ABSTRACT
Unlike the current injectable influenza vaccines, intranasally administered influenza vaccines induce influenza virus-specific IgA antibodies in the local respiratory mucosa as well as IgG antibodies in the systemic circulation. Our previous study showed that after five volunteers underwent intranasal administration with inactivated H3N2 or H5N1 vaccines, their IgA antibodies on the upper respiratory tract were present as monomers, dimers, and multimers (trimers and tetramers). Moreover, the multimers associated with the highest virus neutralizing activity. However, it has remained elusive whether a more practical intranasal vaccination strategy could induce the high-performance IgA multimers in the nasal mucosa. In the present study, volunteers were administered with two doses of the intranasal trivalent whole-virus inactivated influenza vaccine and showed that in nasal wash samples the amount of multimeric IgA correlated positively with virus neutralizing titers, indicating that the multimeric IgA antibodies play an important role in the antiviral activity at the nasal mucosa. Surface plasmon resonance analysis of the binding dynamics of nasal wash derived IgA monomers, dimers, and multimers against recombinant trimeric influenza virus HA showed that sample fractions containing IgA multimers dissociated from HA less well than sample fractions without IgA multimers. Thus, IgA multimers may “stick” to the antigen more tightly than the other structures. In summary, intranasal administration of two doses of multivalent inactivated influenza vaccines induced multimeric IgA. Multimerization of mucosal IgA antibodies conferred higher neutralizing activity against viruses in the nasal mucosa, possibly by increasing their cohesion to virus antigens.
KEYWORDS: Secretory IgA antibody, multimeric SIgA antibody, influenza virus, intranasal inactivated influenza vaccine, upper respiratory mucosa
Introduction
Seasonal influenza is a significant and ongoing global problem since it causes approximately 3–5 million severe cases and 1 million deaths annually.1 The influenza virus targets the epithelial cells that line the respiratory tract. Once it enters the cells, the innate immune system of the human host becomes activated and produces local inflammatory responses. These responses eventually promote the activation of the adaptive immune system, particularly the humoral arm.2 The most important of these humoral responses are virus-specific secretory IgA antibodies, which are located on the surface of the local respiratory tract mucosal epithelium, and IgG antibodies, which circulate in the blood.3 While these immune responses are weak and delayed after the first encounter with the virus, subsequent encounters mobilize a strong and rapid humoral immune response that quickly eliminates the virus. Since vaccines can mimic the first infection with the virus and induce robust humoral immune responses when the actual virus is encountered, they constitute a particularly potent and cost-effective countermeasure against influenza.
The majority of the current vaccines against influenza are detergent-disrupted split virus vaccines that are administered by either intramuscular or subcutaneous injection. However, these vaccines only induce virus-specific IgG antibodies in the circulation; they do not produce the secretory IgA antibodies that coat the upper respiratory tract mucosal surface, which form the first line of defense against the virus and prevent it from infecting the respiratory tract epithelial cells.2-4 This suggests that the protection provided by influenza vaccines could be improved by ensuring that both systemic IgG and local neutralizing IgA responses are generated. Intranasally administered inactivated influenza vaccines have been developed to meet this need5-8 and in 2002, the Food and Drug Administration (FDA) approved a live attenuated vaccine (Flumist, MedImmune Vaccines, Inc., USA) for intranasal administration.9 This vaccine confers strong protective efficacy compared to the parenteral inactivated vaccines. However, due to the safety concerns, it is not licensed for children under the age of two and adults over 49 years of age. Furthermore, it has been recently reported that the effectiveness of this vaccine against the H1N1 pandemic virus strain has fallen significantly.10-12 Thus, an alternative intranasal vaccine that could induce effective mucosal immunity to a broad range of people is in need. In a previous study, it was demonstrated that inactivated whole virus influenza vaccines (WIIVs) that were administered via the intranasal route could induce both sufficient systemic antibody responses (as defined by the European Medicines Agency) and local antibody responses in the nasal mucosa.13 Therefore, WIIV has been a promising candidate of a novel intranasal influenza vaccine.
Recently, we reported the results of our study on the characteristics of the local IgA antibody responses that are induced by intensive (five vaccinations) intranasal vaccination of volunteers with inactivated H3N2 or H5N1 whole-virion influenza vaccines. We found that the IgA collected from the upper respiratory tract was present in the form of monomers, dimers, and multimers (trimers and tetramers). Of these structures, the multimeric IgA structures neutralized the virus most potently.14 However, it is not yet clear whether other intranasal vaccine strategies, such as a more practical intranasal vaccination using multivalent inactivated influenza vaccines can also produce anti-viral IgA multimers contributing to efficient virus neutralization on the upper respiratory mucosa. Studies that characterize the functions of these IgA multimers in detail are warranted because they will not only show precisely how intranasally administered mucosal vaccines lead to protective immunity, but they will also promote the development of better vaccine strategies.
In the present study, the structures and functions of the IgA antibodies on the human upper respiratory tract mucosa that were induced by two doses of intranasal inactivated influenza vaccines were determined. Thus, nasal antibodies were collected from 21 healthy adult volunteers who were vaccinated intranasally with the trivalent whole-virus inactivated influenza vaccine (WIIV), and the molecular architectures of local IgA, their antiviral functions were evaluated.
Results
Neutralizing titers of serum and nasal wash samples collected from intranasal vaccine-administered volunteers
In total, 47 healthy adult volunteers (mean age and range, 36.2 and 18–65 years; 22 males and 25 females) participated in the study. All were immunized intranasally twice at 3 week intervals with the WIIV that was based on three vaccine strains, namely, A/Texas/50/2012 (H3N2), A/California/7/2009 (H1N1)pdm09, and B/Massachusetts/2/2012 (Yamagata-lineage). Serum and nasal wash samples were obtained before the first vaccination, just before the second vaccination 3 weeks later, and 3 weeks after the second vaccination. Each sample was subjected to a neutralization test (NT) assay employing all three vaccine strains as well as A/Victoria/361/2011 (H3N2), which was the vaccine strain that had been selected in the previous season.
Compared to their pre-vaccination serum samples, the vaccinated subjects exhibited significantly higher NT activities in their 6 week serum samples against A/Texas/50/2012 (H3N2), A/California/7/2009 (H1N1)pdm09, and A/Victoria/361/2011 (H3N2), but not B/Massachusetts/2/2012 (Yamagata lineage) (Fig. 1A). By contrast, compared to the pre-vaccination nasal wash samples, the 6 week nasal wash samples had significantly higher NT activities against all four strains, including the influenza B virus (Fig. 1B). The poor serum response to B/Massachusetts/2/2012 (Yamagata lineage) reflects the poor immunogenicity of influenza B viruses in intranasal influenza vaccines. The poor antigenicity of these viruses is also a problem for the current injectable influenza vaccines.15 Notably, the vaccinations affected the nasal antibody responses more profoundly than the systemic antibody responses. In addition, these NT activities of the serum and nasal wash samples had strong positive correlation especially against H3N2 viruses (Fig. 1C), indicating that the NT assays using nasal wash samples standardized based on total protein concentration could fairly evaluate the anti-viral mucosal antibody responses in upper respiratory tract. This suggests that measuring the antibody titers in the upper respiratory mucosa is a sensitive and informative way to evaluate the immune responses induced by intranasal vaccination.
