Letter to the editor
Polymyxins, like colistin (polymyxin E), are a group of cationic antimicrobial cyclic polypeptide, which have been extensively-used as prophylactic feed additives in animal production since the 1960s [1]. A strong association has especially been drawn between antimicrobial use and resistance in poultry and pig farms [2–5]. The advent and rise of multi-drug resistant bacteria has now prompted the re-introduction of colistin as a last-resort treatment option in human medicine [6,7]. However, the recent emergence and diversity of plasmid-borne mobile colistin resistance determinants (mcr-1 to mcr-5) in Enterobacteriaceae has severely challenged its use in a clinical setting [8–12]. In the past two years, mcr-1 has been detected in over 40 countries across 5 of 7 continents worldwide [13–15]. Except rare cases of chromosomally-integrated mcr-1 [16,17], its prevalent transmission relies on the transfer by diversified plasmids of different replication incompatibilities [18,19].
Mechanistically, the mcr-1 gene that encodes phosphoethanolamine transferase mediates the modification of the lipopolysaccharide layer (LPS) of the outer membrane of Gram-negative bacteria through the addition of phosphoethanolamine (PEA) to the 1 (or 4′)-phosphate position of lipid A moieties [20,21]. This reduces the affinity of polymyxin antibiotics to their primary target, the LPS layer. Since its discovery during routine surveillance in China [8], increasingly-accumulated evidence suggested the presence of mcr-1-bearing bacteria in food producing animals and humans across the world. To the best of our knowledge, the leading two types of mcr-1-harboring plasmids referred to IncI2 [8,18,22,23] and IncX4 [24–26], have greatly facilitated global dissemination of mcr-1 colistin resistance [14,27]. In Pakistan, the mcr-1 gene has been identified in Escherichia coli (E. coli) isolates from human [28], wildlife [29] and a broiler suffering from colibacillosis [30]. However, little is known about the prevalence of mcr-1 and its genetic environment in commensal E. coli isolates from poultry in Pakistan.
From December 2016 to January 2017, cloacal swabs from a total of 100 healthy broiler chicken were obtained from four commercial farms (n = 25 each) in the Faisalabad region of Pakistan. To screen the colistin resistant E. coli, all the samples were seeded directly onto MacConkey agar supplemented with 4 μg/ml of colistin and were incubated at 37°C for 24 hours. Of 100 birds, colistin resistant E. coli were found in only 8 (8%) samples. A single colony of E. coli was selected per sample and identified using API 20E biochemical strips (bioMérieux, Marcy l'Etoile, France). The presence of mcr-1 gene was confirmed among all 8 E. coli isolates by conventional PCR as we recently conducted [19,31]. Subsequently, the minimum inhibitory concentration (MIC) of colistin among these strains was tested by micro-dilution according to the guidelines of Clinical and Laboratory Standards Institute [32,33]. The mcr-1-positive E. coli gave MIC of colistin between 2–8 μg/ml (Table 1). Plasmids were extracted from mcr-1-positive E. coli using alkaline lysis method. To elucidate the genetic context of mcr-1 on these plasmids, the conventional multiplex PCR with 7 primer sets was performed (Table S1) as we recently described [3,19]. The plasmids isolated from the different strains had unexpectedly similar PCR profiles with the exception of pPK112 which lacks the tnpA loci (Fig. 1B). Genetic context of mcr-1 shows that all the plasmids lack the insertion element ISApl1 (Fig. 1A), which has been responsible for insertion of mcr-1 in previous studies [34]. Also, these plasmids (Fig. 1A) are identical to the mcr-1-carrying plasmid pE15017 isolated in China [19,22]. These bacteria were subjected to multi-locus sequence typing analyses with the Warwick method (http://mlst.warwick.ac.uk/mlst/). Diversified sequence types were detected, namely, ST10, ST2847, ST155, ST361 and ST6395. Evidently, all strains belonged to different STs with the exception of two strains of ST361 (Table 1). Despite that none of these STs have been reported from Pakistan in the past, ST10 and ST155 have been reported in mcr-1-harboring E. coli isolated from chicken in China [17,35].
Table 1.
