedical Research and Assurance Information Network (BRAIN). All methods and procedures were performed in accordance with the relevant guidelines and regulations. We previously described the study inclusion and exclusion criteria.(9) In brief, we included adult outpatients with cirrhosis. We excluded subjects with Acute kidney injury (AKI) or chronic kidney disease stage 5, transjugular intrahepatic porto-systemic shunt placement, liver or kidney transplantation, any existing condition that could influence steady-state renal function (e.g. acute gastrointestinal bleeding, acute infection, exacerbation of hepatic encephalopathy, new use or dose modification of diuretics, angiotensin converting enzyme inhibitors, angiotensin receptor blockers within 1 week of enrollment).
Study Procedures
We previously described all study procedures in detail except analysis of metabolomic biomarkers.(5, 9, 14) Briefly, during Visit 1, we obtained informed consent, medical history and performed physical examination. Visit 2 procedures included measurement of GFR by iothalamate plasma clearance, renal and metabolomic biomarkers, and other laboratory analysis. Visit 2 was scheduled within 1 to 3 weeks of Visit 1.
Detection, Identification and Measurement of Plasma Metabolites
Non-targeted global metabolomic profiling (15–17) was performed on 103 blinded plasma samples at Metabolon, Inc. (Durham, NC). Following receipt by Metabolon, Inc., plasma samples were immediately stored at −80°C. The automated MicroLab STAR® system from Hamilton Company was used to prepare the samples. Proteins in the samples were precipitated with methanol under vigorous shaking for 2 minutes (Glen Mills GenoGrinder 2000) followed by centrifugation. The subsequent extract was separated into five fractions: two for analysis by two separate reverse phase/ultrahigh performance liquid chromatography (UPLC)/tandem mass spectometry (MS/MS) methods with positive ion mode electrospray ionization (ESI), one for analysis by RP/UPLC-MS/MS with negative ion mode ESI, one for analysis by Hydrophilic Interaction Liquid Chromatography (HILIC)/UPLC-MS/MS with negative ion mode ESI, and one sample was reserved for backup. The organic solvent in the samples was removed using TurboVap® (Zymark), and the sample extracts were stored overnight under nitrogen before preparation for analysis. Metabolite identification and measurement were performed using a Waters ACQUITY UPLC and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer (MS) interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. The Metabolon platform used four different methods mentioned above to identify and measure metabolites in a given sample. The majority of metabolites were measured on multiple platforms, and the platform that had the best characteristics (i.e., least amount of nearby interference, ion suppression, etc.) was reported back. The raw data that were reported back from each platform were the integrated peak areas for the quant ion of the molecule of interest (the units are ‘ion counts’) quantified using area-under-the-curve. Metabolites were identified based on three criteria including the presence of the quant ion of interest within a narrow retention index window for the proposed metabolite, an accurate mass match to the library +/− 5 ppm, and forward and reverse fit matches of the MS/MS ions present in the experimental spectrum and library spectrum. In regards to data quality, values for instrument and process variability were reported as a median relative standard deviation of 4% and 7%, respectively. All samples were processed and run on the same platform day except for aliquots that were run across the HILIC/UPLC-MS/MS platform, and which were collected across two platform run days. For metabolite data collected from the HILIC/UPLC-MS/MS platform, a run-day block normalization was performed in which the areas under the curve were scaled to the median of each run-day block and then multiplied by the global median across both run-day blocks to correct for variation resulting from instrument inter-day tuning differences. For the remaining data streams collected in a single run day, block normalization was not required nor applied.
Data were scaled to set the median equal to 1, and missing values (generally due to the metabolite level falling below the limit of detection of the instrument used) were imputed with the minimum observed value of that metabolite, as described elsewhere. (18, 19) For the entire data analysis, these scaled imputed data were used. Additional necessary data transformations are described below.
Data Analysis
All statistical analyses were performed using SAS Version 9.4 TS level 1M3 W32_7PRO platform (SAS, Cary, NC)(20) and R software(21) at Baylor College of Medicine, Houston, TX.
Identification of Metabolomic Signature
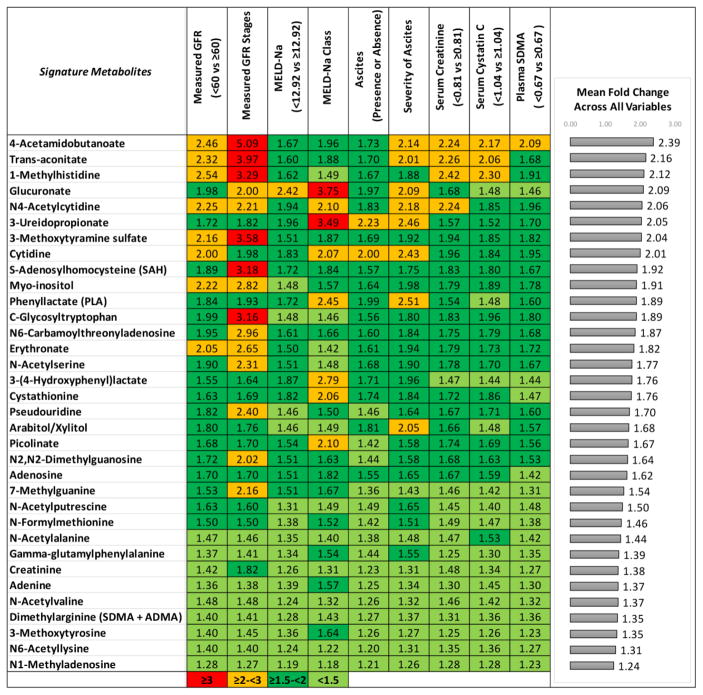
Statistical analysis of metabolomic biomarkers was performed after metabolite values were transformed into log base 2 scale. We preferred log base 2 over log base 10 and natural log transformation as log base 2 scale is commonly used in transcriptomics, metabolomics, and proteomics analysis.(22, 23) Patients were stratified based on low and high liver and kidney disease severity groups for nine clinical and laboratory variables (Table 1). The categories of the variables that defined low vs. high liver and kidney disease severity as shown in Table 1 were categorized as follows: ascites (absent vs. present), severity of ascites (no ascites, diuretic-sensitive and -refractory ascites), measured GFR stages (stage 1: GFR ≥ 90, stage 2: GFR ≥ 60 to < 90, Stage 3: GFR ≥ 30 to <60, Stage 4: GFR ≥ 15 to < 30), measured GFR (< 60 vs. ≥ 60), MELD-Na score classes (6–9, 10–19, 20–40) and above vs. below the median values of GFR biomarkers (serum Cr, cystatin C and Symmetric dimethylarginine (SDMA)), and MELD-Na score. Fold change was defined as the change in the mean value of a metabolite from low disease severity to high disease severity category. We further inferred for each metabolite the mean fold change as the average of all mean fold changes across comparisons for the nine clinical and laboratory variables. We used a t-test to determine statistically significant inducible metabolites between low and high liver and kidney disease severity groups. P values were adjusted for false discovery rate (FDR) using the Benjamini-Hochberg method (79), as implemented in the R statistical system.(21) Metabolites were considered significant for Q value <0.05 and at least 5% change in the mean value. For the categorical clinical variables with more than two categories including measured GFR stages, severity of ascites, and MELD-Na score classes, we considered a metabolite significant if it was significant across a comparison between any pair of patient groups, and if significant across multiple comparisons, the direction of change was consistent between low vs. high disease severity. Furthermore, we selected the highest fold change in the mean value of the metabolite between any pair of groups for this categorical clinical variable. After generating comparison results for each independent clinical variable, we further selected metabolites that were significantly changed across nine clinical and laboratory variables. For statistical rigor, we considered all the metabolites detected by Metabolon, Inc. For further interpretation of results and functional enrichment analysis, we excluded unnamed metabolites and xenobiotic metabolites. After applying these series of filters, we obtained a metabolomic signature comprised of 34 metabolites. For clarity of presentation, we further sorted the 34 metabolites in decreasing order of the mean fold change across all variables (Figure 1).
Table 1.
Categories of Clinical and Laboratory Variables Stratified Based on Low and High Liver and Kidney Disease Severity
| Clinical Variables | Low Disease Severity Groups | vs. | High Disease Severity Groups |
|---|---|---|---|
| Measured GFR (ml/min/1.73m2) | ≥ 60 (N=69) | vs. | < 60 (N=34) |
| Measured GFR Stages* (ml/min/1.73m2) | Stage 1+2 (N=69) | vs. | Stage 3 (N=32) |
| Stage 1+2 (N=69) | vs. | Stage 4 (N=2) | |
| Stage 3 (N=32) | vs. | Stage 4 (N=2) | |
| Creatinine (mg/dL) | < 0.815 (N=51) | vs. | ≥ 0.815 (N=52) |
| Cystatin C (mg/L) | < 1.04 (N=51) | vs. | ≥ 1.04 (N=52) |
| Symmetric Dimethylarginine (micromole/L) | < 0.669 (N=51) | vs. | ≥ 0.669 (N=52) |
| Ascites Status | Absent (N=32) | vs. | Present (N=71) |
| Severity of Ascites | No ascites (N=32) | vs. | Diuretic-Sensitive (N=38) |
| No ascites (N=32) | vs. | Diuretic-Refractory (N=33) | |
| Diuretic-Sensitive (N=38) | vs. | Diuretic-Refractory (N=33) | |
| MELD-Na Score Categories | 6–9 (N=26) | vs. | 10–19 (N=61) |
| 6–9 (N=26) | vs. | 20–40 (N=16) | |
| 10–19 (N=61) | vs. | 20–40 (N=16) | |
| MELD-Na Score | < 12.924 (N=51) | vs. | ≥ 12.924 (N=52) |
Measured GFR stages (stage 1: GFR ≥ 90, stage 2: GFR ≥ 60 to < 90, Stage 3: GFR ≥ 30 to <60, Stage 4: GFR ≥ 15 to < 30)
Figure 1.
Among the 1028 metabolites identified in plasma, 34 were significantly increased and associated with all the nine clinical and laboratory variables indicative of liver and kidney disease severity.
Power Analysis
For quantitative variables where we stratify patients based on the median values, we use a sample number of n=51 per group. Assuming a standard deviation of 75% of the population mean (a reasonably high assumption for human cohorts), we can detect a fold change of 1.25x at the significance level of α=0.05 with a power of 86.65%.
Identification of Enriched Pathways Associated with Metabolomic Signature
To perform integrative analysis such as pathway enrichment, we applied a two-pronged strategy. First, we used the Super-Pathway and Sub-Pathways annotations provided by Metabolon, Inc. for each detected, named metabolite, excluding the xenobiotic metabolites. We determined enriched pathways and processes in the 34 metabolites by employing over-representation analysis and hypergeometric enrichment; significance was assessed at Benjamini-Hochberg (79)-adjusted Q value <0.05. Next, we mined the Human Metabolome Database (HMDB) (24–26) compendium to identify proteins associated with each metabolite, then carried out enrichment analysis using hypergeometric distribution against the compendium of databases KEGG(27), REACTOME(28), BIOCARTA(29), and GENE ONTOLOGY(30), as compiled by the Molecular Signature Database (MSigDB) compendium.(31) The enrichment was carried out using the Python scientific computing libraries (32–34) NumPy (35) and SciPy (32). For clarity of presentation, enriched pathway significance was depicted as bar graphs after conversion to −log10 (Q value) form. To appreciate existing interaction between significant pathways, we inferred the network of enriched pathways by considering as nodes the enriched pathways, and considering common edges whenever two pathways shared common metabolites or common associated proteins. The inferred pathway networks were further visualized using the Cytoscape scientific visualization software package.(36)
Associations between Metabolomic Signature and Measured GFR
After identifying the metabolomic signature, we assessed the association between metabolites and measured GFR using linear regression models. For this analysis, we used natural log transformed values of metabolites and measured GFR. We first sorted data by P values in ascending order and assessed significance at P<0.05. Then, we adjusted P values for FDR using the Benjamini-Hochberg method.(79) Metabolites were considered significant for Q value <0.05.