Figure 1.
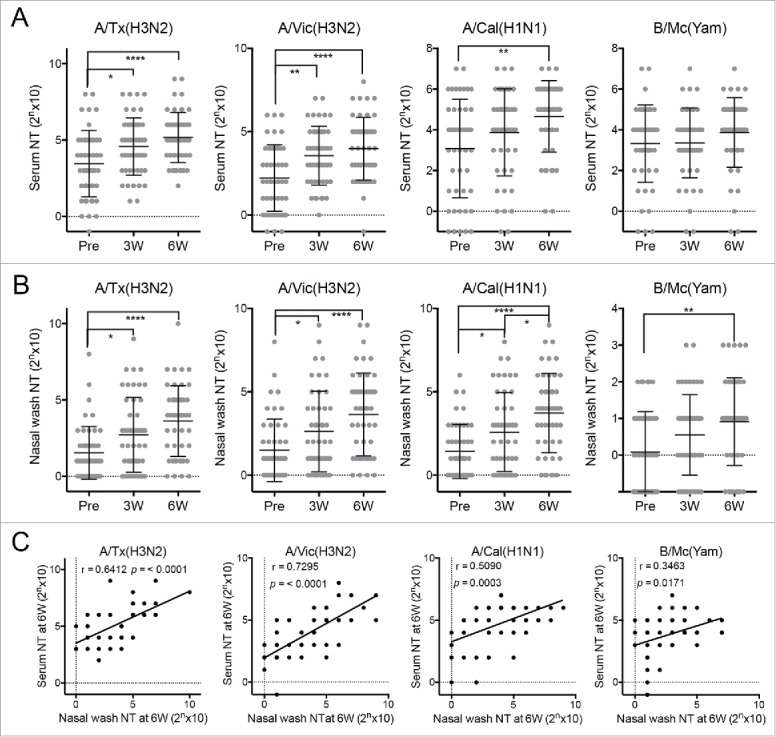
Neutralizing test (NT) titers against four influenza virus strains in (A) serum and (B) nasal wash samples collected from 47 healthy adult volunteers before (Pre) and 3 (3W) and 6 weeks (6W) after intranasal vaccination. A/Tx(H3N2): A/Texas/50/2012 (H3N2) (a vaccine strain); A/Vic(H3N2): A/Victoria/361/2011 (H3N2) (the vaccine strain from the previous year); A/Cal(H1N1): A/California/7/2009 (H1N1)pdm09 (a vaccine strain); B/Mc (Yam): B/Massachusetts/2/2012 (Yamagata-lineage) (a vaccine strain). Each dot represents the NT titer of the individual volunteer-derived samples. The data are expressed as scatter plots with mean ± SD. *p < 0.05, **p < 0.01, ****p < 0.0001, as determined by One-way ANOVA. (C) The correlation between serum and nasal wash neutralizing test (NT) titers at 6 week against various virus strains were examined using Pearson's correlation coefficient (r).
Structural characterization of vaccine-induced human nasal antibodies
The fact that intranasal influenza vaccination had a greater effect on the local antibody responses than on the systemic immune responses suggests that it is important to understand precisely how intranasal influenza vaccines induce protective immunity. We reported recently that the human nasal IgA antibodies that are generated by intranasal influenza virus vaccination have at least five quaternary structures, namely, monomers, dimers, trimers, tetramers, and larger polymeric structures. We also showed that the trimers and other polymeric structures had greater antiviral activity than the monomers and dimers.14 However, volunteers who were enrolled in the previous study were intensely immunized (five doses of 45 μg HA/dose vaccine at three week intervals) with a monovalent antigen. Therefore, it had remained elusive whether the induction of nasal IgA multimers in the upper respiratory tract could also be seen in individuals vaccinated with a more practical intranasal vaccination protocol (e.g., two doses of multivalent inactivated vaccines). To address the issue, the nasal wash IgA antibodies from 21 participants who provided large volumes (500 ml/participant) of nasal wash fluid 3 weeks after the second vaccination were sharply concentrated and purified. Fig. 2A shows that, on average, 67.5% of the immunoglobulins in these samples were IgA. Most of the remaining immunoglobulins were IgG. The IgA molecules were then affinity-purified and separated by size exclusion chromatography. Due to the limited amount of purified nasal IgA antibodies (100 – 400 µg/participant), fractionation by size exclusion chromatography was restricted to just three fractions, namely, Fr1, Fr2, and Fr3 based on the shape of the chromatogram consisting of one main peak accompanied by two shoulders (Fig. 2B). Quantification of the IgA in these fractions showed that the highest levels of IgA were in Fr2 (51.6%), followed by Fr3 (36.5%). Fr1 (11.8%) had the lowest levels of IgA (Fig. 2C). Since dimeric IgA is known to be the main population in upper respiratory tract IgA,16 we assumed that Fr2 mainly contained dimeric IgA.
Figure 2.