Characteristics of mcr-1-positive E. coli isolates from healthy broilers in Faisalabad, Pakistan.
| Strains | Source | Date | MIC (μg/ml) | MLST | Farm no. |
|---|---|---|---|---|---|
| PK102 | cloacal | 27/12/2016 | ≥8 | ST10 | 1 |
| PK103 | cloacal | 27/12/2016 | ≥4 | ST2847 | 1 |
| PK105 | cloacal | 27/12/2016 | ≥8 | ST155 | 1 |
| PK107 | cloacal | 27/12/2016 | ≥8 | New ST | 1 |
| PK109 | cloacal | 27/12/2016 | ≥4 | ST361 | 1 |
| PK110 | cloacal | 30/01/2017 | ≥4 | ST6395 | 3 |
| PK111 | cloacal | 30/01/2017 | ≥2 | ST361 | 3 |
| PK112 | cloacal | 30/01/2017 | ≥8 | New ST | 3 |
Figure 1.
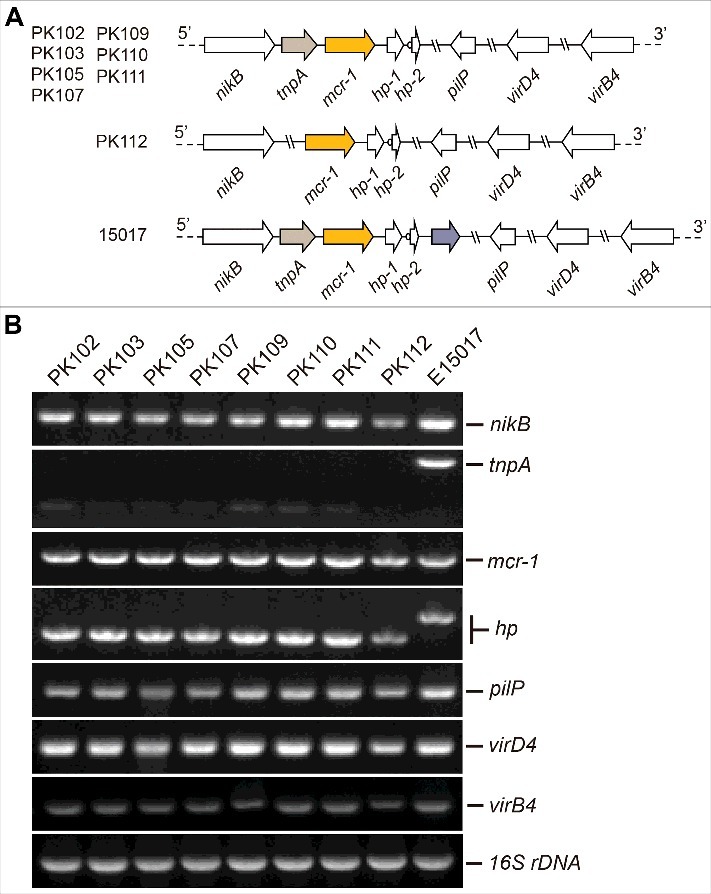
Genetic analyses of mcr-1-harboring plasmids in this study A. Scheme of different mcr-1-bearing plasmids B. PCR assays of mcr-1 and neighboring loci in plasmids 16S rDNA is specific to the E. coli specie.
Subsequently, a representative mcr-1-carrying plasmid pPK105 (Table 1) was subjected to whole genome sequencing using the method of Illumina HiSeq X-ten. The plasmid sequences were annotated by RAST, and the genome maps were drawn with the Circos program. As a result, the genome size of pPK105 was determined to be 60.499 kb (Acc. no.: MG808035, Fig. 2A), encoding hundreds of open reading frames with a GC content of 42.3%. Unlike the prevalent IncX4 type plasmid reported in Pakistan, Plasmid Finder at the web server (https://cge.cbs.dtu.dk/services/PlasmidFinder/) indicated that pPK105 is a member of IncI2-type plasmid family (Fig. 2B). Notably, mcr-1 is the only resistance gene detected in pPK105 (Fig. 2B). It is quite different from the scenarios observed in the other mcr-1-containing Pakistan isolate coharboring ESBL (extended-spectrum β-lactamase gene) and heavy-metal resistance. Although that ISApl1 sequence is missing, pPK105 retains the two conjugative genes vir and pil (Fig. 2). Intriguingly, we noticed that the mcr-1 gene in pPK105 is next to a sdrl gene, a serine-aspartate repeat surface protein known to bind collagen (Fig. 2B). However, its relevance remains unclear. Nevertheless, it has been shown earlier that ISApl1 transposon element is highly unstable in IncI2 plasmid [36].