Associations between Metabolomic Signature and Mortality
Subjects were followed from the procedure date until death, liver transplantation, or the last encounter. We determined survival outcomes by either calling the subjects or reviewing their medical records. Patients were censored at the time of last follow-up or liver transplantation. For mortality analysis, we used natural log transformed values of metabolites. We performed Kaplan-Meier tests (80) to estimate median survival time. To assess the association between each metabolomic biomarker and mortality, we fit separate Cox proportional hazards regression (81) models for each metabolomic biomarker. We further evaluated the metabolites that were significant in univariate Cox models in multivariate Cox models to assess the mortality independent of kidney disease and demographics. We first sorted data by P values in ascending order and assessed significance at P<0.05. Then, we adjusted P values for FDR using the Benjamini-Hochberg method.(79) Metabolites were considered significant for Q value <0.05.
Associations between Metabolomic Biomarkers and Measured GFR Stages, Severity of Ascites and MELD-Na score classes
We assessed significant fold changes for each categorical variable including measured GFR stages, MELD-Na score classes and severity of ascites by taking all 1028 metabolites into account. For measured GFR stages, MELD-Na score classes and severity of ascites categories, significant metabolites were graphically depicted as a heatmap across all the patients after z-score transformation, using the Python Scientific Computing Libraries (32–34) NumPy (35) and Matplotlib (37). Metabolites were considered significant for Q value <0.05. We sorted metabolites by maximum fold change as described above for non-binary clinical variables. We considered a metabolite to be statistically significant if it was significant across a comparison between any pair of subject groups; we further selected the highest fold change in the mean value of the metabolite between any pair of groups for the respective GFR stages (1+2, 3 and 4), MELD-Na score classes (6–9, 10–19, 20–40) and severity of ascites categories (no ascites, diuretic-sensitive and diuretic-refractory ascites). For presentation purposes, the metabolite values were z-score transformed across all subjects; metabolites were further sorted in decreasing order based on the maximum fold change across comparisons between GFR stages, MELD-Na score classes and severity of ascites categories.
Analysis of variance (ANOVA) test was performed to compare differences in myo-inositol/creatine ratio, beta-alanine, betaine and methionine among categorical variables that had more than two categories including GFR stages (1+2, 3 and 4), MELD-Na score classes (6–9, 10–19, 20–40) and severity of ascites (no ascites, diuretic-sensitive and diuretic-refractory ascites). However, we did not impose the additional criteria of consistent changes between low- and high-disease severity.
Study Subjects and Non-Metabolite Variables
T-test and Fisher’s exact tests were performed to compare differences in continuous and categorical variables, respectively between patients with measured GFR ≥ 60 ml/min/1.73m2 and those with GFR < 60 ml/min/1.73m2.
RESULTS
Study Population
We enrolled a total of 134 subjects with cirrhosis of whom 103 completed GFR measurement and other study procedures.(9) Multiple clinical variables, summarized in Table 1 were rigorously collected for each subject. Table 2 shows the demographic, laboratory, and clinical characteristics of the study population. Thirty-three percent of the subjects had a measured GFR < 60 ml/min/1.73m2. The proportion of subjects was significantly different in GFR groups (measured GFR ≥ vs. < 60 ml/min/1.73m2) when they were compared based on severity of ascites (P=0.001) and MELD-Na score (P=0.004). The mean values of GFR biomarkers including serum Cr (P<0.0001), cystatin C (P<0.0001) and plasma SDMA (P<0.0001) were significantly higher among patients with GFR < 60 ml/min/1.73m2 compared with those with ≥ 60 ml/min/1.73m2. There was no significant difference at the P<0.05 level in age, gender, race, etiology of cirrhosis, diabetes, the presence of proteinuria or glomerular disease between patients with measured GFR < 60 and ≥ 60 ml/min/1.73m2.
Table 2.
Characteristics of 103 Subjects with Cirrhosis Stratified by Measured GFR
| Characteristics | GFR ≥ 60 ml/min/1.73m2 | GFR < 60 ml/min/1.73m2 | P Value | |
|---|---|---|---|---|
|
| ||||
| N (%) | N (%) | |||
|
| ||||
| 69 (67) | 34 (33) | |||
|
| ||||
| Gender | Male | 40 (58) | 18 (53) | 0.676 |
| Female | 29 (42) | 16 (47) | ||
|
| ||||
| Race | Non-Black | 53 (77) | 24 (71) | 0.630 |
| Black | 16 (23) | 10 (29) | ||
|
| ||||
| Etiology of Cirrhosis | Hepatitis C | 28 (41) | 14 (41) | 0.466 |
| Hepatitis B | 3 (4) | 0 (0) | ||
| Alcohol | 18 (26) | 12 (35) | ||
| Nonalcoholic fatty liver disease | 13 (19) | 5 (15) | ||
| Primary biliary cholangitis | 2 (3) | 0 (0) | ||
| Primary sclerosing cholangitis | 1 (1) | 1 (3) | ||
| Autoimmune hepatitis | 3 (4) | 0 (0) | ||
| Sarcoidosis | 0 (0) | 1 (3) | ||
| Sickle cell disease | 0 (0) | 1 (3) | ||
| Hemochromatosis | 1 (1) | 0 (0) | ||
|
| ||||
| Ascites | No ascites | 29 (42) | 3 (9) | 0.001 |
| Diuretic-sensitive | 23 (33) | 15 (44) | ||
| Diuretic-refractory | 17 (25) | 16 (47) | ||
|
| ||||
| MELD-Na score | 6–9 | 24 (35) | 2 (6) | 0.004 |
| 10–19 | 36 (52) | 25 (74) | ||
| 20–40 | 9 (13) | 7 (21) | ||
|
| ||||
| Proteinuria (> 0.5 g/24 hours) | Yes | 4 (6) | 3 (9) | 0.686 |
|
| ||||
| Glomerular disease (spot urine protein/creatinine ratio ≥ 0.2) | Yes | 11 (17) | 8 (24) | 0.428 |
|
| ||||
| Diabetes | Yes | 21 (30) | 9 (26) | 0.819 |
|
| ||||
| Hypothyroidism | Yes | 7 (10) | 4 (12) | 1.000 |
|
| ||||
| Mean (SD) | Mean (SD) | P Value | ||
|
| ||||
| Age (yr) | 53.55 (9.06) | 56.53 (8.15) | 0.112 | |
| Weight (kg) | 87.23 (18.9) | 76.45 (17.92) | 0.007 | |
| Height (m) | 1.69 (0.09) | 1.68 (0.1) | 0.456 | |
| Body-surface area (m2) | 1.97 (0.21) | 1.85 (0.23) | 0.012 | |
| MELD-Na score | 12.25 (5.11) | 16.12 (4.82) | 0.0004 | |
| Total bilirubin (mg/dL) | 2.01 (2.12) | 3.41 (5.88) | 0.193 | |
| Prothrombin time (sec) | 17.24 (3.53) | 17.11 (2.74) | 0.844 | |
| International normalized ratio | 1.37 (0.37) | 1.35 (0.29) | 0.768 | |
| Serum albumin (g/dL) | 3.08 (0.51) | 3.07 (0.61) | 0.937 | |
| BUN (mg/dl) | 11.75 (6.93) | 19.94 (9.28) | <0.0001 | |
| Serum sodium (mmol/L) | 137.09 (3.95) | 136.68 (3.19) | 0.603 | |
| Measured GFR (ml/min/1.73m2) | 96.48 (31.74) | 47.38 (9.25) | <0.0001 | |
| Cr (mg/dL) | 0.74 (0.2) | 1.22 (0.35) | <0.0001 | |
| Cystatin C (mg/L) | 0.96 (0.27) | 1.61 (0.43) | <0.0001 | |
| SDMA (micromole/L) | 0.65 (0.19) | 1.07 (0.43) | <0.0001 | |
SD=Standard deviation
Metabolomic Signature
Identification of Metabolomic Signature
We identified 1028 metabolites; of which 771 were named and 257 were unnamed. Among the 1028 metabolites identified in plasma, 34 were significantly increased and associated with all of the nine clinical and laboratory variables indicative of liver and kidney disease severity. We displayed fold changes of these 34 metabolites in Figure 1. The highest mean fold change (2.39) was observed for 4-acetamidobutanoate when subjects with low liver and kidney disease severity were compared with those with high disease severity across nine clinical variables. The lowest significant mean fold change (1.24) occurred with N1-methyladenosine.
Although creatine (precursor of Cr), beta-alanine, betaine and methionine were not among the 34 signature metabolites, we further assessed differences in these metabolites and in myo-inositol/creatine ratio among GFR stages, MELD-Na score classes and categories of severity of ascites to provide a more accurate mechanistic insight to explain significant fold changes in the levels of myo-inositol, 3-ureidopropionate and S-adenosylhomocysteine in the metabolomic signature. Mean and P values for these metabolites were as follows: Mean myo-inositol/creatine ratio was 0.85, 1.33 and 2.48 in subjects without ascites, diuretic-sensitive ascites and diuretic-refractory ascites, respectively (P=0.0005); mean myo-inositol/creatine ratio was 0.84, 2.99, and 2.86 in subjects with measured GFR stages 1+2, 3 and 4, respectively (P<0.0001); mean myo-inositol/creatine ratio was 0.68, 1.85, and 1.79 in subjects with MELD-Na score 6–9, 10–19, and 20–40, respectively (P=0.015), mean beta-alanine level was 1.03, 0.97 and 1.45 in subjects without ascites, diuretic-sensitive ascites, and diuretic-refractory ascites, respectively (P=0.001); mean betaine level was 0.87, 1.14 and 1.75 in subjects with MELD-Na score 6–9, 10–19, and 20–40, respectively (P=0.003); mean methionine level was 0.88, 1.04 and 1.69 in subjects with MELD-Na score 6–9, 10–19, and 20–40, respectively (P=0.001).