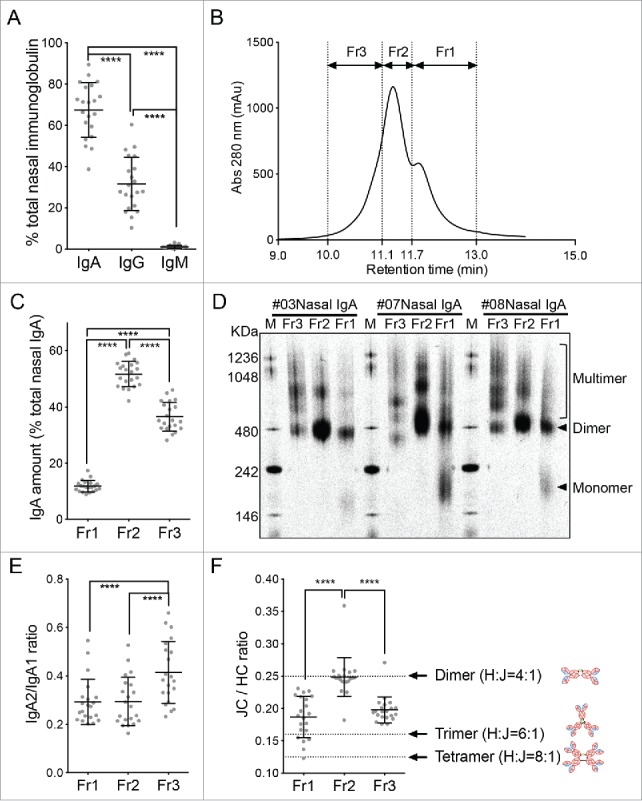
Biochemical characterization of the human nasal wash-derived IgA samples. (A) Relative proportions of IgA, IgG, and IgM in the total immunoglobulin of the concentrated nasal washes collected from 21 individuals 3 weeks after the second vaccination. These 21 subjects were selected from the original cohort of 47 because they provided large amounts of wash samples. Each dot represents an individual volunteer-derived sample. (B) The purified nasal wash IgA samples of the 21 individuals were subjected to size exclusion chromatography and fractionated into fractions Fr1, Fr2, and Fr3 based on the three peaks on the chromatogram. A representative chromatogram is shown. (C) Relative proportions of the total nasal IgA that is contained in the Fr1, Fr2, and Fr3 fractions. (D) Blue native-PAGE analysis of the Fr1, Fr2, and Fr3 fractions from three representative individuals (#03, #07, and #08). Arrowheads indicate monomers and dimers while the square bracket indicates multimers. (E) IgA2/IgA1 ratio in the Fr1, Fr2, and Fr3 fractions from the 21 individuals, as determined by IgA subunit quantification by LC-MS. (F) J chain (JC)/ IgA heavy chain (HC) ratio of the Fr1, Fr2, and Fr3 fractions of the 21 volunteers, as determined by IgA subunit quantification by LC-MS. Dotted lines indicate the theoretical JC/HC values of the dimers (0.25), trimers (0.167), and tetramers (0.125). The data are expressed as scatter plots with the mean ± SD. **p < 0.01, ***p < 0.001, ****p < 0.0001, as determined by One-way ANOVA.
To determine the molecular architecture(s) of the IgA molecules in the three fractions, we subjected the fractions obtained from three of the 21 volunteers (subjects #03, #07, and #08) to blue native-PAGE analysis. This analysis showed that Fr1 contained monomers, dimers, and multimers that were larger than dimers. By contrast, Fr2 and Fr3 did not contain monomers and were mostly composed of dimers and multimers. However, the frequency of each IgA structure within each fraction differed depending on the individual (Fig. 2D). Therefore, for the purpose of evaluating the relative abundance of each IgA molecular architecture, we here established a LC-MS method by selecting two tryptic marker peptides for each IgA subunit so that respective subunits can be separately quantified even between IgA1 and IgA2 heavy chains (Table 1). In addition, we used stable isotope-labeled peptides corresponding to the marker peptides as internal standards to achieve highly accurate quantification. All fractionated IgA samples collected from the 21 individuals were subjected to IgA subunit quantification by LC-MS. Heavy chain (HC) analysis of the IgA samples showed that Fr3 contained significantly more IgA2 subclass HC than Fr1 and Fr2 (Fig. 2E). Furthermore, the mean J chain (JC):HC (total IgA heavy chain) abundance ratio of Fr2 was approximately 0.25; since that is the theoretical JC:HC ratio of dimers, it seems that Fr2 largely contained dimers. Since Fr1 had a smaller mean ratio (0.19) than Fr2, suggesting that Fr1 is a mixture of dimers and monomers not containing JC. By contrast, the mean JC/HC ratio of Fr3 (0.20) was between the theoretical JC/HC ratios of dimers (0.25) and trimers/tetramers (0.125–0.167) (Fig. 2F). This suggests that Fr3 is a mixture of dimers and multimers. Two additional facts suggest that Fr3 may be enriched in multimeric IgA compared to the other two fractions: 1) Fr3 has more IgA2 than the other two fractions (Fig. 2E), and subclass IgA2 forms multimers more efficiently than subclass IgA117; and 2) Fr3 had a markedly lower JC/HC ratio than both Fr1 and Fr2 (Fig. 2F).
Table 1.
Marker peptides used to quantitate IgA subunits.
| Maker | Peptide sequence | m/z | z |
|---|---|---|---|
| HC (IgA1 specific)_1 | NFPPSQDASGDLYTTSSQLTLPATQCLAGK | 1056.8360 ± 0.02 | 3 |
| HC (IgA1 specific)_1-IS | NFPPSQDASGDLYTTSSQLTLPATQCLAGK | 1059.5074 ± 0.02 | 3 |
| HC (IgA1 specific)_2 | DASGVTFTWTPSSGK | 770.8603 ± 0.02 | 2 |
| HC (IgA1 specific)_2-IS | DASGVTFTWTPSSGK | 774.8674 ± 0.02 | 2 |
| HC (IgA2 specific)_1 | NFPPSQDASGDLYTTSSQLTLPATQCPDGK | 1066.1555 ± 0.02 | 3 |
| HC (IgA2 specific)_1-IS | NFPPSQDASGDLYTTSSQLTLPATQCPDGK | 1068.8269 ± 0.02 | 3 |
| HC (IgA2 specific)_2 | DASGATFTWTPSSGK | 756.8446 ± 0.02 | 2 |
| HC (IgA2 specific)_2-IS | DASGATFTWTPSSGK | 760.8517 ± 0.02 | 2 |
| HC (IgA common)_1 | WLQGSQELPR | 607.3126 ± 0.02 | 2 |
| HC (IgA common)_1-IS | WLQGSQELPR | 612.3167 ± 0.02 | 2 |
| HC (IgA common)_2 | YLTWASR | 448.7276 ± 0.02 | 2 |
| HC (IgA common)_2-IS | YLTWASR | 453.7317 ± 0.02 | 2 |
| J_1 | SSEDPNEDIVER | 695.3028 ± 0.02 | 2 |
| J_1-IS | SSEDPNEDIVER | 700.3069 ± 0.02 | 2 |
| J_2 | CYTAVVPLVYGGETK | 828.9115 ± 0.02 | 2 |
| J_2-IS | CYTAVVPLVYGGETK | 832.9185 ± 0.02 | 2 |
K: 13C615N2-lysine. R: 13C615N4-arginine. C: carbamidomethylated cysteine.
Relationship between IgA molecular architecture and virus-neutralizing activity
Since it has been revealed previously that polymerization of human mucosal IgA antibodies associates with enhanced antiviral activity,14 we hypothesized that the nasal wash samples of the 21 volunteers that have the largest proportions of multimeric IgA relative to total immunoglobulin (i.e., IgA + IgG + IgM) will also have the highest antiviral activity against at least some of the four influenza strains. To test this, we performed various correlation analyses (Table 2). First, there were no significant correlations between the nasal IgG proportions in total nasal immunoglobulin and either the serum NT or nasal wash NT activities (Fig. 3A). Second, there were no significant correlations between the total IgA proportions in total nasal immunoglobulin and either serum NT or nasal wash NT activities (Fig. 3B). We then assessed the relationship between the proportion of the IgA of the nasal wash samples that is in the Fr1, Fr2, and Fr3 fractions, and the serum and nasal wash NT activities of each individual in order to reveal which IgA fraction contributed most to virus neutralization activity in nasal mucosa. The Fr1 and Fr2 IgA proportions did not correlate with either serum or nasal wash NT activities (Fig. 3C and D). The Fr3 proportion (which has abundant multimers) also did not correlate significantly with serum NT activities (Fig. 3E, left plot). However, it did correlate positively with nasal wash NT activities against virus strain A/Victoria/361/2011 (H3N2) (Fig. 3E, right plot). Thus, a relatively high abundance of Fr3 IgA antibodies associated with enhanced NT activity the total nasal antibodies. This suggests that IgA multimers play an important role in the antiviral activity at the human upper respiratory mucosa.