Figure 2.
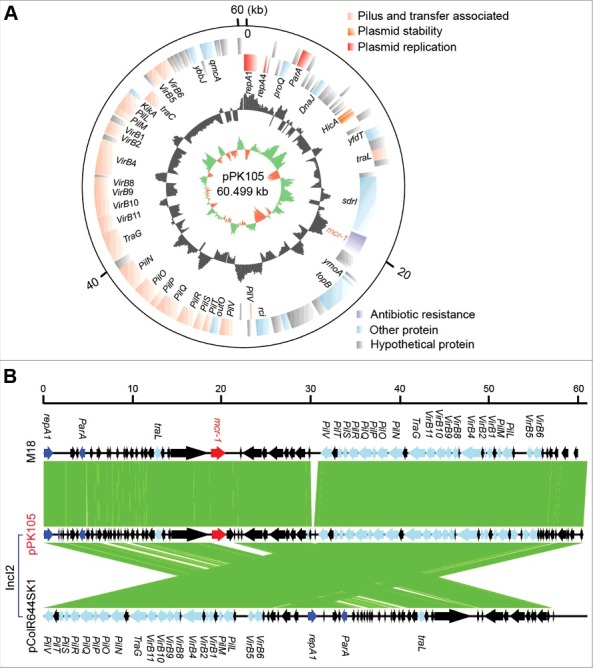
The representative mcr-1-bearing plasmid in a colistin resistant E. coli isolate PK105 A. Diagram for the mcr-1-positive plasmid (pPK105) that exists in the E. coli isolate PK105 B. Colinear comparison of the IncI2-type plasmid pPK105 with two closely-related plasmids mcr-1-postive M18 and mcr-1-negative pColR644SK1.
For further investigation, the chemical modification of lipid A by the mcr-1 protein product MCR-1, the bacterial LPS was extracted as we conducted earlier [20,37] and then subjected to matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) [38]. In comparison to E. coli MG1655 (m/z 1796.29, Fig. 3A), a shift in the predominant lipid A species (m/z 1920.136, Fig. 3A) was observed in the mcr-1-expressing PK105 strain (Δmass is close to 123) corresponding to the addition of a PEA moiety (Fig. 3B). This new peak corresponds to a single modification that may occur at either the 1 or the 4′ position (Fig. 3B). This highlighted that surface remodeling by the mcr-1-encoding phosphoethanolamine transferase contributes to the resultant colistin resistance [14].
Figure 3.
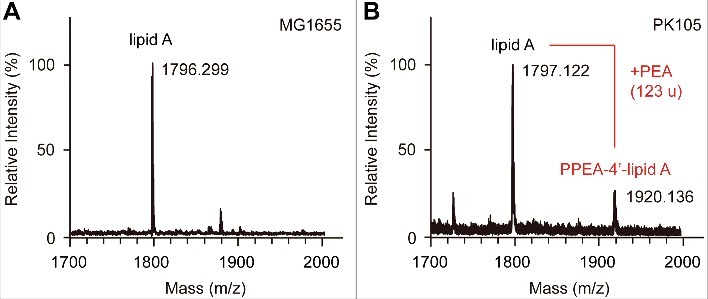
MALDI-TOF MS analyses of lipid A pools of E. coli strains with (or without) mcr-1 A. MS profile of the LPS-lipid A in the negative control strain E. coli MG1655 B. MS spectrum of the LPS-lipid A in PK105, a representative strain of E. coli carrying mcr-1. A single modification may occur at the 1 (or 4′) position. The position indicated here is suggestive [20].