Identification of Enriched Pathways Associated with Metabolomic Signature
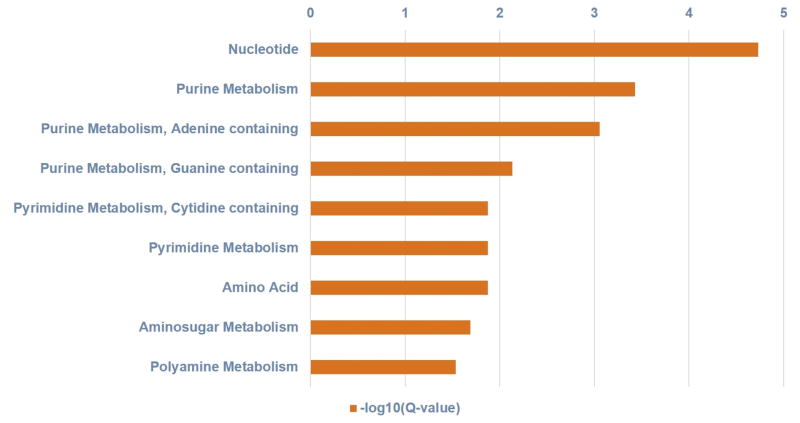
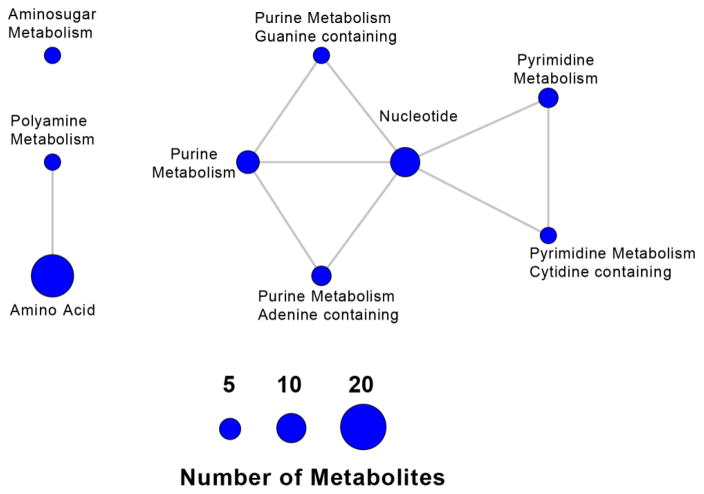
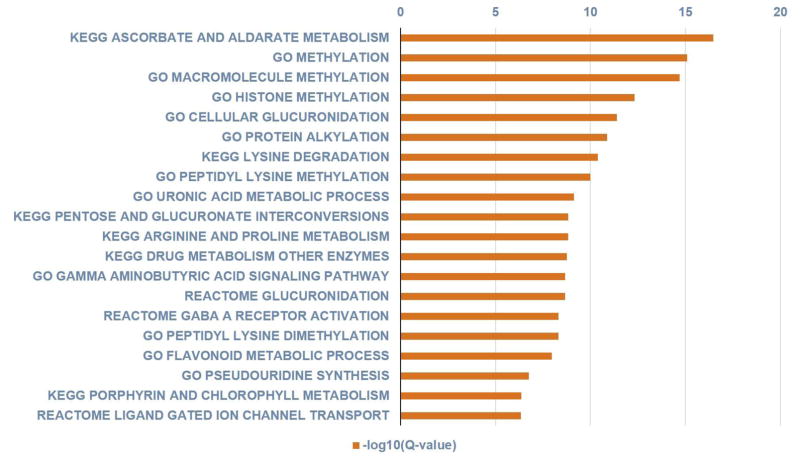
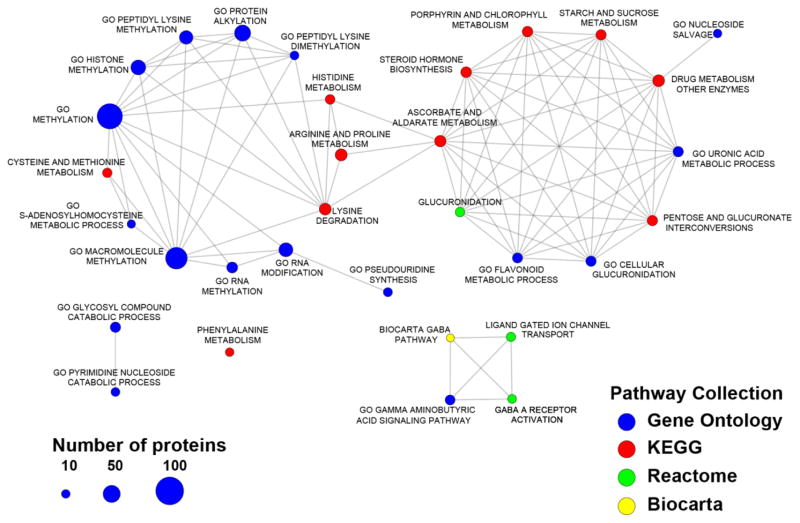
Using the metabolites/genes association compiled in the HMDB, we further analyzed pathways enriched in the 34 metabolites. Figs 2 and 3 show the metabolite-based enriched pathways, in descending order, nucleotide, purine, pyrimidine, amino acid, amino sugar and polyamine metabolisms that were significantly associated with the metabolomic signature (P values ranged from 2.07E-06 to 0.02919; Q values ranged from 1.86E-05 to 0.02919) and their associated pathway network, respectively. Figures 4 and 5 show the 20 most significantly associated protein-based enriched pathways (P values ranged from 1.09E-18 to 7.61E-05; Q values ranged from 3.49E-17 to 7.61E-05) and their associated pathway network, respectively.
Figure 2.
Figure shows the metabolite-based enriched pathways, in descending order, nucleotide, purine, pyrimidine, amino acid, amino sugar and polyamine metabolisms that were significantly associated with the metabolomic signature (hypergeometric distribution; Q values ranged from 1.86E-05 to 0.02919).
Figure 3.
Pathway network representation of the metabolite-based enriched pathways for the 34 metabolites associated with low vs. high liver and kidney disease severity. Nodes correspond to significant pathways, with node size associated with the number of significant metabolites in each pathway; edges indicate common metabolites between two pathways. We identified significant groups of processes including a) polyamine metabolism and amino acid; b) purine metabolism, pyrimidine metabolism, and nucleotides.
Figure 4.
Figure shows the 20 most significantly associated protein-based enriched pathways in descending order of significance (hypergeometric distribution; Q values ranged from 3.49E-17 to 7.61E-05).
Figure 5.
Network of enriched pathways for the 34 metabolites associated with low vs. high hepatic and kidney disease severity, based on functionally-related proteins. Nodes correspond to significant pathways, with node size associated with the number of associated proteins in each pathway; edges indicate common proteins between two pathways. We identified significant groups of processes including a) methylation; b) glucuronidation; c) GABA-ergic pathways.
Associations between Metabolomic Signature and Measured GFR
All metabolites were significantly associated with measured GFR (Table 3). The 10 metabolites that showed the strongest association with measured GFR based on R–square value, in descending order, were erythronate (R-square=0.571, P<0.0001), N6-carbamoylthreonyladenosine (R-square=0.539, P<0.0001), 1-methylhistidine (R-square=0.539, P<0.0001), pseudouridine (R-square=0.535, P<0.0001), N-acetylserine (R-square=0.528, P<0.0001), Cr (R-square=0.525, P<0.0001), 7-methylguanine (R-square=0.484, P<0.0001), N2-N2-dimethylguanosine (R-square=0.481, P<0.0001), C-glycosyltryptophan (R-square=0.473, P<0.0001) and myo-inositol (R-square=0.464, P<0.0001). When controlled for age, gender and race, erythronate (R=0.594, P<0.0001) and N6-carbamoylthreonyladenosine (R=0.591, P<0.0001) showed stronger associations with measured GFR compared to Cr (R=0.588, P<0.0001).
Table 3.
Associations between Metabolomic Signature and Measured GFR in 103 Patients with Cirrhosis in Univariate Regression Models*
| Metabolites | R -Square | P Value | Q Value | Super Pathway | Sub Pathway |
|---|---|---|---|---|---|
| erythronate* | 0.571 | <0.0001 | 0.0001 | Carbohydrate | Aminosugar Metabolism |
| N6-carbamoylthreonyladenosine* | 0.539 | <0.0001 | 0.0001 | Nucleotide | Purine Metabolism, Adenine containing |
| 1-methylhistidine | 0.539 | <0.0001 | 0.0001 | Amino Acid | Histidine Metabolism |
| pseudouridine | 0.535 | <0.0001 | 0.0001 | Nucleotide | Pyrimidine Metabolism, Uracil containing |
| N-acetylserine | 0.528 | <0.0001 | 0.0001 | Amino Acid | Glycine, Serine and Threonine Metabolism |
| creatinine | 0.525 | <0.0001 | 0.0001 | Amino Acid | Creatine Metabolism |
| 7-methylguanine | 0.484 | <0.0001 | 0.0001 | Nucleotide | Purine Metabolism, Guanine containing |
| N2,N2-dimethylguanosine | 0.481 | <0.0001 | 0.0001 | Nucleotide | Purine Metabolism, Guanine containing |
| C-glycosyltryptophan | 0.473 | <0.0001 | 0.0001 | Amino Acid | Tryptophan Metabolism |
| myo-inositol | 0.464 | <0.0001 | 0.0001 | Lipid | Inositol Metabolism |
| 3-methoxytyramine sulfate | 0.433 | <0.0001 | 0.0001 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| trans-aconitate | 0.406 | <0.0001 | 0.0001 | Energy | TCA Cycle |
| N-formylmethionine | 0.395 | <0.0001 | 0.0001 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| 4-acetamidobutanoate | 0.394 | <0.0001 | 0.0001 | Amino Acid | Polyamine Metabolism |
| N-acetylvaline | 0.382 | <0.0001 | 0.0001 | Amino Acid | Leucine, Isoleucine and Valine Metabolism |
| N-acetylalanine | 0.379 | <0.0001 | 0.0001 | Amino Acid | Alanine and Aspartate Metabolism |
| N1-methyladenosine | 0.365 | <0.0001 | 0.0001 | Nucleotide | Purine Metabolism, Adenine containing |
| S-adenosylhomocysteine | 0.364 | <0.0001 | 0.0001 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| dimethylarginine (SDMA + ADMA) | 0.343 | <0.0001 | 0.0001 | Amino Acid | Urea cycle; Arginine and Proline Metabolism |
| N6-acetyllysine | 0.311 | <0.0001 | 0.0001 | Amino Acid | Lysine Metabolism |
| arabitol/xylitol | 0.310 | <0.0001 | 0.0001 | Carbohydrate | Pentose Metabolism |
| cytidine | 0.239 | <0.0001 | 0.0001 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| N-acetylputrescine | 0.234 | <0.0001 | 0.0001 | Amino Acid | Polyamine Metabolism |
| N4-acetylcytidine | 0.232 | <0.0001 | 0.0001 | Nucleotide | Pyrimidine Metabolism, Cytidine containing |
| adenosine | 0.222 | <0.0001 | 0.0001 | Nucleotide | Purine Metabolism, Adenine containing |
| adenine | 0.156 | <0.0001 | 0.0001 | Nucleotide | Purine Metabolism, Adenine containing |
| 3-methoxytyrosine | 0.156 | <0.0001 | 0.0001 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| glucuronate | 0.141 | <0.0001 | 0.0001 | Carbohydrate | Aminosugar Metabolism |
| picolinate | 0.136 | 0.0001 | 0.0001 | Amino Acid | Tryptophan Metabolism |
| cystathionine | 0.124 | 0.0003 | 0.0003 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| phenyllactate | 0.121 | 0.0003 | 0.0003 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| gamma-glutamylphenylalanine | 0.120 | 0.0003 | 0.0003 | Peptide | Gamma-glutamyl Amino Acid |
| 3-(4-hydroxyphenyl)lactate | 0.109 | 0.0007 | 0.0007 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| 3-ureidopropionate | 0.102 | 0.0010 | 0.0010 | Nucleotide | Pyrimidine Metabolism, Uracil containing |
Log transformed values of metabolites were used in the linear regression models. For calculation of Q-values, P-values < 0.0001 were considered as 0.0001.
The metabolites in this table were first sorted by their P-value and then Q-values were determined. The Q-value assigns the false discovery rate for the selected list of metabolites. All metabolites in this table have a false discovery rate below 0.1%.
When controlled for age, gender and race, erythronate (R=0.594, P<0.0001) and N6-carbamoylthreonyladenosine (R=0.591, P<0.0001) showed stronger associations with measured GFR compared to Cr (R=0.588, P<0.0001).
Associations between Metabolomic Signature and Mortality
Median follow-up was 1.9 years. At last follow-up time, 52 subjects were alive, 34 subjects had died, and 17 subjects had undergone liver transplantation. Median survival was 4.4 years. Table 4 shows the Cox models that assessed the association between 34 metabolites and mortality. Among 34 metabolites, glucuronate (P=0.0004), 3-ureidopropionate (P=0.0015), S-adenosylhomocysteine (P=0.0018), trans-aconitate (P=0.0025), 3-(4-hydroxyphenyl)lactate (P=0.0032), arabitol/xylitol (P=0.0080), 3-methoxytyramine sulfate (P=0.0155), phenyllactate (P=0.0167), N-formylmethionine (P=0.0253), erythronate (P=0.0447), 4-acetamidobutanoate (P=0.0465), adenosine (P=0.0472), and 7-methylguanine (P=0.0486) significantly predicted mortality (Table 4). Among these metabolites, hazard ratio for S-adenosylhomocysteine, glucuronate, trans-aconitate, 3-ureidopropionate, 3-(4-hydroxyphenyl)lactate, 3-methoxytyramine sulfate, arabitol/xylitol, N-formylmethionine, phenyllactate and 7-methylguanine remained significant after controlling for measured GFR and demographics (Table 5).