Table 2.
Correlation analyses between the nasal immunoglobulin proportions and the serum NT or nasal wash NT activities.
| Correlations between nasal immunoglobulin and serum NT, Pearson's correlation coefficient (P value) |
Correlations between the immunoglobulin and nasal NT, Pearson's correlation coefficient (P value) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Nasal immunoglobulin isotype | A/Cal(H1N1) | A/Tx(H3N2) | A/Vic(H3N2) | B/Mc(Yam) | A/Cal(H1N1) | A/Tx(H3N2) | A/Vic(H3N2) | B/Mc(Yam) |
| IgA_Fr3 | 0.30 (0.18) | 0.17 (0.47) | 0.20 (0.39) | 0.05 (0.84) | 0.38 (0.08) | 0.34 (0.14) | 0.46 (0.03)* | 0.08 (0.73) |
| IgA_Fr2 | 0.13 (0.59) | -0.13 (0.58) | -0.06 (0.79) | 0.01 (0.96) | 0.23 (0.31) | 0.01 (0.96) | 0.07 (0.77) | 0.29 (0.20) |
| IgA_Fr1 | -0.09 (0.71) | -0.09 (0.69) | 0.01 (0.95) | 0.05 (0.83) | 0.25 (0.27) | -0.11 (0.64) | -0.04 (0.86) | 0.10 (0.68) |
| IgA_total | 0.19 (0.41) | -0.03 (0.91) | 0.04 (0.86) | 0.03 (0.89) | 0.33 (0.14) | 0.13 (0.58) | 0.22 (0.33) | 0.23 (0.31) |
| IgG | -0.20 (0.39) | 0.00 (0.99) | -0.07 (0.77) | -0.05 (0.84) | -0.36 (0.11) | -0.15 (0.51) | -0.25 (0.28) | -0.24 (0.30) |
*p < 0.05.
Figure 3.
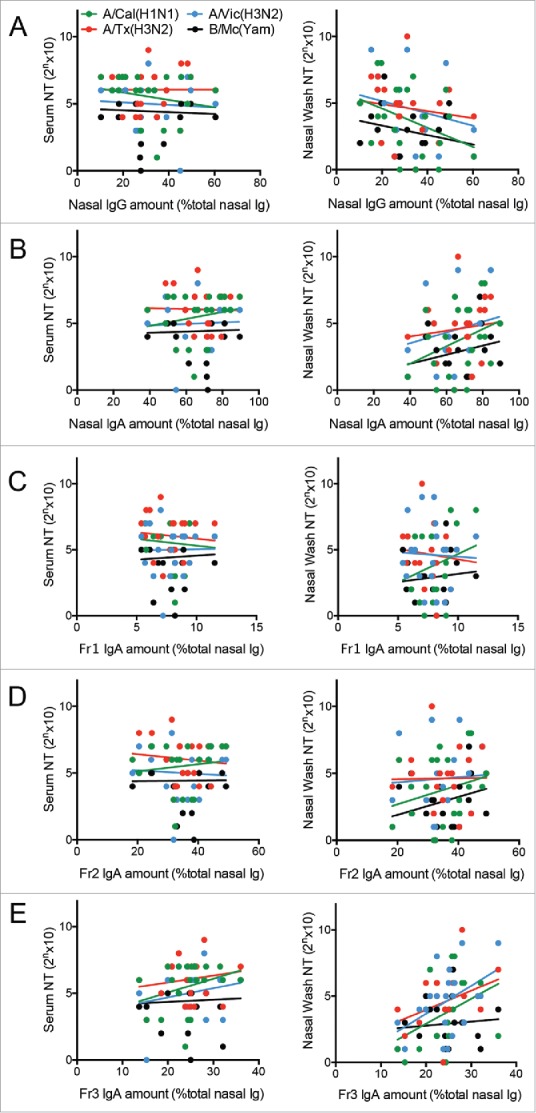
Correlation analyses assessing the relationship between serum and nasal wash neutralizing test (NT) titers against various virus strains and (A) the proportion of total nasal immunoglobulin (Ig) that is IgG, (B) the proportion of total nasal Ig that is IgA, (C) the proportion of total nasal IgA that is in Fr1, (D) the proportion of total nasal IgA that is in Fr2, and (E) the proportion of total nasal IgA that is in Fr3. Each dot represents an IgA sample from each of the 21 subjects who provided large amounts of nasal wash sample. The viruses that were tested were A/California/7/2009 (H1N1)pdm09 (green, A/Cal(H1N1)), A/Victoria/210/2009 (H3N2) (blue, A/Vic(H3N2)), A/Texas/50/2012 (H3N2) (red, A/Tx(H3N2)), and B/Massachusetts/2/2012 (Yamagata-lineage) (black, B/Mc (Yam)). The only positive correlation was that between the proportion of total nasal IgA that is in Fr3 and the nasal wash NT titers against A/Victoria/361/2011 (H3N2).