The discovery of the mcr-1 gene, prompted a shift in focus from chromosomal mutations causing colistin-resistance to a transmissible plasmid-borne colistin resistance determinant. In addition to clinical isolates in humans, antimicrobial surveillance programs in Europe have identified mcr-1 in commensal bacterial populations from broilers, pigs and turkeys [39]. This study shows a similar threatening scenario in the Faisalabad region of Pakistan where high rates of mcr-1 positive E. coli were identified in healthy broilers. Retrospectively, the discovery of diverse clonal backgrounds of E. coli harboring the plasmid-borne mcr-1 is similar to scenarios observed earlier in Chinese poultry.
Similarly, data on global population structure of mcr-1-positive E. coli showed large diversity in STs but limited plasmid types, particularly with regional spread of IncHI2 plasmids in Europe and IncI2 in Asia [14]. This indicated the possible spread of a single mcr-1 colistin resistance gene across large geographical distances. It seems likely that farm animals act as a reservoir for the genetic diversity of mcr-1 [40].
Supplementary Material
Funding Statement
This work was supported by National Key R&D Program of China (2017YFD0500202, YF), National Key Basic Research Program of China (2016YFC1200100, YF), and National Natural Science Foundation of China (31570027 & 81772142, YF). Dr. Feng is a recipient of the “Young 1000 Talents” Award.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
YF and MM designed and supervised this project; YF, MM, JL, SL, SS, RW, and J-X L performed experiments; YF, JL, MM, and SL analyzed the data and prepared figures; YF, MM, SS and SL drafted this manuscript.
References
- [1].Li J, Nation RL, Turnidge JD, et al.. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6:589–601. [DOI] [PubMed] [Google Scholar]
- [2].Nguyen NT, Nguyen HM, Nguyen CV, et al.. Use of colistin and other critical antimicrobials on pig and chicken farms in Southern Vietnam and its association with resistance in commensal escherichia coli bacteria. Appl Environ Microbiol. 2016;82:3727–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li Z, Tan C, Lin J, et al.. Diversified variants of the mcr-1-carrying plasmid reservoir in the swine lung microbiota. Sci China Life Sci. 2016;59:971–3. [DOI] [PubMed] [Google Scholar]
- [4].Gao R, Li Y, Lin J, et al.. Unexpected complexity of multidrug resistance in the mcr-1-harbouring Escherichia coli. Sci China Life Sci. 2016;59:732–4. [DOI] [PubMed] [Google Scholar]
- [5].Zhang H, Seward CH, Wu Z, et al.. Genomic insights into the ESBL and MCR-1-producing ST648 Escherichia coli with multi-drug resistance. Sci Bull (Beijing). 2016;61:875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Falagas ME, Kasiakou SK. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis. 2005;40:1333–41. [DOI] [PubMed] [Google Scholar]
- [7].Caniaux I, van Belkum A, Zambardi G, et al.. MCR: modern colistin resistance. Eur J Clin Microbiol Infect Dis. 2017;36:415–420. [DOI] [PubMed] [Google Scholar]
- [8].Liu YY, Wang Y, Walsh TR, et al.. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8. [DOI] [PubMed] [Google Scholar]
- [9].Yin W, Li H, Shen Y, et al.. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. 2017;8:e00543–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carattoli A, Villa L, Feudi C, et al.. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22:pii 30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Borowiak M, Fischer J, Hammerl JA, et al.. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72:3317–3324. [DOI] [PubMed] [Google Scholar]
- [12].Xavier BB, Lammens C, Ruhal R, et al.. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27). doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- [13].Sun J, Zhang H, Liu YH, et al.. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018; pii: S0966-842X(18)30042-8. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- [14].Feng Y. Transferability of MCR-1/2 polymyxin resistance: Complex dissemination and genetic mechanism. ACS Infect Dis. 2018;4:291–300. [DOI] [PubMed] [Google Scholar]
- [15].Wang X, Zhang H, Sun J, et al.. The MCR-1 colistin resistance: A new challenge to global public health. Chin Sci Bull. 2017;62:1018–1029. [Google Scholar]
- [16].Sun J, Li XP, Fang LX, et al.. Co-occurence of mcr-1 in the chromosome and IncHI2 plasmid: persistence of colistin resistance in Escherichia coli. Int J Antimicrob Agents. 2018; pii: S0924-8579(18)30010-4. doi: 10.1016/j.ijantimicag.2018.01.007. [DOI] [PubMed] [Google Scholar]