Table 4.
Associations between Metabolomic Signature and Mortality in 103 Patients with Cirrhosis in Univariate Cox Regression Models
| Metabolites | Hazard Ratio | P Value | Q Value | Super Pathway | Sub Pathway |
|---|---|---|---|---|---|
| MELD-Na Score | 1.16 | <0.0001 | 0.0035 | ||
| glucuronate | 2.34 | 0.0004 | 0.0070 | Carbohydrate | Aminosugar Metabolism |
| 3-ureidopropionate | 2.45 | 0.0015 | 0.0158 | Nucleotide | Pyrimidine Metabolism, Uracil containing |
| S-adenosylhomocysteine | 2.73 | 0.0018 | 0.0158 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| trans-aconitate | 2.08 | 0.0025 | 0.0175 | Energy | TCA Cycle |
| 3-(4-hydroxyphenyl)lactate | 2.63 | 0.0032 | 0.0187 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| arabitol/xylitol | 2.42 | 0.0080 | 0.0400 | Carbohydrate | Pentose Metabolism |
| 3-methoxytyramine sulfate | 2.13 | 0.0155 | 0.0649 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| phenyllactate | 1.56 | 0.0167 | 0.0649 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| N-formylmethionine | 3.00 | 0.0253 | 0.0886 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| erythronate | 2.05 | 0.0447 | 0.1215 | Carbohydrate | Aminosugar Metabolism |
| 4-acetamidobutanoate | 1.62 | 0.0465 | 0.1215 | Amino Acid | Polyamine Metabolism |
| adenosine | 2.17 | 0.0472 | 0.1215 | Nucleotide | Purine Metabolism, Adenine containing |
| 7-methylguanine | 2.66 | 0.0486 | 0.1215 | Nucleotide | Purine Metabolism, Guanine containing |
| N-acetylputrescine | 1.97 | 0.0592 | 0.1310 | Amino Acid | Polyamine Metabolism |
| N6-carbamoylthreonyladenosine | 1.99 | 0.0613 | 0.1310 | Nucleotide | Purine Metabolism, Adenine containing |
| adenine | 2.18 | 0.0672 | 0.1310 | Nucleotide | Purine Metabolism, Adenine containing |
| N-acetylserine | 1.95 | 0.0682 | 0.1310 | Amino Acid | Glycine, Serine and Threonine Metabolism |
| N2,N2-dimethylguanosine | 2.12 | 0.0711 | 0.1310 | Nucleotide | Purine Metabolism, Guanine containing |
| creatinine | 3.24 | 0.1027 | 0.1797 | Amino Acid | Creatine Metabolism |
| cytidine | 1.45 | 0.1471 | 0.2351 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| N1-methyladenosine | 3.05 | 0.1504 | 0.2351 | Nucleotide | Purine Metabolism, Adenine containing |
| pseudouridine | 1.80 | 0.1545 | 0.2351 | Nucleotide | Pyrimidine Metabolism, Uracil containing |
| myo-inositol | 1.49 | 0.1727 | 0.2519 | Lipid | Inositol Metabolism |
| gamma-glutamylphenylalanine | 1.74 | 0.1940 | 0.2714 | Peptide | Gamma-glutamyl Amino Acid |
| N4-acetylcytidine | 1.33 | 0.2016 | 0.2714 | Nucleotide | Pyrimidine Metabolism, Cytidine containing |
| picolinate | 1.46 | 0.2400 | 0.3111 | Amino Acid | Tryptophan Metabolism |
| dimethylarginine (SDMA + ADMA) | 2.02 | 0.2562 | 0.3203 | Amino Acid | Urea cycle; Arginine and Proline Metabolism |
| N-acetylalanine | 1.68 | 0.2926 | 0.3470 | Amino Acid | Alanine and Aspartate Metabolism |
| N-acetylvaline | 1.74 | 0.2974 | 0.3470 | Amino Acid | Leucine, Isoleucine and Valine Metabolism |
| C-glycosyltryptophan | 1.27 | 0.4804 | 0.5424 | Amino Acid | Tryptophan Metabolism |
| cystathionine | 1.17 | 0.5012 | 0.5482 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| N6-acetyllysine | 1.43 | 0.5508 | 0.5842 | Amino Acid | Lysine Metabolism |
| 3-methoxytyrosine | 0.76 | 0.5928 | 0.6102 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| 1-methylhistidine | 1.01 | 0.9740 | 0.9740 | Amino Acid | Histidine Metabolism |
Log transformed values of metabolites were used in the Cox proportional hazards regression models. Patients were censored at the time of last follow-up or liver transplantation. For calculation of Q-values, P-values < 0.0001 were considered as 0.0001.
The metabolites in this table were first sorted by their P-value and then Q-values were determined. The Q-value assigns the false discovery rate for the selected list of metabolites. Twenty-two of the metabolites are significantly associated with mortality at the Q-value level of 25%.
Table 5.
Mortality Prediction of 10 Metabolomic Biomarkers in the Signature in 103 Patients with Cirrhosis in Multivariate Cox Regression Models*
| Metabolites | Beta Coefficient for the Metabolite | Hazard Ratio | 95% Hazard Ratio Confidence Limits | P Value | Q Value | Super Pathway | Sub Pathway | |
|---|---|---|---|---|---|---|---|---|
| S-adenosylhomocysteine | 1.578 | 4.84 | 2.05 | 11.47 | 0.0003 | 0.0030 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| glucuronate | 0.952 | 2.59 | 1.50 | 4.47 | 0.0006 | 0.0030 | Carbohydrate | Aminosugar Metabolism |
| trans-aconitate | 0.864 | 2.37 | 1.38 | 4.08 | 0.0018 | 0.0053 | Energy | TCA Cycle |
| 3-ureidopropionate | 0.988 | 2.69 | 1.43 | 5.04 | 0.0021 | 0.0053 | Nucleotide | Pyrimidine Metabolism, Uracil containing |
| 3-(4-hydroxyphenyl)lactate | 1.038 | 2.82 | 1.38 | 5.80 | 0.0047 | 0.0094 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| 3-methoxytyramine sulfate | 1.184 | 3.27 | 1.33 | 8.03 | 0.0098 | 0.0154 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| arabitol/xylitol | 0.959 | 2.61 | 1.25 | 5.46 | 0.0108 | 0.0154 | Carbohydrate | Pentose Metabolism |
| N-formylmethionine | 1.590 | 4.90 | 1.35 | 17.79 | 0.0156 | 0.0195 | Amino Acid | Methionine, Cysteine, SAM and Taurine Metabolism |
| phenyllactate | 0.485 | 1.62 | 1.09 | 2.42 | 0.0176 | 0.0196 | Amino Acid | Phenylalanine and Tyrosine Metabolism |
| 7-methylguanine | 1.795 | 6.02 | 1.15 | 31.53 | 0.0336 | 0.0336 | Nucleotide | Purine Metabolism, Guanine containing |
Log transformed values of metabolites were used in the Cox proportional hazards regression models. Patients were censored at the time of last follow-up or liver transplantation. Cox models were controlled for measured GFR, age, gender and race.
The metabolites in this table were first sorted by their P-value and then Q-values were determined. The Q-value assigns the false discovery rate for the selected list of metabolites. All metabolites in this table are significantly associated with mortality at the Q-value level of 3.36%.
Among the metabolites with a P value < 0.05 shown in Table 4, hazard ratio for only S-adenosylhomocysteine, glucuronate, trans-aconitate, 3-ureidopropionate, 3-(4-hydroxyphenyl) lactate, 3-methoxytyramine sulfate, arabitol/xylitol, N-formylmethionine, phenyllactate and 7-methylguanine remained significant after controlling for measured GFR and demographics.
Metabolites Associated with Measured GFR Stages, MELD-Na Score Classes and Severity of Ascites
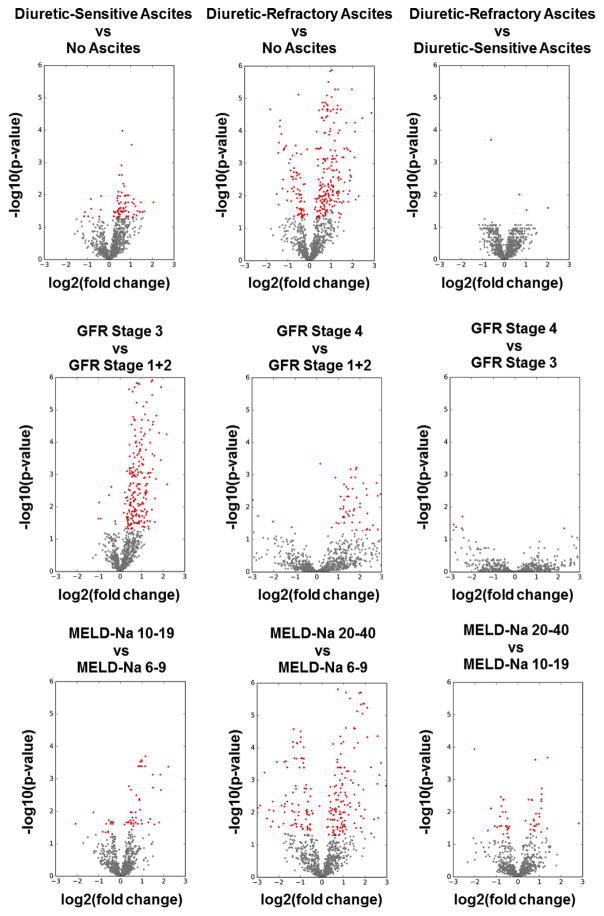
In addition to the metabolomic signature that consisted of 34 metabolites that significantly increased in both high liver and kidney disease severity groups, we identified several metabolites that significantly increased or decreased in either high liver (high MELD-Na score, diuretic-refractory ascites) or high kidney disease severity groups (reduced GFR) in patients with cirrhosis (Figures 6, 7, 8, and 9). We used volcano plots to assess the robustness of association of metabolites with the clinical variables including GFR stages, MELD-Na score classes, and severity of ascites (Figure 6). Volcano plots show all significant and non-significant metabolites across these clinical variables. As shown in Figures 7, 8, and 9, for GFR stages, MELD-Na score classes, and severity of ascites, metabolites can reliably separate patient samples based on their clinical profile.
Figure 6.
Volcano plots for the statistical analysis of metabolites vs clinical variables. Shown in the figure are the scatterplots of P value as a function of fold change for severity of ascites, GFR stages, and MELD-Na score classes. Overall, the plots show robust separation of metabolite values with clinical variables. Red color: significant metabolites. Gray color: detected metabolites, not significant.
Figure 7.