Functional characterization of vaccine-induced human nasal IgA antibodies
From our previous14 and present study, it can be said that IgA polymerization is associated with enhancement of anti-viral activities of IgA antibodies. However, the mechanism underlying this phenomenon remains poorly understood. The question is, do antibody clones with higher antiviral ability simply tend to polymerize and thus polymerization itself has no effect on antibody function? Or does polymerization in fact increase the antiviral efficacy of the antibody clones? We previously used virus antigen (hemagglutinin [HA])-specific ELISA to show that multimeric IgA bound more strongly to virus antigen than the dimeric and monomeric IgA molecules.14 While ELISA is a simple and easy way of evaluating the total binding activity of a sample of interest, it cannot clearly quantitate the antigen-specific antibody clones in the samples or determine the quality of their antibody functions. Therefore, we used surface plasmon resonance (SPR) analysis to characterize the HA-binding of the IgA molecules in the three fractions in detail. Since SPR assays using polyclonal antibody samples require substantial amounts of IgA, we purified the Fr1, Fr2, and Fr3 IgA fractions from the ten volunteers mentioned above who provided large amounts of nasal IgA (subjects #03, #07, #08, #10, #14, #15, #16, #25, #26, and #30). Fig. 4A shows their relative levels of monomers, dimers, and multimers on the basis of their blue native-PAGE data shown in Fig. 2D. In these three representative subjects (#03, #07, and #08), Fr2 mainly consisted of dimers (with some multimers), Fr3 mainly consisted of multimers (with some dimers), and Fr1 consisted of dimers (with some monomers and multimers) (Fig. 4A). These fractionated IgA antibodies containing different structures were subjected to SPR analysis and the binding dynamics against the recombinant trimeric HA of A/Victoria/361/2011 (H3N2) were examined. SPR analysis clearly showed that Fr2 (mainly dimers with some multimers) and Fr3 (enriched with multimers) dissociated from HA less well than Fr1 (mainly dimers with some monomers and multimers) (Fig. 4B). For statistical comparison, dissociation efficiency was defined as the reciprocal of the area under the dissociation curve (AUC) of the SPR sensorgram. As a result, the dissociation efficiencies of Fr2 and Fr3 were significantly lower than that of Fr1, indicating that the dissociation rate of IgA multimers (included in Fr2 and Fr3) was slower than monomers and dimers (the main components of Fr1) (Fig. 4C). Thus, as the molecular size of IgA increases, the avidity of the IgA molecule for HA improves. This is likely to enhance overall antibody function.
Figure 4.
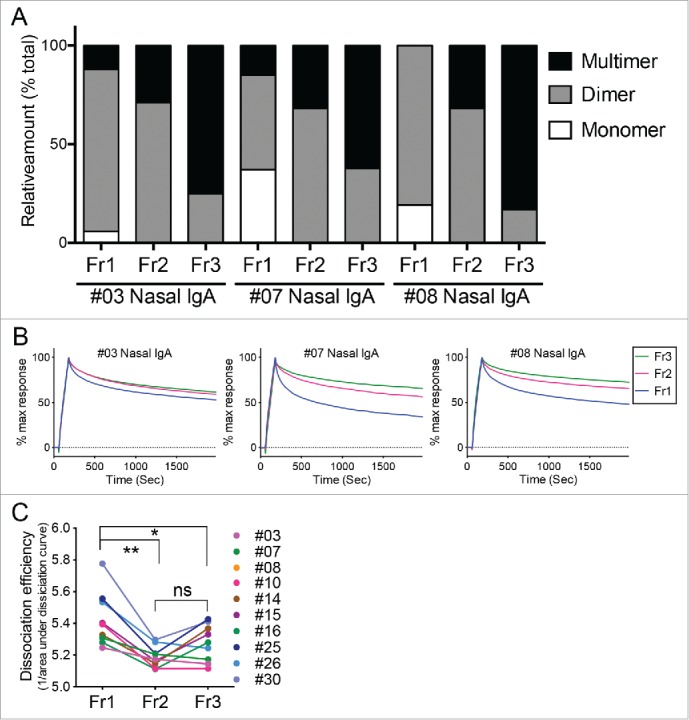
Binding dynamics of human nasal IgA samples to HA. (A) Relative proportions of IgA monomers, dimers, and multimers in the Fr1, Fr2, and Fr3 fractions from three representative individuals (#03, #07, and #08). These calculations were performed on the basis of density quantification of the bands on blue native-PAGE gels (see Fig. 2D). (B) These Fr1, Fr2, and Fr3 IgA samples were subjected to surface plasmon resonance analysis of their binding dynamics to the recombinant trimeric hemagglutinin from A/Victoria/361/2011 (H3N2). Sensorgrams were y-axis adjusted (0 = baseline, 100 = binding) to allow comparisons between samples in terms of the dissociation rate of IgA from HA. (C) Fr1, Fr2 and Fr3 IgA samples derived from 10 volunteers were subjected to SPR analysis. Dissociation efficiency of Fr2 and Fr3, which were defined as the reciprocal of area under the dissociation curve (AUC) of SPR response, were significantly lower than that of Fr1. The data are expressed as scatter plots with mean ± SD. *p < 0.05, **p < 0.01, as determined by paired t-test. ns, not significant.
Discussion
This study evaluated the induction of multimeric IgA antibodies in the human upper respiratory mucosa by two doses of the intranasal trivalent WIIV and its significance in antiviral function. This is the first study to characterize in detail the nasal IgA samples from a relatively large group of individuals who were vaccinated by a practical intranasal vaccine strategy using multivalent WIIV. We found that multimeric IgA antibodies play an important role in the antiviral mechanism induced by just two doses of multivalent WIIV in humans.
Mucosal IgA is a first-line defense against viruses that blocks infection before it even starts.2-4 This effect of intranasal vaccines is one of the advantages of these vaccines that should never be overlooked. However, efficacy evaluation and approval of vaccines is currently done by hemagglutination inhibition tests (HAI) using serum collected from vaccinated individuals.18 This method can measure the systemic magnitude of antibody response against recent vaccination or infection history.19 However, the level of antibody response induced in the local mucosal tissues of the upper respiratory-tract cannot be evaluated by this method. In the current study, we found that compared to the systemic antibody responses, the intranasal trivalent WIIV administration more profoundly elevated the local mucosal antibody responses against various influenza virus strains, including an influenza B virus strain (Fig. 1). This strongly indicates that evaluation of the mucosal IgA populations in the upper respiratory tract as well as their antiviral effects is essential in order to develop new and improved intranasal vaccines.
In our previous study,14 we have shown that intranasal administration of either H3N2 or H5N1 WIIV generates monomeric, dimeric, and multimeric IgA antibodies against influenza viruses at the human upper respiratory tract mucosa. However, volunteers who were enrolled in the previous study were intensely immunized (five doses of 45 μg HA/dose vaccine at three week intervals), and the purified IgA samples were pooled prior to purification, which made it impossible to evaluate binding kinetics of IgA antibodies of each individual using SPR. Therefore, it had been still remained unclear whether the induction of functional IgA multimers in the upper respiratory mucosa is an universal phenomenon in individuals vaccinated with more practical doses and formulations of intranasal inactivated influenza vaccines. In the current study, volunteers were vaccinated intranasally with the trivalent WIIV (only two doses of 45 μg HA/dose for each strain at three week intervals), and nasal wash samples were individually processed following collection from volunteers. As a result, it was confirmed that IgA multimers with high anti-viral activity were induced in volunteers who were administered with the intranasal trivalent influenza vaccine, indicating the generality of this phenomenon (Fig. 4). Furthermore, our analysis of the nasal wash samples of 21 vaccinated volunteers showed that the relative abundance of these different architectures of nasal IgA antibodies in the 21 purified nasal wash samples varied depending on the individual (Fig. 2D and Fig. 4A). Correlation analyses then showed that the proportion of multimeric IgA in the 21 nasal wash samples relative to total immunoglobulin correlated positively and significantly with their NT activity against one of the strains being tested in the NT assay (Fig. 3). This suggests that the efficient induction of IgA multimers may increase antiviral titers, thereby leading to higher vaccine efficacy. Moreover, the frequency of IgA multimers on the nasal mucosa could serve as an index of the immune responses that are induced by intranasal vaccines.