- [17].Zhou HW, Zhang T, Ma JH, et al.. Occurrence of plasmid- and chromosome-encoded mcr-1 in water-borne Enterobacteriaceae in China. Antimicrob Agents Chemother. 2017;61(8). pii. e00017-17. doi: 10.1128/AAC.00017-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang Q, Sun J, Li J, et al.. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome. 2017;5:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ye H, Li Y, Li Z, et al.. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio. 2016;7:e00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xu Y, Lin J, Cui T, et al.. Mechanistic insights into transferable polymyxin resistance among gut bacteria. J Biol Chem. 2018;293:4350–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sun J, Xu Y, Gao R, et al.. Deciphering MCR-2 colistin resistance. mBio. 2017;8:pii e00625-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gao R, Hu Y, Li Z, et al.. 2016 Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog. 2016;12:e1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang Y, Tian GB, Zhang R, et al.. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 2017;17:390–399. [DOI] [PubMed] [Google Scholar]
- [24].Wang Q, Sun J, Ding Y, et al.. Genomic insights into mcr-1-positive plasmids carried by colistin-resistant Escherichia coli isolates from inpatients. Antimicrob Agents Chemother. 2017;61:pii e00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sun J, Fang LX, Wu Z, et al.. Genetic analysis of the IncX4 plasmids: Implications for a unique pattern in the mcr-1 acquisition. Sci Rep. 2017;7:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li R, Xie M, Zhang J, et al.. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2017;72:393–401. [DOI] [PubMed] [Google Scholar]
- [27].Matamoros S, van Hattem JM, Arcilla MS, et al.. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep. 2017;7:15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mohsin M, Raza S, Roschanski N, et al.. Description of the first Escherichia coli clinical isolate harboring the colistin resistance gene mcr-1 from the indian subcontinent. Antimicrob Agents Chemother. 2017;61:pii e01945-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mohsin M, Raza S, Roschanski N, et al.. First description of plasmid-mediated colistin-resistant extended-spectrum beta-lactamase-producing Escherichia coli in a wild migratory bird from Asia. Int J Antimicrob Agents. 2016;48:463–4. [DOI] [PubMed] [Google Scholar]
- [30].Azam M, Ehsan I, Sajjad Ur R, et al.. Detection of the colistin resistance gene mcr-1 in avian pathogenic Escherichia coli in Pakistan. J Glob Antimicrob Resist. 2017;11:152–153. [DOI] [PubMed] [Google Scholar]
- [31].Wang Q, Li Z, Lin J, et al.. Complex dissemination of the diversified mcr-1-harbouring plasmids in Escherichia coli of different sequence types. Oncotarget. 2016;7:82112–82122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Galani I, Kontopidou F, Souli M, et al.. Colistin susceptibility testing by Etest and disk diffusion methods. Int J Antimicrob Agents. 2008;31:434–9. [DOI] [PubMed] [Google Scholar]
- [33].Bakthavatchalam YD, Veeraraghavan B. Challenges, issues and warnings from CLSI and EUCAST working group on polymyxin susceptibility testing. J Clin Diagn Res. 2017;11:DL03–DL04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Snesrud E, McGann P, Chandler M. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio. 2018;9(1). pii. e02381-17. doi: 10.1128/mBio.02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang YQ, Li YX, Song T, et al.. Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob Agents Chemother. 2017;61:pii e01204-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Snesrud E, Ong AC, Corey B, et al.. Analysis of serial isolates of mcr-1-positive Escherichia coli reveals a highly active ISApl1 transposon. Antimicrob Agents Chemother. 2017;61:pii e00056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xu Y, Wei W, Lei S, et al.. An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance. mBio. 2018;9:e02317–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu YY, Chandler CE, Leung LM, et al.. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrob Agents Chemother. 2017;61:pii e00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Perrin-Guyomard A, Bruneau M, Houee P, et al.. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Euro Surveill. 2016;21:6–8. [DOI] [PubMed] [Google Scholar]
- [40].Monte DF, Mem A, Fernandes MR, et al.. Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob Agents Chemother. 2017;61:pii e02718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