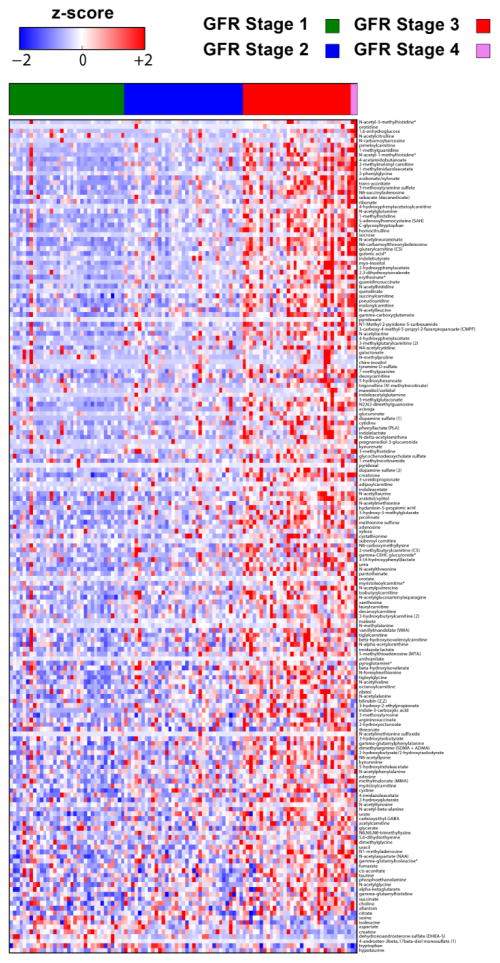
Figure shows the heatmap of the metabolites that significantly changed across GFR stages.
Figure 8.
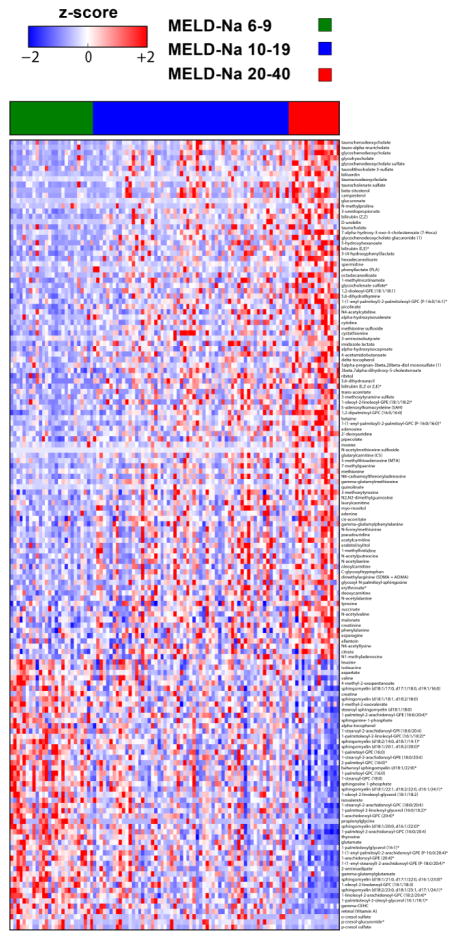
Figure shows the heatmap of the metabolites that significantly changed across MELD-Na score classes. While mean value of several metabolite levels shown in the first 2/3 of the heat map increased in subjects with MELD-Na score 20–40 compared with those with lower MELD-Na scores, mean values of several other metabolites (e.g. sphingomyelins, glycerophosphocholines and glycerophosphoethanolamines) in the last 1/3 of the heat map decreased.
Figure 9.
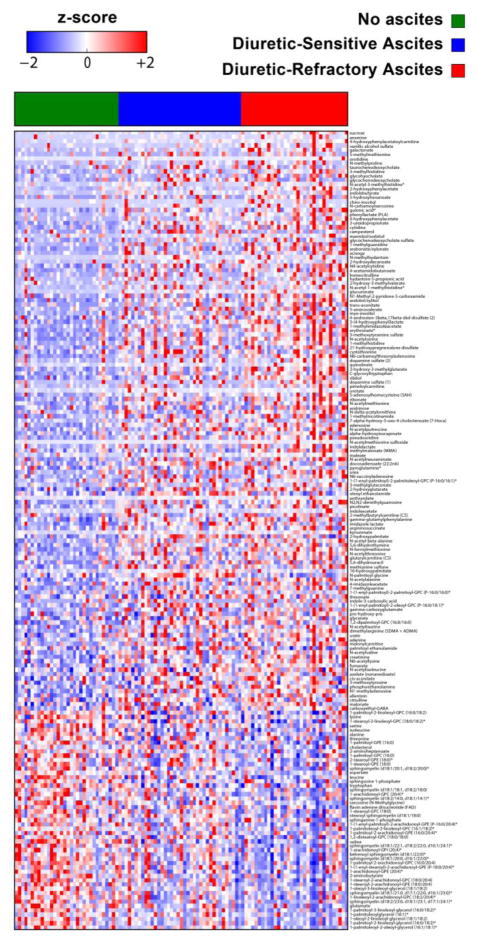
Figure shows the heatmap of the metabolites that significantly changed across severity of ascites. While mean value of several metabolite levels shown in the first 2/3 of the heat map significantly increased in subjects with diuretic-refractory ascites compared with those without ascites and with diuretic-sensitive ascites, mean value of several other metabolites (e.g. sphingomyelins, glycerophosphocholines and glycerophosphoethanolamines) in the last 1/3 of the heat map decreased.
In addition to volcano plots, we used heatmaps to show all metabolites that significantly increased or decreased across the clinical variables including GFR stages, MELD-Na score classes and severity of ascites. Figure 7 shows the heatmap of the metabolites that significantly changed across GFR stages. Figure 8 shows the heatmap of the metabolites that significantly changed across MELD-Na score classes. While mean value of several metabolite levels shown in the first 2/3 of the heatmap increased in subjects with MELD-Na score 20–40 compared with those with lower MELD-Na scores, mean values of several other metabolites (e.g. sphingomyelins, glycerophosphocholines [GPC], glycerophosphoethanolamines [GPE]) in the last 1/3 of the heatmap decreased (e.g. mean sphingomyelin [d18:1/21:0, d17:1/22:0, d16:1/23:0] levels were 1.50, 1.03 and 0.63 in subjects with MELD-Na score classes 6–9, 10–19 and 20–40, respectively, P<0.0001; mean 1-palmitoyl-2-arachidonoyl-GPC (16:0/20:4) levels were 1.21, 0.98, and 0.66 in subjects with MELD-Na score classes 6–9, 10–19 and 20–40, respectively, P<0.0001; mean 1-arachidonoyl-GPE (20:4) levels were 1.35, 1.01, 0.66, P<0.0001). Figure 9 shows the heatmap of the metabolites that significantly changed across severity of ascites. While mean value of several metabolite levels shown in the first 2/3 of the heatmap significantly increased in subjects with diuretic-refractory ascites compared with those without ascites and with diuretic-sensitive ascites, mean value of several other metabolites in the last 1/3 of the heatmap decreased (e.g. mean sphingomyelin [d18:1/21:0, d17:1/22:0, d16:1/23:0] levels were 1.36, 1.03 and 0.89 in subjects without ascites, with diuretic-sensitive ascites and diuretic-refractory ascites, respectively, P=0.0007; mean 1-palmitoyl-2-arachidonoyl-GPC (16:0/20:4) levels were 1.18, 0.97, and 0.82 in subjects without ascites, with diuretic-sensitive ascites and diuretic-refractory ascites, respectively, P=0.0002); mean 1-arachidonoyl-GPE (20:4) levels were 1.24, 1.04, and 0.86 in subjects without ascites, with diuretic-sensitive ascites and diuretic-refractory ascites, respectively, P=0.004).
DISCUSSION
GFR was accurately measured in 103 subjects with cirrhosis using the non-radiolabeled iothalamate plasma clearance method and 1028 metabolites were simultaneously evaluated. This exploration identified 34 unique metabolites which were significantly increased across nine important clinical and laboratory variables in subjects with high liver and kidney disease severity compared with those with low disease severity (Figure 1). To our knowledge, this preliminary report is the first to describe a distinct metabolomic signature of hepatorenal dysfunction and its associated mortality in subjects with cirrhosis. This broad metabolomic signature was significantly associated with 9 distinct metabolite-based pathways (Figures 2 and 3) and multiple protein-based enriched pathways (Figures 4 and 5) that are elaborated upon below. We also identified several metabolites that significantly increased or decreased in patients with high liver disease severity (high MELD-Na score, diuretic-refractory ascites) or high kidney disease severity (Figures 6, 7, 8, and 9). This preliminary report will serve as a foundation to refine these findings into a concise metabolomic profile that could be utilized clinically to make important management decisions.
This study found that the pathway with the strongest association with the metabolomic signature of hepatorenal dysfunction was ascorbate and aldarate metabolism. Signature metabolites associated with this pathway included 4-acetamidobutanoate, glucuronate, gamma-glutamylphenylalanine and myo-inositol. Among all metabolites, 4-acetamidobutanoate, which is urea cycle product (24–26) had the highest significant mean fold change when subjects with low liver and kidney disease severity were compared with those with high liver and kidney disease severity across nine clinical variables (Figure 1). It has been previously reported that plasma 4-acetamidobutanoate is elevated in patients with reduced renal function.(38) Our findings showed that 4-acetamidobutanoate was associated with several key metabolic pathways including ascorbate and aldarate metabolism, arginine and proline metabolism, histidine metabolism, lysine degradation and phenylalanine metabolism. 4-acetamidobutanoate, is also reported to be a gamma-aminobutyric acid (GABA) derivative.(24–26) GABA is an inhibitory neurotransmitter of the nervous system.(39) In cirrhosis, there is increased GABAergic tone resulting in hepatic encephalopathy.(39) Enhanced GABAergic tone (39) may also explain increased levels of 4-acetamidobutanoate in high liver and kidney disease severity groups (Figures 1, 7, 8, and 9) in patients with cirrhosis. However, lack of sufficient data related to a relationship between 4-acetamidobutanoate and GABA limits further interpretation.
Glucuronate was most significantly associated with mortality; subjects with high plasma glucuronate level were 2.34 times more likely to die than those with low glucuronate levels (P=0.0004) (Table 4). Glucuronate is a carboxylic acid used extensively in phase II conjugation reactions in the liver and, to a lesser extent, in other organs (24–26) and is an intermediate product in the catabolism of myo-inositol.(40) The increased risk of death associated with increased levels of glucuronate (Tables 4 and 5) could correlate with altered glucuronidation in the liver (41) and/or increased beta-glucuronidase activity by the gut microbiota (e.g. Echerichia coli).(42) A study conducted using healthy and cirrhotic human livers showed the rate of glucuronidation was reduced in cirrhosis and in turn, impacted drug clearance.(41)
We found significant fold changes in the mean levels of myo-inositol when we compared the subjects with high liver and kidney disease severity to low disease severity. Serum and urinary myo-inositol levels have long been advocated as a sensitive and specific indicator of kidney function (43) and may be important in the setting of hepatorenal dysfunction, too. Myo-inositol is a cyclic polyalcohol that plays a key role in the synthesis of several secondary messengers. (24–26, 40) It is normally catabolized by the kidney.(24–26, 40) It is synthesized from D-glucose and has osmotic properties.(40, 44) Glucuronate is an intermediate product in the catabolism of myo-inositol.(40) We conclude that increased levels of glucuronate levels in subjects with high liver and kidney disease severity can be explained by increased breakdown of excess myo-inositol, which is consistent with findings by Tsalik et al.(38) who showed that plasma glucuronate levels were elevated in patients with renal dysfunction and consistent with findings from Suhre et al.(45) who reported the myo-inositol to be increased in urine samples of renal transplant patients with acute cellular rejection compared to control subjects. As the catabolism of myo-inositol occurs only in the kidney (40), significantly elevated plasma myo-inositol levels in our subjects with high disease severity can be an indicator of severe kidney injury.
We also found significantly elevated mean plasma myo-inositol/creatine ratio in subjects with high liver and kidney disease severity compared with low liver and kidney disease severity and this ratio was directly proportional to the severity of ascites (surrogate marker of portal hypertension), measured GFR stages and MELD-Na score categories. Higher myo-inositol levels observed in subjects with high liver and kidney disease severity compared with those with low disease severity might be either secondary to renal dysfunction due to impaired myo-inositol catabolism as mentioned above or myo-inositol efflux to extracellular compartment secondary to hyperammonemia. By performing proton MR spectroscopy in 14 subjects with cirrhosis without overt encephalopathy, Shawcross et al.(44) showed lower cerebral myo-inositol/creatine (precursor of creatinine) ratio in subjects with cirrhosis who showed deterioration in memory test compared to those who did not. Based on those results, the authors suggested that myo-inositol efflux from astrocytes buffered increased hyperammonemia.(44) Therefore, the deterioration in memory test and development of hepatic encephalopathy in hyperammonemia was because of the impaired capacity of the brain to maintain the osmotic equilibrium by sufficient myo-inositol efflux from astrocytes rather than the degree of hyperammonemia per se (44). Our findings are in line with this study.