To determine why the multimers associated with better nasal wash antiviral activities, we assessed the binding dynamics of the various molecular architectures of the nasal IgA antibodies against recombinant trimeric influenza virus HA. Since this analysis required large amounts of IgA, only ten volunteer samples could be analyzed. Nevertheless, the analysis showed that the Fr2 and F3 fraction that contained IgA multimers were less able to dissociate from HA than the fractions (Fr1) that consisted of dimers and monomers (Fig. 4B). This is consistent with our recent study assessing the binding of multimeric IgA to HA by using atomic force microscopy.14 This study showed that tetramers, for example, have four radial regions protruding from a central region; consequently, the molecule has a four-leafed clover shape. When the round HA molecule binds, it associates with the radial regions of the IgA architecture and is passed continuously, albeit at intervals, from one radial region to the other. This, together with our current observations, suggests that IgA multimers “stick” more potently to the antigen than dimers and monomers, making it harder for the IgA to dissociate from its antigen after binding. However, since the current assay was conducted with human polyclonal IgA, it remains possible that higher avidity antibody clones are those that tend to form multimers and that the association between IgA polymerization and high antiviral activity is merely accidental. To determine the true impact of IgA polymerization on IgA molecule avidity, it will be necessary to compare monomer, dimers, and multimers of monoclonal IgA species that possess identical Fab regions.
In summary, the current study confirmed that although the efficiency differs by individual, IgA multimers, which possess high anti-viral activities could be generally induced in the human upper respiratory mucosa by only two doses of intranasal trivalent WIIV. Analysis of purified nasal IgA samples collected from intranasal trivalent WIIV recipients showed that polymerization enhances the antiviral activity of IgA on the human upper respiratory tract mucosa. Our SPR analyses suggest that this is mediated by increased cohesion of the polymeric particles to the virus antigen.
Materials and methods
Ethics
The study protocol and other relevant study documentation for human studies were reviewed and approved by the medical research ethics committee of the National Institute of Infectious Diseases for the use of human subjects (UMIN Clinical Trials Registry (UMIN-CTR) trial ID: UMIN000013013). The study was conducted during 2013–2014 A(H1N1pdm09) predominant influenza season at 2 sites in Japan. All participating subjects consented in writing to participate in the study before enrollment and after being informed of the nature of the study, its risks, and its benefits. All participants have been fully anonymized.
Human participants and vaccination protocol
In total, 47 healthy adults of both sexes were recruited. The participants were vaccinated intranasally with the trivalent whole-virus inactivated influenza vaccine (WIIV; 45 μg HA/dose for each strain) twice at 3 week intervals. The vaccines were prepared from purified viruses of the reassortant vaccine candidates X-179A, X-223, and BX-51B, which were derived from A/Texas/50/2012 (H3N2), A/California/7/2009 (H1N1)pdm09, and B/Massachusetts/2/2012 (Yamagata-lineage), respectively. The vaccine (lot #NIFEN-003) in this study was an experimental vaccine, and manufactured according to GMP guidelines and appropriately released by the Research Foundation for Microbial Disease of Osaka University (BIKEN, Kanonji, Kagawa, Japan). The vaccine was prepared from purified viruses, which were sedimented through a linear sucrose gradient and treated with formalin by the method of Davenport et al19, and then mixed with carboxy-vinyl polymer (Toko Pharmaceutical Industrial Co.,Ltd., Toyama, Japan) as an excipient to increase viscosity. Intranasal administration was conducted by spraying 0.25 ml vaccine into each nostril (0.5 ml total) by using an atomizer (Toko Pharmaceutical Industrial Co.,Ltd., Toyama, Japan). The procedure of vaccine administration was reviewed and approved by the medical research ethics committee of the National Institute of Infectious Diseases for the use of human subjects.
Serum and nasal wash sample collection
Serum and nasal wash fluids were collected before vaccination, before the second intranasal vaccination (i.e., 3 weeks after the first vaccination), and 3 weeks after the second vaccination (i.e., 6 weeks after the first vaccination). The nasal wash samples were collected by washing the nasal cavity several times with sterile saline (Otsuka Normal Saline, Ref: 1326, NaCl 4.5g/500ml, pH 6.4, Otsuka Pharmaceutical Co., Ltd., Tokushima, Japan) by using nose irrigation devices (Nasal Rinser Nasaline, ENTpro AB, Stockholm, Sweden) according to the manufacturer's instructions. Approximately 100 ml nasal wash fluid was collected per nasal washing session. In addition, 21 participants agreed to undergo nasal washing for five consecutive days starting 3 weeks after the second vaccination (i.e., at the 6 week time point). Thus, 100 ml nasal wash sample was collected each day. All five samples were pooled, yielding a total of 500 ml nasal wash fluid per individual. The protocol for collection of nasal wash samples was reviewed and approved by the medical research ethics committee of the National Institute of Infectious Diseases for the use of human subjects. These nasal wash samples were filtered and concentrated as described previously.13 The total protein concentrations in these nasal wash samples were measured by using a BCA Protein Assay Kit (Thermo Fisher Scientific). The concentration of IgG, IgA, and IgM in all 21 samples was measured by using AlphaLISA IgG, IgA, and IgM kits (PerkinElmer Japan), respectively. The pre-immunization, 3 week, and 6 week serum and nasal wash samples, and the 21 concentrated nasal wash samples were stored at -80°C until use.
Neutralization test (NT) assays
The serum and nasal wash NT titers of the samples that were collected before and after vaccination were examined using microneutralization assays as described previously13 by using the wild-type virus strains of A/Texas/50/2012 (H3N2), A/Victoria/361/2011 (H3N2), A/California/7/2009 (H1N1) pdm09, and B/Massachusetts/2/2012 (Yamagata-lineage) (a kind gift from the Influenza Virus Research Center, NIID). The viruses were propagated in the allantoic cavity of 10-day-old embryonated chicken eggs, and purified from the allantoic fluid. The TCID50 (50% infectious dose in tissue culture) of the virus was estimated using previously described methods.13 In brief, 10-fold serial dilutions of allantoic fluid containing the virus were inoculated onto Madin-Darby canine kidney cells (MDCK: ATCC No.CCL-34) in a 96-well culture plate and incubated for 3 days at 37°C in a humidified 5% CO2 atmosphere. The cytopathic effect observed in the virus-containing wells was evaluated under a microscope and the TCID50 calculated. Total protein concentrations of nasal wash samples were adjusted to 1 mg/ml based on measurements by a BCA Protein Assay Kit (Thermo Fisher Scientific). These standardized nasal wash samples were subjected to NT assays. Two-fold serial dilutions of serum and standardized nasal wash samples were mixed with an equal volume of diluent containing an influenza virus equivalent of 100 TCID50 and added to the wells of a 96-well plate containing a monolayer culture of MDCK cells. Four control wells containing virus or diluent alone were included on each plate. The plates were incubated for 3 or 4 days at 37°C in a humidified 5% CO2 atmosphere. All wells were observed for the presence or absence of cytopathic effects, and then fixed with 10% formalin phosphate buffer for more than 5 min at room temperature and stained with Naphthol blue black. The titers were recorded as the reciprocal of the highest dilution without cytopathic effect.