We found significant fold changes in 3-(4-hydroxyphenyl)lactate, phenyllactate and gamma-glutamylphenylalanine levels in subjects with high liver and kidney disease severity compared with low liver and kidney disease severity. Additionally, increased 3-(4-hydroxyphenyl)lactate and phenyllactate levels were associated with substantial mortality [(3-(4-hydroxyphenyl) lactate, HR=2.63, P=0.0032; phenyllactate, HR=1.56, P=0.0167] (Table 4). Consistent with a previous report of impaired function of phenylalanine hydroxylase in cirrhosis (46), our findings suggest that dysfunction of the phenylalanine hydroxylase pathway exists and results in increased phenylalanine concentrations. Phenylalanine is transformed to tyrosine via phenylalanine hydroxylase which is a rate-limiting enzyme in phenylalanine catabolism.(24–26, 46) In phenylketonuria, which presents with intellectual disabilities and neurological symptoms, there is a deficiency of phenylalanine hydroxylase and therefore, phenylalanine is converted into phenylpyruvic acid instead of tyrosine. (24–26, 47) Phenylpyruvic acid is a precursor of phenyllactate. (24–26) High levels of 3-(4-hydroxyphenyl)lactate and phenyllactate observed in our subjects in high disease severity categories suggest dysfunction of phenylalanine hydroxylase resulting in increased phenylalanine concentrations. Indeed, impaired function of phenylalanine hydroxylase in cirrhosis was previously reported.(46) We also found significant fold changes in the mean values of gamma-glutamylphenylalanine levels when subjects with high liver and kidney disease severity were compared with those with low liver and kidney disease severity. Increased gamma-glutamylphenylalanine levels in subjects with high liver and kidney disease severity could be due to excessive phenylalanine accumulation. Gamma-glutamylphenylalanine is a gamma-glutamyl amino acid that is possibly synthesized from glutathione and phenylalanine via transpeptidation reaction by gamma-glutamyl transpeptidase (GGT).(48) Indeed, in 1970’s, Orlowski and Wilk (48) showed that administration of L-phenylalanine resulted in detection of gamma-glutamylphenylalanine in the liver. Excessive phenylalanine accumulated due to phenylalanine hydroxylase dysfunction would saturate large neutral amino acid transporter (LNAAT) that moves large neutral amino acids across the blood-brain barrier and thereby prevent passage of large neutral amino acids to brain.(47) In 1978, Morgan et al.(49) showed that lower plasma ratio of valine+leucine+isoleucine to phenylalanine+tyrosine correlated with the severity of the liver disease independent of hepatic encephalopathy. McPhail et al.(50) found significantly higher phenylalanine levels in subjects with decompensated cirrhosis who did not survive compared to those with decompensated cirrhosis who survived and healthy controls. As such, we conclude that the accumulation of 3-(4-hydroxyphenyl)lactate and phenyllactate could be an important predictors of mortality and points to specific hypotheses to be tested.
S-adenosylhomocysteine levels were found to be significantly elevated in this study’s subjects with high liver and kidney disease severity and these subjects were 2.73 times more likely to die than subjects with low S-adenosylhomocysteine levels (P=0.0018) (Table 4). This finding suggested that transmethylation reactions were impaired because elevated levels of S-adenosylhomocysteine were shown to inhibit transmethylation reactions.(51–53) The liver is one of the principal organs for methylation reactions producing the large amounts of the S-adenosyl-L-methionine, the key methyl donor.(51–53) DNA methylation is one of the critical steps in protecting against overall genomic hypomethylation, a feature known to predispose to malignant transformation.(51, 52) Inhibition of transmethylation can also occur if the ratio of S-adenosylmethionine: S-adenosylhomocysteine is reduced(51, 54) and may increase hepatic tumor necrosis factor-alpha levels, caspase-8 activity, and contribute to cell death.(54) The pathway analysis showed that methylation was among the five most significant pathways associated with the metabolomic signature (Figure 4).
We also found significantly elevated methionine levels in subjects with MELD-Na between 20 and 40 which suggests down-regulation of MAT1A or dysfunction of the enzyme methionine adenosyltransferase (MAT) (P=0.001). The MAT which synthesizes S-adenosylmethionine from methionine and Adenosine triphosphate (ATP) is the gene product of MAT1A.(51, 52) Interestingly, Lu et al.(51, 55) showed that MAT1A knockout mice developed non-alcoholic steatohepatitis and hepatocellular carcinoma. Considering these findings and previous experimental model studies, supplementation of S-adenosylmethionine in decompensated cirrhosis might be considered.
We found elevated mean betaine (P=0.003) and methionine (P=0.001) levels in subjects with higher MELD-Na scores. We also found increased S-adenosylhomocysteine levels in subjects with high liver and kidney disease severity and this can also be due to down-regulation of the enzyme betaine homocysteine methyltransferase (BHMT). BHMT was shown to be down-regulated in bile duct ligation-induced cirrhosis in rats (56) and down-regulation of BHMT may result in accumulation of betaine and homocysteine, reduction in hepatic S-adenosylmethionine levels, hypomethylated DNA, and altered transcriptional activity.(56) Betaine functions as a methyl donor and methylates homocysteine to generate methionine, a process that produces N, N-dimethylglycine.(27, 51, 56–60) Increased betaine and methionine levels in subjects with cirrhosis at increased risk of death suggests the possibility that BHMT levels, its activity, or that of another methionine cycle enzyme may be altered. In addition to down-regulation of BHMT, up-regulation of BHMT was also reported in cirrhosis.(61) Up-regulation of BHMT may result in depletion of betaine in cirrhosis.(57, 61, 62) Several studies showed a beneficial effect of supplementary betaine in animal models of cirrhosis.(61–63) Betaine supplementation can reduce S-adenosylhomocysteine levels, and increase S-adenosylmethionine.(61, 62) Together, our findings support significant alterations in methylation pathways at multiple enzymatic levels in advanced cirrhosis (Figures 4 and 5) and merit further investigations.
This study is also the first to report the signature metabolites that were independent predictors of measured GFR in cirrhosis (Table 3). We found that all the metabolites in the signature significantly predicted measured GFR (Table 3). Among these metabolites, erythronate, N6-carbamoylthreonyladenosine, 1-methylhistidine, pseudouridine, and N-acetylserine showed stronger associations with measured GFR compared to Cr (Table 3). Erythronate, which is an organic acid that originates from different sources including glycated proteins, ascorbic acid breakdown or oxidation of N-acetyl-D-glucosamine (24–26) showed the strongest association with measured GFR among all metabolites with or without controlling for age, sex and race (P<0.0001). A metabolome-wide study conducted in a general population by Sekula et al.(64) identified significant associations between pseudouridine, erythronate, n-acetylalanine, and myo-inositol and estimated GFR. Similarly, a plasma metabolomic profile of subjects with diabetes mellitus type II showed that pseudouridine and myo-inositol accurately discriminated those who progressed to end-stage renal disease.(65) Tsalik et al.(38) showed that increased plasma concentrations of erythronate, pseudouridine, and N6-carbamoylthreonyladenosine were associated with reduced renal function. Elevated pseudouridine levels were previously reported in uremia.(66) In regards to N6-carbamoylthreonyladenosine, the carbamylation, a form of post-translational modification of amino acids or proteins, was shown to occur frequently in chronic renal failure. (67) In a study conducted in 158 subjects with diabetes mellitus type I, chronic kidney disease stage 3 and proteinuria, N6-carbamoylthreonyladenosine, pseudouridine, and N-acetylserine were shown to predict development of end-stage renal disease.(68) Our results are in line with the findings of these studies (Table 3).
The metabolic signature showed increased levels of several N-acetylated amino acids including N-acetylserine, N-acetylvaline, N-acetylalanine, and N-acetylputrescine in subjects with high liver and kidney disease severity compared with low liver and kidney disease severity. Our results also showed that N-acetylserine, N-acetylalanine, N-acetylvaline, and N-acetylputrescine were independent predictors of GFR. Sekula et al.(64) previously reported that N-acetylalanine was associated with reduced estimated GFR; our findings have confirmed that using measured GFR by iothalamate plasma clearance. Increased urinary excretion of N-acetylated amino acids were reported in patients with a deficiency of aminoacylase 1 (ACY1) that hydrolyzes N-acetylated proteins.(69) ACY1-deficient individuals were reported to have psychomotor delays.(69) It remains to be determined whether there is ACY1 dysfunction in cirrhosis and whether increased concentration of N-acetylated amino acids play a role or contributes in hepatic encephalopathy.
This study is also the first to report several metabolomic predictors of mortality in cirrhosis independent of kidney function (Tables 4 and 5). As mentioned above, glucuronate, 3-(4-hydroxyphenyl)lactate and S-adenosylhomocysteine were significantly associated with mortality independent of kidney disease which itself is a strong predictor of mortality in patients with and without cirrhosis.(70, 71) 3-Ureidopropionate was another metabolite that strongly predicted mortality in cirrhosis independent of kidney disease. Beta-ureidopropionase is a bidirectional enzyme that catalyzes the conversion of 3-ureidopropionate to beta-alanine and/or; beta-alanine to 3-ureidopropionate.(24–26) Deficiency of beta-ureidopropionase, an inborn error in the pyrimidine metabolism, results in elevated 3-ureidopropionate levels and presents with mental and psychomotor retardation.(72) Beta-alanine functions as a GABAA receptor agonist and may increase in hepatic encephalopathy.(73, 74) Based on these findings, significant fold changes in the mean value of 3-ureidopropionate levels in high liver and kidney disease severity categories can be either secondary to dysfunction of the beta-ureidopropionase enzyme or elevated beta-alanine levels in subjects with high liver and kidney disease severity categories or a combination of these two factors. We found increased beta-alanine levels in subjects with cirrhosis and diuretic-refractory ascites compared with those without ascites and with diuretic-sensitive ascites (P=0.001). These findings suggest that increased beta-alanine levels could be the source of elevation in 3-ureidopropionate levels in subjects with high liver and kidney disease severity categories.
When all the metabolites were analyzed in comparison to MELD-Na score classes (Figure 8) and severity of ascites (Figure 9), we observed significantly and concordantly decreased levels of several sphingomyelins, glycerophosphocholines and glycerophosphoethanolamines in subjects with high MELD-Na scores and diuretic-refractory ascites compared with those with lower MELD-Na scores and lower severity of ascites and this may be due to either reduced synthesis or increased breakdown of these molecules. Sphingomyelins play important roles in both membrane stabilization and lipid signaling.(75, 76) In our cohort, the association between increased levels of sphingomyelin and decreased risk of death may suggest a better capacity for maintaining lipid signaling (ceramide utilization), membrane stability, and reduced endoplasmic reticulum stress.(75, 76) Similarly, Zheng et al.(77), reported negative correlation between sphingolipids and MELD score in patients with hepatitis B cirrhosis while another study by Grammaticos et al.(78) showed negative correlation between serum concentrations of long-chain and very long-chain ceramides and severity of cirrhosis. Consistent with our findings, McPhail et al.(50) reported a negative correlation between lysophosphatidylcholines and phosphatidylcholines and mortality, and attributed this to hepatocyte death.