IgA purification and fractionation
IgA was purified from the 21 concentrated nasal wash samples by affinity chromatography using CaptureSelect IgA Affinity Matrix (Thermo Fisher Scientific). The purified antibodies were then filtered with Cosmo spin filter H (Nacalai Tesque, Inc.) to remove precipitates or debris. The purified IgA samples (100 – 400 µg/participant) were then fractionated by size exclusion chromatography analysis. Thus, the antibodies were passed through an Agilent Bio SEC-5 1000 Å (7.8 × 300 mm) column (Agilent Technologies) that was coupled to an Agilent 1260 Infinity Bio-inert HPLC system (Agilent Technologies). All analyses were performed with a flow rate of 1 ml/min. Phosphate-buffered saline (pH 7.4) served as the eluent. The purified IgA was separated into three fractions, namely, Fr1, Fr2, and Fr3. These peaks correspond to the three peaks that were identified in the chromatogram. The amount of IgA in each fraction was calculated by using the volume and the concentration of each fraction, as determined by using an AlphaLISA IgA kit (PerkinElmer Japan). Thereafter, each fraction was concentrated to a final volume of approximately 50 μl with Amicon Ultra-4, 30K Centrifugal Filter Units (Merck Millipore). These fractionated samples were used in the analyses. Thus, they were analyzed by blue native-polyacrylamide gel electrophoresis (PAGE) by using NativePAGE 4–16% (wt/vol) Bis-Tris Gels (Thermo Fisher Scientific). NativeMark (Invitrogen) served as a molecular-weight standard.
Quantification of IgA subunits
To investigate the IgA subtypes in the three IgA fractions, mass spectrometry (LC-MS) was performed. Thus, stable isotope-labeled peptides (summarized in Table 1) were custom synthesized by Anygen. The internal standard peptides (30 pmol each) were mixed with 2 μg of each IgA sample dissolved in PBS. After adding Tris-HCl (pH 7.6) and CaCl2 (final concentrations of 100 mM and 1 mM, respectively), the samples were reduced with 5 mM dithiothreitol (Wako) at 56°C for 30 min. Thereafter, they were alkylated with 25 mM iodoacetamide (Wako) at room temperature for 30 min. The samples were then digested with sequencing-grade modified trypsin (Promega; 1:20 enzyme/substrate ratio) at 37°C for 16 h. Formic acid was added to a final concentration of 1%. After filtration through a 0.45 µm filter, the peptide solutions were analyzed by electrospray LC-MS using an ultra-high resolution quadrupole time-of-flight mass spectrometer maXis II (Bruker Daltonics) coupled to a Shimadzu Prominence UFLC-XR system (Shimadzu). Peptide separation was performed by using a BIOshell A160 Peptide C18 HPLC Column (5 µm particle size, L × I.D. 150 mm × 2.1 mm; Supelco) at a flow rate of 500 µl/min and a column temperature of 80°C with a binary gradient as follows: 98% solvent A (0.1% formic acid) for 2 min, linear gradient of 2–50% solvent B (100% acetonitrile) for 4 min, 90% solvent B for 2 min, and 98% solvent A for 2 min. The MS scan was performed over an m/z range of 50–2500 with a frequency of 2 Hz by using otofControl version 4.0 (Bruker Daltonics). The absolute amounts of the specific IgA subunits were estimated by comparing the peak area ratio of selected marker peptides to that of the corresponding internal standard peptides.
Recombinant trimeric HA production
To express trimeric HA proteins, the expression vector that encodes the extracellular HA domain of A/Victoria/361/2011 (H3N2) with a thrombin site, a T4 fibritin foldon trimerization domain, and a hexa-histidine affinity tag fused to the C-terminus was constructed according to a previous report.20 The trimeric HA proteins were expressed by using the Expi293 Expression System (ThermoFisher Scientific), according to the manufacturer's instructions. Five days post-transfection, the cell medium was centrifuged at 2,000 × g, filtered, and purified on a HisTrap excel column (GE Healthcare). The purified HA proteins were concentrated by using Amicon Ultracell (Millipore) centrifugation units with a cut-off of 30 kDa and the buffer was changed to phosphate-buffered saline (pH 7.4). The HA proteins were stored at -80°C until use.
Surface plasmon resonance (SPR) assay
The SPR assay was performed by using Biacore X100 (GE Healthcare Japan). Thus, the recombinant trimeric HA of A/Victoria/361/2011 (H3N2) with a C-terminal His-tag was immobilized on the surface of Sensor Chip NTA (GE Healthcare Japan) by using the NTA reagent kit (GE Healthcare Japan) according to the manufacturer's instructions. After trimeric HA immobilization (5 µg/ml for 60 seconds), the molecular interaction of HA with fractionated IgA (the concentration of IgA was adjusted to 500 µg/ml) was analyzed with a contact time of 180 seconds and a dissociation time of 1800 seconds. To compare the degree of IgA dissociation from HA of the three fractions, sensorgrams obtained from the analyses of the three fractions were y = axis adjusted (0 = baseline, 100 = binding). Area under the dissociation curve (AUC) of SPR response (from 181 to 1981 seconds following analyte injection) was calculated with a baseline y = -1000 using the Prism statistical software package (version 7.0b; GraphPad Software, Inc.). The dissociation efficiency was defined as the reciprocal of the AUC. Paired t-test was used for statistical analysis. The threshold for statistical significance was set at 5% (p < 0.05).
Statistical analyses
All data analyses, graphical representations, and correlation and linear regression analyses were performed by using Prism statistical software package (version 7.0b; GraphPad Software, Inc.). One-way ANOVA followed by Tukey's multiple comparison test was used to analyze each dataset except for datasets of SPR analysis. Correlation analyses were performed by Pearson's correlation tests. The threshold for statistical significance was set at 5% (p < 0.05).
Funding Statement
This work was supported in part by the Emerging/Re-emerging Infectious Diseases Project of Japan from the Japan Agency for Medical Research and Development, AMED, and in part by grants for Scientific Research from the Ministry of Education, Culture, Science, Sports and Technology (MEXT) of Japan <number 17K08386>.
Disclosure of potential conflicts of interest
Yuki Taga and Kiyoko Ogawa-Goto are employees of Nippi, Inc.
Acknowledgments
We thank Mr. T Tanimoto, Dr. Y Gomi, Dr. S Manabe, Mr. T Ishikawa, and Dr. Y Okuno at BIKEN for supplying the inactivated whole virus influenza vaccines; Mr. T Miyazaki and Mr. T Kamishita at Toko Yakuhin Kogyo Co., Ltd. for valuable suggestion on vaccination procedures of intranasal vaccine.
References
- 1.Vaccines against influenza WHO position paper – November 2012 Wkly Epidemiol Rec. 2012;87:461–76. PMID:23210147. [PubMed] [Google Scholar]
- 2.Murphy BR, Clements ML. The systemic and mucosal immune response of humans to influenza A virus. Curr Top Microbiol Immunol. 1989;146:107–16. PMID:2659262. [DOI] [PubMed] [Google Scholar]
- 3.Tamura S, Funato H, Hirabayashi Y, Kikuta K, Suzuki Y, Nagamine T, Aizawa C, Nakagawa M, Kurata T. Functional role of respiratory tract haemagglutinin-specific IgA antibodies in protection against influenza. Vaccine. 1990;8:479–85. doi: 10.1016/0264-410X(90)90250-P. PMID:2251874. [DOI] [PubMed] [Google Scholar]
- 4.Hasegawa H, Ichinohe T, Tamura S, Kurata T. Development of a mucosal vaccine for influenza viruses: preparation for a potential influenza pandemic. Expert Rev Vaccines. 2007;6:193–201. doi: 10.1586/14760584.6.2.193. PMID:17408369. [DOI] [PubMed] [Google Scholar]
- 5.Maassab HF. Adaptation and growth characteristics of influenza virus at 25 degrees c. Nature. 1967;213:612–4. doi: 10.1038/213612a0. PMID:6040602. [DOI] [PubMed] [Google Scholar]
- 6.Waldman RH, Wood SH, Torres EJ, Small PA Jr. Influenza antibody response following aerosal administration of inactivated virus. Am J Epidemiol. 1970;91:574–85. doi: 10.1093/oxfordjournals.aje.a121170. PMID:5445644. [DOI] [PubMed] [Google Scholar]
- 7.Wright PF, Murphy BR, Kervina M, Lawrence EM, Phelan MA, Karzon DT. Secretory immunological response after intranasal inactivated influenza A virus vaccinations: evidence for immunoglobulin A memory. Infect Immun. 1983;40:1092–5. PMID:6852914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura SI, Asanuma H, Ito Y, Hirabayashi Y, Suzuki Y, Nagamine T, Aizawa C, Kurata T, Oya A. Superior cross-protective effect of nasal vaccination to subcutaneous inoculation with influenza hemagglutinin vaccine. Eur J Immunol. 1992;22:477–81. doi: 10.1002/eji.1830220228. PMID:1537382. [DOI] [PubMed] [Google Scholar]
- 9.CDC Recommendations of the Advisory Committee on Immunization Practices (ACIP): Prevention and Control of Seasonal Influenza with Vaccines. MMWR,2009;58:1–52. [PubMed] [Google Scholar]
- 10.Caspard H, Gaglani M, Clipper L, Belongia EA, McLean HQ, Griffin MR, Talbot HK, Poehling KA, Peters TR, Veney N, et al. Effectiveness of live attenuated influenza vaccine and inactivated influenza vaccine in children 2–17 years of age in 2013–2014 in the United States. Vaccine. 2016;34:77–82. doi: 10.1016/j.vaccine.2015.11.010. PMID:26589519. [DOI] [PubMed] [Google Scholar]
- 11.Gaglani M, Pruszynski J, Murthy K, Clipper L, Robertson A, Reis M, Chung JR, Piedra PA, Avadhanula V, Nowalk MP, et al. Influenza Vaccine Effectiveness Against 2009 Pandemic Influenza A(H1N1) Virus Differed by Vaccine Type During 2013–2014 in the United States. J Infect Dis. 2016;213:1546–56. doi: 10.1093/infdis/jiv577. PMID:26743842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penttinen PM, Friede MH. Decreased effectiveness of the influenza A(H1N1)pdm09 strain in live attenuated influenza vaccines: an observational bias or a technical challenge? Euro Surveill. 2016;21:30350. doi: 10.2807/1560-7917.ES.2016.21.38.30350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ainai A, Tamura S, Suzuki T, van Riet E, Ito R, Odagiri T, Tashiro M, Kurata T, Hasegawa H. Intranasal vaccination with an inactivated whole influenza virus vaccine induces strong antibody responses in serum and nasal mucus of healthy adults. Hum Vaccin Immunother. 2013;9:1962–70. doi: 10.4161/hv.25458. PMID:23896606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T, Kawaguchi A, Ainai A, Tamura S, Ito R, Multihartina P, Setiawaty V, Pangesti KN, Odagiri T, Tashiro M, et al. Relationship of the quaternary structure of human secretory IgA to neutralization of influenza virus. Proc Natl Acad Sci U S A. 2015;112:7809–14. doi: 10.1073/pnas.1503885112. PMID:26056267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kishida N, Fujisaki S, Yokoyama M, Sato H, Saito R, Ikematsu H, Xu H, Takashita E, Tashiro M, Takao S, et al. Evaluation of influenza virus A/H3N2 and B vaccines on the basis of cross-reactivity of postvaccination human serum antibodies against influenza viruses A/H3N2 and B isolated in MDCK cells and embryonated hen eggs. Clin Vaccine Immunol. 2012;19:897–908. doi: 10.1128/CVI.05726-11. PMID:22492743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woof JM, Russell MW. Structure and function relationships in IgA. Mucosal Immunol. 2011;4:590. doi: 10.1038/mi.2011.39. PMID:21937984. [DOI] [PubMed] [Google Scholar]
- 17.Crago SS, Kutteh WH, Moro I, Allansmith MR, Radl J, Haaijman JJ, Mestecky J. Distribution of IgA1-, IgA2-, and J chain-containing cells in human tissues. J Immunol. 1984;132:16–8. PMID:6418796. [PubMed] [Google Scholar]
- 18.US Department of Health and Human Services Food and Drug Administration Center for Biologics Evaluation and Research. Guidance for Industry: Clinical Data Needed to Support the Licensure of Pandemic Influenza Vaccines. In: Office of Communication TaMA, Rockville, Maryland: , 2007. [Google Scholar]
- 19.Davenport FM, Hennessy AV, Brandon FM, Webster RG, Barrett CD Jr., Lease GO. COMPARISONS OF SEROLOGIC AND FEBRILE RESPONSES IN HUMANS TO VACCINATION WITH INFLUENZA A VIRUSES OR THEIR HEMAGGLUTININS. J Lab Clin Med. 1964;63:5–13. [PubMed] [Google Scholar]
- 20.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–70. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]