Our exploratory study had two significant strengths including direct measurement of GFR and use of a robust metabolomic platform (Metabolon, Inc. (Durham, NC)), but had several limitations to be subsequently addressed in subsequent studies that will include and larger sample size, internal and external validations cohorts. None of the biomarkers have yet to be serially tested; therefore, it is difficult to ascribe a high degree of confidence to any isolated analytical outcome separate from the multivariate signature. Furthermore, the metabolomic signature described above may not apply to other etiologies of liver disease, beyond our population with cirrhosis studied herein, as it is not yet known how these biomarkers would change. We excluded subjects with cirrhosis with AKI and chronic kidney disease stage 5 because the iothalamate GFR measurements required steady-state renal function that could not be adjusted for. The requirement for steady-state renal function for GFR measurements limited our ability to enroll more subjects with higher MELD-Na scores. Whereas the study was appropriately powered for quantitative clinical variables split over the median value, for categorical variables, some categories were comprised of relatively few patients; a larger cohort might provide additional insights into informative metabolites.
In conclusion, we identified a unique metabolomic signature associated with hepatorenal dysfunction in cirrhosis. Specific pathways including ascorbate and aldarate metabolism, methylation and cellular glucuronidation showed the strongest association with this metabolic signature. Metabolites including 4-acetamidobutanoate, erythronate, pseudouridine, glucuronate, S-adenosylhomocysteine, myo-inositol, glutamylphenylalanine, 3-(4-hydroxyphenyl)lactate, phenyllactate and ureidopropionate along with other metabolites in the signature provide mechanistic insights into altered metabolism in hepatorenal dysfunction in cirrhosis. This study provides the rationale to target several of these newly identified metabolomic biomarkers, either alone or in combination to develop better evaluative approaches and new therapeutics in patients with cirrhosis and renal dysfunction.
BRIEF COMMENTRY.
Background
There is an unmet need to developing accurate biomarkers to conveniently assess renal function and extent of kidney damage in patients with cirrhosis, because serum creatinine alone is insufficient to give true clinical assessments. Specifically, accurate estimation of glomerular filtration rate, differentiation of hepatorenal syndrome from acute tubular necrosis, identification of response to vasoconstrictive treatment, and prediction of recovery of renal function remain major challenges in managing cirrhosis.
Translational Significance
This study provides the rationale to target several of the newly identified metabolites, to develop better evaluative approaches and new therapeutics in patients with cirrhosis and renal dysfunction.
Acknowledgments
Funding
The project described was supported in part by award 5 K23 DK089008-05 from the National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases (to Ayse L. Mindikoglu, M.D., M.P.H.) and its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the NIH. This project was also supported in part by the University of Maryland School of Medicine, Department of Medicine funds, University of Maryland Clinical Translational Science Institute, University of Maryland General Clinical Research Center, and NIH Public Health Service grant P30DK056338, which funds the Texas Medical Center Digestive Diseases Center and its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the NIH. Cristian Coarfa, Ph.D. was partially supported by Cancer Prevention and Research Institute of Texas (CPRIT) awards RP170295 and RP170005, and by the NIH/NIDDK award R01DK111522. Antone Opekun, M.S., PA-C was partially supported by an unrestricted institutional grant from DR and GL Laws.
The authors thank Heather L. Rebuck, MT (ASCP), CLS (NCA) and Sharon Y. Huang, MT for analysis of blood samples and University of Maryland General Clinical Research Center Staff.
All authors have read the journal’s authorship agreement and that the manuscript has been reviewed by and approved by all named authors.
All authors have read the journal’s policy on disclosure of potential conflicts of interest.
Footnotes
Meeting Material:
Preliminary results of this study were presented as an abstract at Digestive Disease Week®, Chicago, IL, May 6–9, 2017: Mindikoglu AL, Opekun AR, Coarfa C, Putluri N, Devaraj S, Sheikh-Hamad D, Vierling JM, Goss JA, Rana A, Sood GK, Jalal PK, Inker LA, Mohney RP, Tighiouart H, Christenson RH, Dowling TC, Weir MR, Seliger SL, Hutson WR, Howell C, Raufman JP, Magder LS. Robust Metabolomic Signature is Associated with Altered Renal Hemodynamics in Patients with Cirrhosis. Gastroenterology 2017;152:5, S1044.
Author Contributions:
A.L.M. designed and conducted the study, drafted the manuscript, analyzed data and performed critical review and revision of the manuscript for important intellectual content.
A.R.O. critically reviewed and revised the manuscript for important intellectual content.
T.C.D. analyzed GFR data and critically reviewed the manuscript for important intellectual content.
L.S.M. analyzed data and critically reviewed the manuscript for important intellectual content.
C.C. performed metabolomic and pathway statistical analyses, constructed volcano plots, heatmaps and pathway network, critically reviewed and revised the manuscript for important intellectual content.
All other authors critically reviewed the manuscript for important intellectual content.
Disclosures:
Ayse L. Mindikoglu, M.D., M.P.H. A provisional patent application (serial no: 62/442,479) is filed with the US patent office on 01/05/2017 (Metabolomic Markers to Predict Mortality in Patients with Cirrhosis). A second provisional patent application (serial no. 62/586,966) is filed with the U.S. Patent and Trademark Office on November 16, 2017, entitled “Metabolomic Biomarkers of Hepatorenal Dysfunction and Mortality in Patients with Cirrhosis”.
Lesley A. Inker, M.D., M.S. reports funding to Tufts Medical Center for research and contracts with the National Institutes of Health, National Kidney Foundation, Pharmalink, Gilead Sciences, Otsuka, Reata, for consulting to Tricida Inc and has a provisional patent: Coresh, Inker and Levey filed 8/15/2014 – “Precise estimation of glomerular filtration rate from multiple biomarkers” PCT/US2015/044567. Tufts Medical Center, John Hopkins University and Metabolon, Inc. have a collaboration agreement to develop a product to estimate GFR from a panel of markers.
Robert P. Mohney, Ph.D. is an employee of Metabolon, Inc. and, as such, has affiliations with or financial involvement with Metabolon, Inc., Tufts Medical Center, and John Hopkins University have a collaboration agreement to develop a product to estimate GFR from a panel of markers. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. None of the other authors have disclosures to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mindikoglu AL, Pappas SC. New Developments in Hepatorenal Syndrome. Clin Gastroenterol Hepatol. 2018;16(2):162–177. doi: 10.1016/j.cgh.2017.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim WR, Biggins SW, Kremers WK, Wiesner RH, Kamath PS, Benson JT, et al. Hyponatremia and mortality among patients on the liver-transplant waiting list. N Engl J Med. 2008;359(10):1018–26. doi: 10.1056/NEJMoa0801209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malinchoc M, Kamath PS, Gordon FD, Peine CJ, Rank J, ter Borg PC. A model to predict poor survival in patients undergoing transjugular intrahepatic portosystemic shunts. Hepatology. 2000;31(4):864–71. doi: 10.1053/he.2000.5852. [DOI] [PubMed] [Google Scholar]
- 4.Francoz C, Prie D, Abdelrazek W, Moreau R, Mandot A, Belghiti J, et al. Inaccuracies of creatinine and creatinine-based equations in candidates for liver transplantation with low creatinine: impact on the model for end-stage liver disease score. Liver Transpl. 2010;16(10):1169–77. doi: 10.1002/lt.22128. [DOI] [PubMed] [Google Scholar]
- 5.Mindikoglu AL, Dowling TC, Weir MR, Seliger SL, Christenson RH, Magder LS. Performance of chronic kidney disease epidemiology collaboration creatinine-cystatin C equation for estimating kidney function in cirrhosis. Hepatology. 2014;59(4):1532–42. doi: 10.1002/hep.26556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francoz C, Nadim MK, Baron A, Prie D, Antoine C, Belghiti J, et al. Glomerular filtration rate equations for liver-kidney transplantation in patients with cirrhosis: validation of current recommendations. Hepatology. 2014;59(4):1514–21. doi: 10.1002/hep.26704. [DOI] [PubMed] [Google Scholar]
- 7.Nair S, Verma S, Thuluvath PJ. Pretransplant renal function predicts survival in patients undergoing orthotopic liver transplantation. Hepatology. 2002;35(5):1179–85. doi: 10.1053/jhep.2002.33160. [DOI] [PubMed] [Google Scholar]
- 8.Inker LA, Schmid CH, Tighiouart H, Eckfeldt JH, Feldman HI, Greene T, et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med. 2012;367(1):20–9. doi: 10.1056/NEJMoa1114248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mindikoglu AL, Dowling TC, Magder LS, Christenson RH, Weir MR, Seliger SL, et al. Estimation of Glomerular Filtration Rate in Patients With Cirrhosis by Using New and Conventional Filtration Markers and Dimethylarginines. Clin Gastroenterol Hepatol. 2016;14(4):624–32e2. doi: 10.1016/j.cgh.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cholongitas E, Ioannidou M, Goulis I, Chalevas P, Ntogramatzi F, Athanasiadou Z, et al. Comparison of creatinine and cystatin formulae with 51 Chromium-ethylenediaminetetraacetic acid glomerular filtration rate in patients with decompensated cirrhosis. J Gastroenterol Hepatol. 2017;32(1):191–8. doi: 10.1111/jgh.13446. [DOI] [PubMed] [Google Scholar]
- 11.Kalim S, Rhee EP. An overview of renal metabolomics. Kidney Int. 2017;91(1):61–9. doi: 10.1016/j.kint.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss RH, Kim K. Metabolomics in the study of kidney diseases. Nat Rev Nephrol. 2011;8(1):22–33. doi: 10.1038/nrneph.2011.152. [DOI] [PubMed] [Google Scholar]
- 13.Tzoulaki I, Ebbels TM, Valdes A, Elliott P, Ioannidis JP. Design and analysis of metabolomics studies in epidemiologic research: a primer on -omic technologies. Am J Epidemiol. 2014;180(2):129–39. doi: 10.1093/aje/kwu143. [DOI] [PubMed] [Google Scholar]
- 14.Mindikoglu AL, Dowling TC, Wong-You-Cheong JJ, Christenson RH, Magder LS, Hutson WR, et al. A pilot study to evaluate renal hemodynamics in cirrhosis by simultaneous glomerular filtration rate, renal plasma flow, renal resistive indices and biomarkers measurements. Am J Nephrol. 2014;39(6):543–52. doi: 10.1159/000363584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans AM, Bridgewater BR, Liu Q, Mitchell MW, Robinson RJ, Dai H, et al. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics. 2014;4(2):1. [Google Scholar]
- 16.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2(1):9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dehaven CD, Evans AM, Dai H, Lawton KA. In: Software Techniques for Enabling High-Throughput Analysis of Metabolomic Datasets, Metabolomics. Roessner Ute., editor. InTech; 2012. Available from: https://mts.intechopen.com/books/metabolomics/software-techniques-for-enabling-high-throughput-analysis-on-metabolomic-datasets. [DOI] [Google Scholar]
- 18.Armitage EG, Godzien J, Alonso-Herranz V, Lopez-Gonzalvez A, Barbas C. Missing value imputation strategies for metabolomics data. Electrophoresis. 2015;36(24):3050–60. doi: 10.1002/elps.201500352. [DOI] [PubMed] [Google Scholar]
- 19.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109(7):2625–9. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.SAS software. Http://www.Sas.Com/. The data analysis for this paper was generated using SAS software, Version 9.2 of the SAS System for Windows. Copyright © 2002–2008 SAS Institute Inc. SAS and all other SAS Institute Inc. product or service names are registered trademarks or trademarks of SAS Institute Inc., Cary, NC, USA.
- 21.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. URL https://www.R-project.org/ [Google Scholar]
- 22.Kettner NM, Voicu H, Finegold MJ, Coarfa C, Sreekumar A, Putluri N, et al. Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell. 2016;30(6):909–24. doi: 10.1016/j.ccell.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.York B, Li F, Lin F, Marcelo KL, Mao J, Dean A, et al. Pharmacological inhibition of CaMKK2 with the selective antagonist STO-609 regresses NAFLD. Sci Rep. 2017;7(1):11793. doi: 10.1038/s41598-017-12139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37(Database issue):D603–10. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35(Database issue):D521–6. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. [Accessed on March 28, 2016];KEGG: Kyoto Encyclopedia of Genes and Genomes. Available at http://www.genome.jp/kegg/
- 28.Reactome. [Accessed on July 14]; Available at http://www.reactome.org/PathwayBrowser/
- 29.Biocarta. [Accessed on July 14]; Available at https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways.
- 30.Gene Ontology Consortium. [Accessed on July 14]; Available at http://www.geneontology.org/
- 31.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones E, Oliphant E, Peterson P, et al. SciPy: open source Scientific tools for Python [Online] 2001 Available at: http://www.scipy.org/. Accessed June 28, 2017.
- 33.Millman KJ, Aivazis M. Python for scientists and engineers. Comput Sci Eng. 2011;13:9–12. doi: 10.1109/MCSE.2011.36. [DOI] [Google Scholar]
- 34.Oliphant TE. Python for scientific computing. Comput Sci Eng. 2007;9:10–20. doi: 10.1109/MCSE.2007.58. [DOI] [Google Scholar]
- 35.van der Walt S, Colbert SC, Varoquaux G. The NumPy Array: a structure for efficient numerical computation. Comput Sci Eng. 2011;13:22–30. doi: 10.1109/MCSE.2011.37. [DOI] [Google Scholar]
- 36.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9:90–5. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 38.Tsalik EL, Willig LK, Rice BJ, van Velkinburgh JC, Mohney RP, McDunn JE, et al. Renal systems biology of patients with systemic inflammatory response syndrome. Kidney Int. 2015;88(4):804–14. doi: 10.1038/ki.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones EA, Mullen KD. Theories of the pathogenesis of hepatic encephalopathy. Clin Liver Dis. 2012;16(1):7–26. doi: 10.1016/j.cld.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Croze ML, Soulage CO. Potential role and therapeutic interests of myo-inositol in metabolic diseases. Biochimie. 2013;95(10):1811–27. doi: 10.1016/j.biochi.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Furlan V, Demirdjian S, Bourdon O, Magdalou J, Taburet AM. Glucuronidation of drugs by hepatic microsomes derived from healthy and cirrhotic human livers. J Pharmacol Exp Ther. 1999;289(2):1169–75. [PubMed] [Google Scholar]
- 42.Adams MR, Grubb SM, Hamer A, Clifford MN. Colorimetric enumeration of Escherichia coli based on beta-glucuronidase activity. Appl Environ Microbiol. 1990;56(7):2021–4. doi: 10.1128/aem.56.7.2021-2024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitkanen E. Changes in serum and urinary myo-inositol levels in chronic glomerulonephritis. Clin Chim Acta. 1976;71(3):461–8. doi: 10.1016/0009-8981(76)90097-8. [DOI] [PubMed] [Google Scholar]
- 44.Shawcross DL, Balata S, Olde Damink SW, Hayes PC, Wardlaw J, Marshall I, et al. Low myo-inositol and high glutamine levels in brain are associated with neuropsychological deterioration after induced hyperammonemia. Am J Physiol Gastrointest Liver Physiol. 2004;287(3):G503–9. doi: 10.1152/ajpgi.00104.2004. [DOI] [PubMed] [Google Scholar]
- 45.Suhre K, Schwartz JE, Sharma VK, Chen Q, Lee JR, Muthukumar T, et al. Urine Metabolite Profiles Predictive of Human Kidney Allograft Status. J Am Soc Nephrol. 2016;27(2):626–36. doi: 10.1681/ASN.2015010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishii T, Furube M, Hirano S, Takatori K, Iida K, Kajiwara M. Evaluation of 13C-phenylalanine and 13C-tyrosine breath tests for the measurement of hepatocyte functional capacity in patients with liver cirrhosis. Chem Pharm Bull (Tokyo) 2001;49(12):1507–11. doi: 10.1248/cpb.49.1507. [DOI] [PubMed] [Google Scholar]
- 47.Pietz J, Kreis R, Rupp A, Mayatepek E, Rating D, Boesch C, et al. Large neutral amino acids block phenylalanine transport into brain tissue in patients with phenylketonuria. J Clin Invest. 1999;103(8):1169–78. doi: 10.1172/JCI5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orlowski M, Wilk S. Intermediates of the gamma-glutamyl cycle in mouse tissues. Influence of administration of amino acids on pyrrolidone carboxylate and gamma-glutamyl amino acids. Eur J Biochem. 1975;53(2):581–90. doi: 10.1111/j.1432-1033.1975.tb04101.x. [DOI] [PubMed] [Google Scholar]
- 49.Morgan MY, Milsom JP, Sherlock S. Plasma ratio of valine, leucine and isoleucine to phenylalanine and tyrosine in liver disease. Gut. 1978;19(11):1068–73. doi: 10.1136/gut.19.11.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McPhail MJ, Shawcross DL, Lewis MR, Coltart I, Want EJ, Antoniades CG, et al. Multivariate metabotyping of plasma predicts survival in patients with decompensated cirrhosis. J Hepatol. 2016;64:1058–67. doi: 10.1016/j.jhep.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu SC, Mato JM. Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol. 2005;35(3):227–34. doi: 10.1016/j.alcohol.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 52.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92(4):1515–42. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finkelstein JD. Methionine metabolism in mammals. J Nutr Biochem. 1990;1(5):228–37. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- 54.Song Z, Zhou Z, Uriarte S, Wang L, Kang YJ, Chen T, et al. S-adenosylhomocysteine sensitizes to TNF-alpha hepatotoxicity in mice and liver cells: a possible etiological factor in alcoholic liver disease. Hepatology. 2004;40(4):989–97. doi: 10.1002/hep.20412. [DOI] [PubMed] [Google Scholar]
- 55.Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A. 2001;98(10):5560–5. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forestier M, Banninger R, Reichen J, Solioz M. Betaine homocysteine methyltransferase: gene cloning and expression analysis in rat liver cirrhosis. Biochim Biophys Acta. 2003;1638(1):29–34. doi: 10.1016/s0925-4439(03)00037-1. [DOI] [PubMed] [Google Scholar]
- 57.Craig SA. Betaine in human nutrition. Am J Clin Nutr. 2004;80(3):539–49. doi: 10.1093/ajcn/80.3.539. [DOI] [PubMed] [Google Scholar]
- 58.Avila MA, Berasain C, Torres L, Martin-Duce A, Corrales FJ, Yang H, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33(6):907–14. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 59.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barak AJ, Beckenhauer HC, Junnila M, Tuma DJ. Dietary betaine promotes generation of hepatic S-adenosylmethionine and protects the liver from ethanol-induced fatty infiltration. Alcohol Clin Exp Res. 1993;17(3):552–5. doi: 10.1111/j.1530-0277.1993.tb00798.x. [DOI] [PubMed] [Google Scholar]
- 62.Barak AJ, Beckenhauer HC, Mailliard ME, Kharbanda KK, Tuma DJ. Betaine lowers elevated s-adenosylhomocysteine levels in hepatocytes from ethanol-fed rats. J Nutr. 2003;133(9):2845–8. doi: 10.1093/jn/133.9.2845. [DOI] [PubMed] [Google Scholar]
- 63.Erman F, Balkan J, Cevikbas U, Kocak-Toker N, Uysal M. Betaine or taurine administration prevents fibrosis and lipid peroxidation induced by rat liver by ethanol plus carbon tetrachloride intoxication. Amino Acids. 2004;27(2):199–205. doi: 10.1007/s00726-004-0105-5. [DOI] [PubMed] [Google Scholar]
- 64.Sekula P, Goek ON, Quaye L, Barrios C, Levey AS, Romisch-Margl W, et al. A Metabolome-Wide Association Study of Kidney Function and Disease in the General Population. J Am Soc Nephrol. 2016;27(4):1175–88. doi: 10.1681/ASN.2014111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int. 2014;85(5):1214–24. doi: 10.1038/ki.2013.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dzurik R, Lajdova I, Spustova V, Opatrny K., Jr Pseudouridine excretion in healthy subjects and its accumulation in renal failure. Nephron. 1992;61(1):64–7. doi: 10.1159/000186836. [DOI] [PubMed] [Google Scholar]
- 67.Gillery P, Jaisson S. Post-translational modification derived products (PTMDPs): toxins in chronic diseases? Clin Chem Lab Med. 2014;52(1):33–8. doi: 10.1515/cclm-2012-0880. [DOI] [PubMed] [Google Scholar]
- 68.Niewczas MA, Mathew AV, Croall S, Byun J, Major M, Sabisetti VS, et al. Circulating Modified Metabolites and a Risk of ESRD in Patients With Type 1 Diabetes and Chronic Kidney Disease. Diabetes Care. 2017;40(3):383–90. doi: 10.2337/dc16-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sass JO, Mohr V, Olbrich H, Engelke U, Horvath J, Fliegauf M, et al. Mutations in ACY1, the gene encoding aminoacylase 1, cause a novel inborn error of metabolism. Am J Hum Genet. 2006;78(3):401–9. doi: 10.1086/500563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cholongitas E, Senzolo M, Patch D, Shaw S, O’Beirne J, Burroughs AK. Cirrhotics admitted to intensive care unit: the impact of acute renal failure on mortality. Eur J Gastroenterol Hepatol. 2009;21(7):744–50. doi: 10.1097/MEG.0b013e328308bb9c. [DOI] [PubMed] [Google Scholar]
- 71.Deo R, Fyr CL, Fried LF, Newman AB, Harris TB, Angleman S, et al. Kidney dysfunction and fatal cardiovascular disease--an association independent of atherosclerotic events: results from the Health, Aging, and Body Composition (Health ABC) study. Am Heart J. 2008;155(1):62–8. doi: 10.1016/j.ahj.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 72.van Kuilenburg AB, Meinsma R, Beke E, Assmann B, Ribes A, Lorente I, et al. beta-Ureidopropionase deficiency: an inborn error of pyrimidine degradation associated with neurological abnormalities. Hum Mol Genet. 2004;13(22):2793–801. doi: 10.1093/hmg/ddh303. [DOI] [PubMed] [Google Scholar]
- 73.Sergeeva OA. GABAergic transmission in hepatic encephalopathy. Arch Biochem Biophys. 2013;536(2):122–30. doi: 10.1016/j.abb.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 74.Tiedje KE, Stevens K, Barnes S, Weaver DF. Beta-alanine as a small molecule neurotransmitter. Neurochem Int. 2010;57(3):177–88. doi: 10.1016/j.neuint.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 75.Slotte JP. Biological functions of sphingomyelins. Prog Lipid Res. 2013;52(4):424–37. doi: 10.1016/j.plipres.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 76.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139–50. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 77.Zheng SJ, Qu F, Li JF, Zhao J, Zhang JY, Liu M, et al. Serum sphingomyelin has potential to reflect hepatic injury in chronic hepatitis B virus infection. Int J Infect Dis. 2015;33:149–55. doi: 10.1016/j.ijid.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 78.Grammatikos G, Ferreiros N, Waidmann O, Bon D, Schroeter S, Koch A, et al. Serum Sphingolipid Variations Associate with Hepatic Decompensation and Survival in Patients with Cirrhosis. PLoS One. 2015;10(9):e0138130. doi: 10.1371/journal.pone.0138130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple hypothesis testing. Journal of the Royal Statistical Society, Series B. 1995;57:289–300. [Google Scholar]
- 80.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of the American Statistical Association. 1958;53:457–481. [Google Scholar]
- 81.Cox DR. Regression Models and Life-Tables. Journal of the Royal Statistical Society, Series B. 1972;34(2):187–220. [Google Scholar]